Abstract
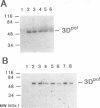
RNA-dependent RNA polymerases contain a highly conserved region of amino acids with a core segment composed of the amino acids YGDD which have been hypothesized to be at or near the catalytic active site of the molecule. Six mutations in this conserved YGDD region of the poliovirus RNA-dependent RNA polymerase were made by using oligonucleotide site-directed DNA mutagenesis of the poliovirus cDNA to substitute A, C, M, P, S, or V for the amino acid G. The mutant polymerase genes were expressed in Escherichia coli, and the purified RNA polymerases were tested for in vitro enzyme activity. Two of the mutant RNA polymerases (those in which the glycine residue was replaced with alanine or serine) exhibited in vitro enzymatic activity ranging from 5 to 20% of wild-type activity, while the remaining mutant RNA polymerases were inactive. Alterations in the in vitro reaction conditions by modification of temperature, metal ion concentration, or pH resulted in no significant differences in the activities of the mutant RNA polymerases relative to that of the wild-type enzyme. An antipeptide antibody directed against the wild-type core amino acid segment containing the YGDD region of the poliovirus polymerase reacted with the wild-type recombinant RNA polymerase and to a limited extent with the two enzymatically active mutant polymerases; the antipeptide antibody did not react with the mutant RNA polymerases which did not have in vitro enzyme activity. These results are discussed in the context of secondary-structure predictions for the core segment containing the conserved YGDD amino acids in the poliovirus RNA polymerase.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALTIMORE D., EGGERS H. J., FRANKLIN R. M., TAMM I. Poliovirus-induced RNA polymerase and the effects of virus-specific inhibitors on its production. Proc Natl Acad Sci U S A. 1963 Jun;49:843–849. doi: 10.1073/pnas.49.6.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. C., Lawson M. A., Semler B. L., Ehrenfeld E. Effects of mutations in poliovirus 3Dpol on RNA polymerase activity and on polyprotein cleavage. J Virol. 1989 Nov;63(11):4866–4874. doi: 10.1128/jvi.63.11.4866-4874.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Baron M. H., Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc Natl Acad Sci U S A. 1977 Sep;74(9):3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Van Dyke T. A. Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J Virol. 1979 Oct;32(1):155–161. doi: 10.1128/jvi.32.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Hirashima A. Interference with viral infection by defective RNA replicase. J Virol. 1987 Dec;61(12):3946–3949. doi: 10.1128/jvi.61.12.3946-3949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Purifoy D. J., Powell K. L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. 1987 Jun 25-Jul 1Nature. 327(6124):716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Air G. M., Webster R. G., Smith-Gill S. J. Epitopes on protein antigens: misconceptions and realities. Cell. 1990 May 18;61(4):553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- Marcy A. I., Hwang C. B., Ruffner K. L., Coen D. M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among alpha-like DNA polymerases is involved in substrate recognition. J Virol. 1990 Dec;64(12):5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C. D., Dasgupta A. Antibody to a synthetic nonapeptide corresponding to the NH2 terminus of poliovirus genome-linked protein VPg reacts with native VPg and inhibits in vitro replication of poliovirus RNA. J Virol. 1983 Nov;48(2):429–439. doi: 10.1128/jvi.48.2.429-439.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C. D., Warren B., Lentz M. R. Expression of enzymatically active poliovirus RNA-dependent RNA polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6050–6054. doi: 10.1073/pnas.84.17.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S. J., Palant O., Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989 Jan;63(1):216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C., Ivanoff L. A., Bienkowska-Szewczyk K., Butt B., Petteway S. R., Jr, Rothstein M. A., Ehrenfeld E. Formation of poliovirus RNA polymerase 3D in Escherichia coli by cleavage of fusion proteins expressed from cloned viral cDNA. Virology. 1987 Dec;161(2):348–356. doi: 10.1016/0042-6822(87)90127-9. [DOI] [PubMed] [Google Scholar]
- Rothstein M. A., Richards O. C., Amin C., Ehrenfeld E. Enzymatic activity of poliovirus RNA polymerase synthesized in Escherichia coli from viral cDNA. Virology. 1988 Jun;164(2):301–308. doi: 10.1016/0042-6822(88)90542-9. [DOI] [PubMed] [Google Scholar]
- Rueckert R. R., Wimmer E. Systematic nomenclature of picornavirus proteins. J Virol. 1984 Jun;50(3):957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler B. L., Dorner A. J., Wimmer E. Production of infectious poliovirus from cloned cDNA is dramatically increased by SV40 transcription and replication signals. Nucleic Acids Res. 1984 Jun 25;12(12):5123–5141. doi: 10.1093/nar/12.12.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. A., Flanegan J. B. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J Virol. 1980 Sep;35(3):732–740. doi: 10.1128/jvi.35.3.732-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Walgate R. Armadillos fight leprosy. Nature. 1981 Jun 18;291(5816):527–527. doi: 10.1038/291527a0. [DOI] [PubMed] [Google Scholar]