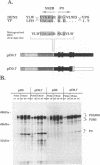
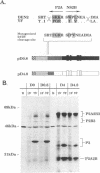
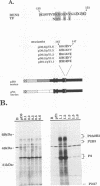
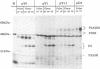
Abstract
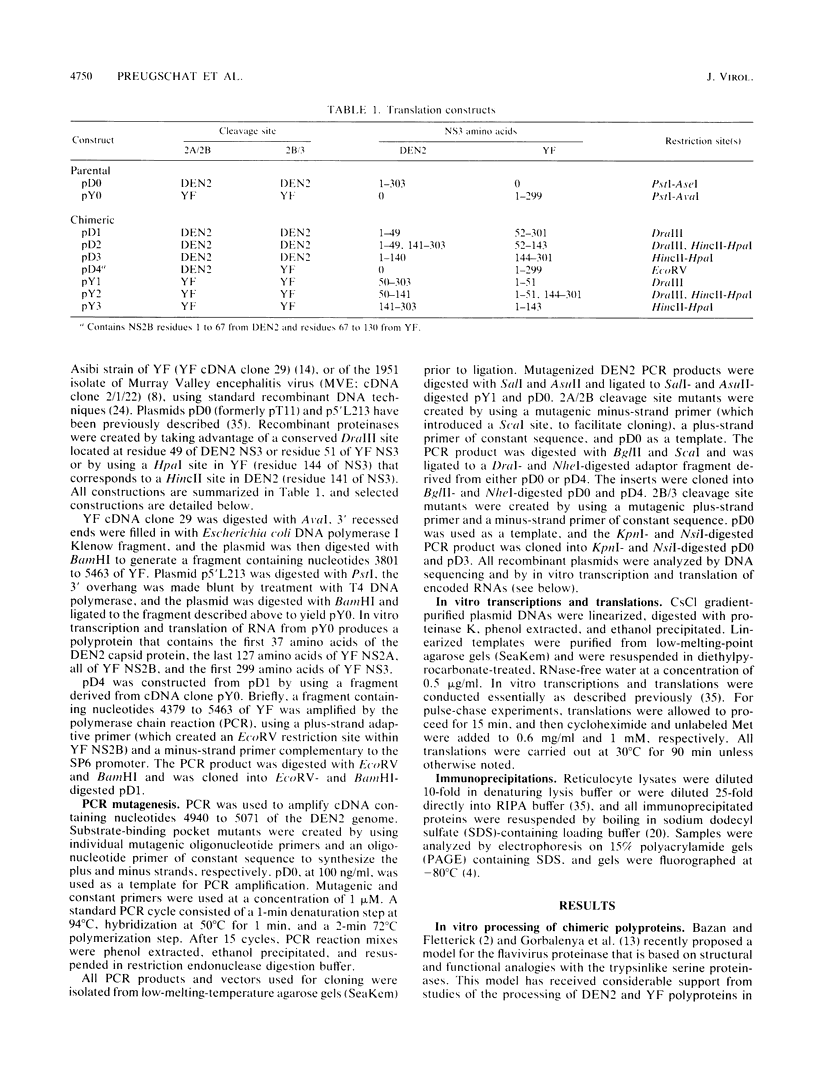
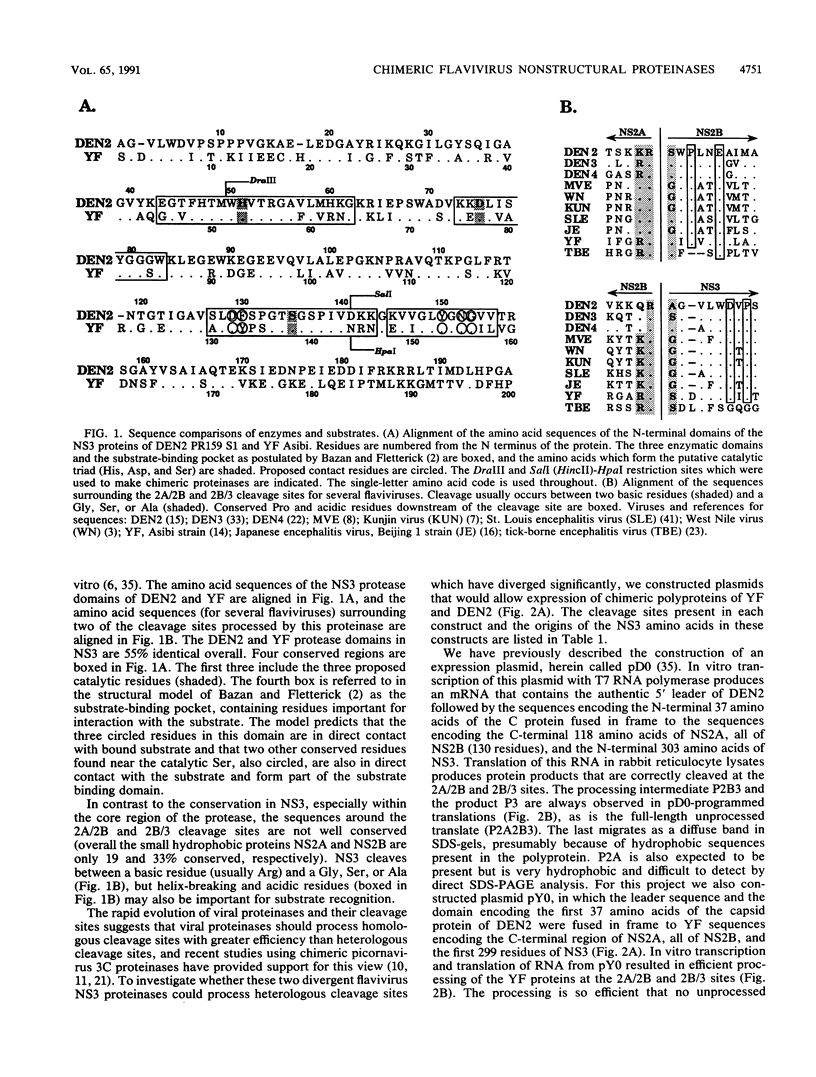
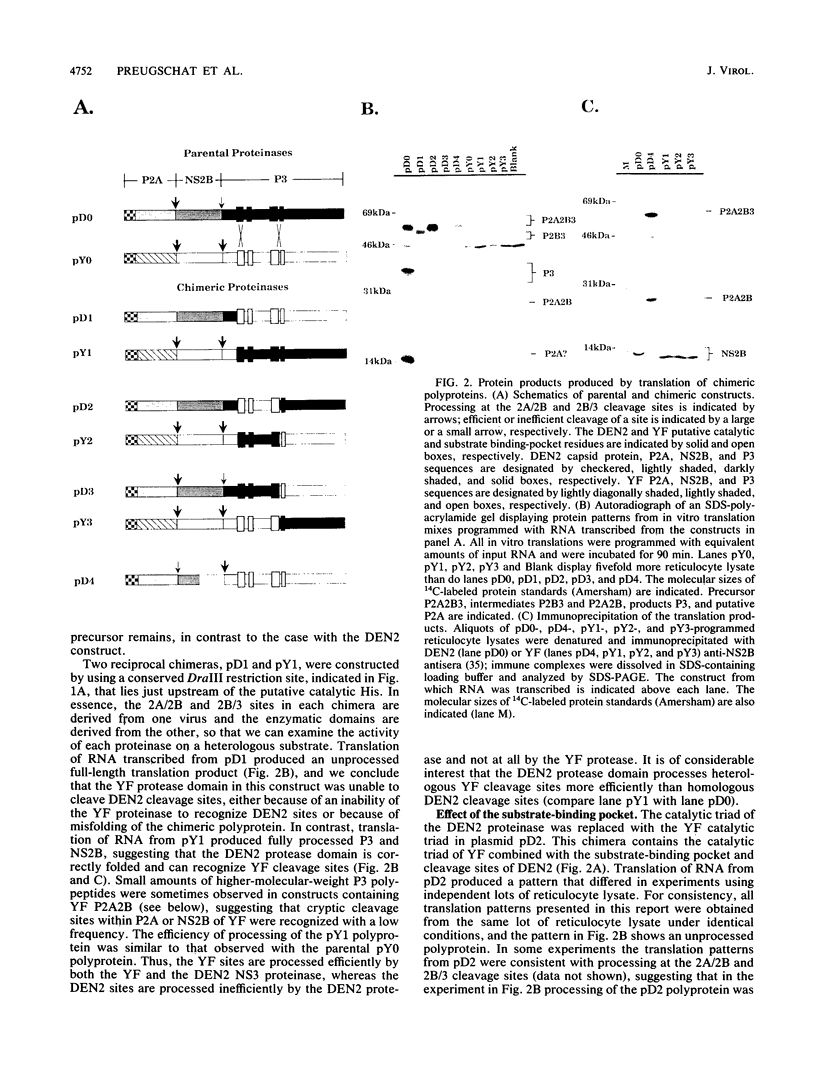
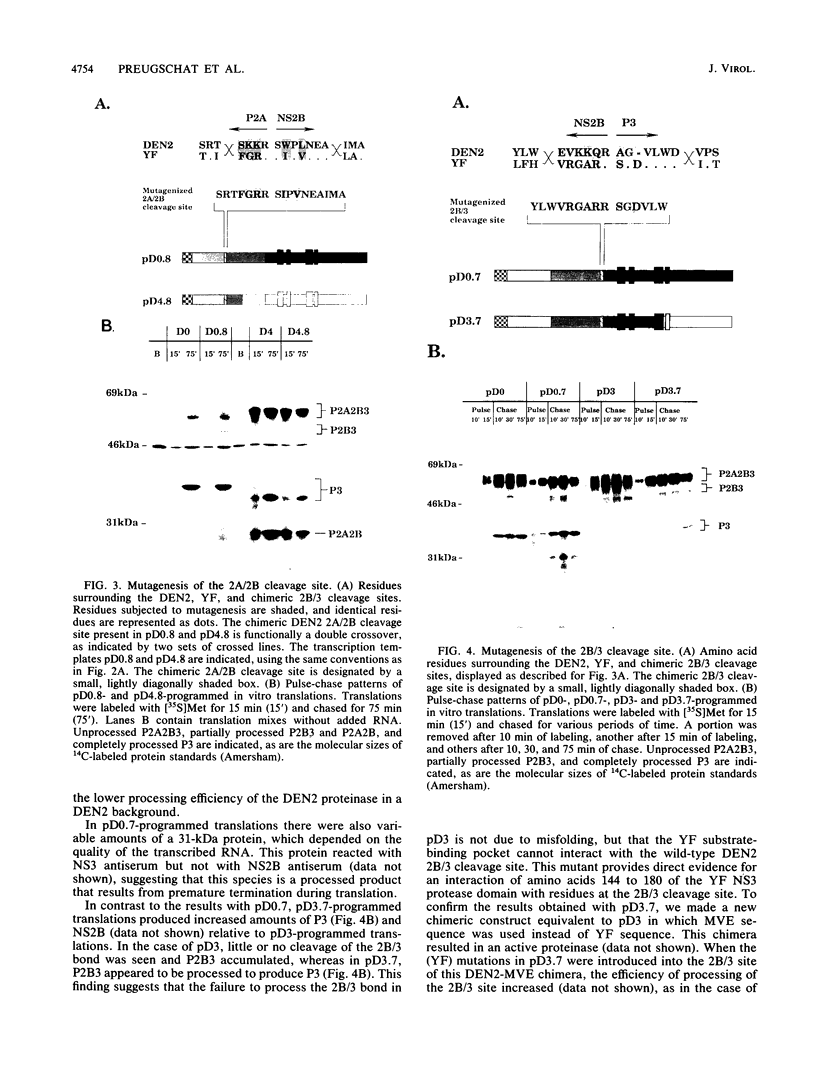
The proteins of flaviviruses are translated as a single long polyprotein which is co- and posttranslationally processed by both cellular and viral proteinases. We have studied the processing of flavivirus polyproteins in vitro by a viral proteinase located within protein NS3 that cleaves at least three sites within the nonstructural region of the polyprotein, acting primarily autocatalytically. Recombinant polyproteins in which part of the polyprotein is derived from yellow fever virus and part from dengue virus were used. We found that polyproteins containing the yellow fever virus cleavage sites were processed efficiently by the yellow fever virus enzyme, by the dengue virus enzyme, and by various chimeric enzymes. In contrast, dengue virus cleavage sites were cleaved inefficiently by the dengue virus enzyme and not at all by the yellow fever virus enzyme. Studies with chimeric proteinases and with site-directed mutants provided evidence for a direct interaction between the cleavage sites and the proposed substrate-binding pocket of the enzyme. We also found that the efficiency and order of processing could be altered by site-directed mutagenesis of the proposed substrate-binding pocket.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5' noncoding region. J Virol. 1990 Feb;64(2):607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989 Aug;171(2):637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Castle E., Leidner U., Nowak T., Wengler G., Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986 Feb;149(1):10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia G., Parker M. D., Speight G., Byrne M. E., Westaway E. G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988 Jan;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Dalgarno L., Trent D. W., Strauss J. H., Rice C. M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. Comparison of the encoded polypeptides with yellow fever virus structural and non-structural proteins. J Mol Biol. 1986 Feb 5;187(3):309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- Dewalt P. G., Blair W. S., Semler B. L. A genetic locus in mutant poliovirus genomes involved in overproduction of RNA polymerase and 3C proteinase. Virology. 1990 Feb;174(2):504–514. doi: 10.1016/0042-6822(90)90104-y. [DOI] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C. S., Dalrymple J. M., Strauss J. H., Rice C. M. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc Natl Acad Sci U S A. 1987 Apr;84(7):2019–2023. doi: 10.1073/pnas.84.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Galler R., Hunkapiller T., Dalrymple J. M., Strauss J. H., Strauss E. G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988 Jan;162(1):167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Nomoto A., Watanabe K., Mori T., Takezawa T., Aizawa C., Takegami T., Hiramatsu K. Molecular cloning and complete nucleotide sequence of the genome of Japanese encephalitis virus Beijing-1 strain. Virus Genes. 1988 Jun;1(3):305–317. doi: 10.1007/BF00572709. [DOI] [PubMed] [Google Scholar]
- Jones P. T., Dear P. H., Foote J., Neuberger M. S., Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. 1986 May 29-Jun 4Nature. 321(6069):522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- Kotler M., Katz R. A., Danho W., Leis J., Skalka A. M. Synthetic peptides as substrates and inhibitors of a retroviral protease. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4185–4189. doi: 10.1073/pnas.85.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräusslich H. G., Ingraham R. H., Skoog M. T., Wimmer E., Pallai P. V., Carter C. A. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc Natl Acad Sci U S A. 1989 Feb;86(3):807–811. doi: 10.1073/pnas.86.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Dasmahapatra B., Semler B. L. Species-specific substrate interaction of picornavirus 3C proteinase suballelic exchange mutants. J Biol Chem. 1990 Sep 15;265(26):15920–15931. [PubMed] [Google Scholar]
- Mackow E., Makino Y., Zhao B. T., Zhang Y. M., Markoff L., Buckler-White A., Guiler M., Chanock R., Lai C. J. The nucleotide sequence of dengue type 4 virus: analysis of genes coding for nonstructural proteins. Virology. 1987 Aug;159(2):217–228. doi: 10.1016/0042-6822(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Mandl C. W., Heinz F. X., Stöckl E., Kunz C. Genome sequence of tick-borne encephalitis virus (Western subtype) and comparative analysis of nonstructural proteins with other flaviviruses. Virology. 1989 Nov;173(1):291–301. doi: 10.1016/0042-6822(89)90246-8. [DOI] [PubMed] [Google Scholar]
- Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989 Aug;63(8):3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas M. T., Chen C. Y., Hitzeman R. A., Riggs A. D. Active human-yeast chimeric phosphoglycerate kinases engineered by domain interchange. Science. 1986 Aug 15;233(4765):788–790. doi: 10.1126/science.3526552. [DOI] [PubMed] [Google Scholar]
- McQuade T. J., Tomasselli A. G., Liu L., Karacostas V., Moss B., Sawyer T. K., Heinrikson R. L., Tarpley W. G. A synthetic HIV-1 protease inhibitor with antiviral activity arrests HIV-like particle maturation. Science. 1990 Jan 26;247(4941):454–456. doi: 10.1126/science.2405486. [DOI] [PubMed] [Google Scholar]
- Miller M., Jaskólski M., Rao J. K., Leis J., Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989 Feb 9;337(6207):576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- Moore M. L., Bryan W. M., Fakhoury S. A., Magaard V. W., Huffman W. F., Dayton B. D., Meek T. D., Hyland L., Dreyer G. B., Metcalf B. W. Peptide substrates and inhibitors of the HIV-1 protease. Biochem Biophys Res Commun. 1989 Mar 15;159(2):420–425. doi: 10.1016/0006-291x(89)90008-9. [DOI] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Nicklin M. J., Harris K. S., Pallai P. V., Wimmer E. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J Virol. 1988 Dec;62(12):4586–4593. doi: 10.1128/jvi.62.12.4586-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak T., Färber P. M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989 Apr;169(2):365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Osatomi K., Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990 Jun;176(2):643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- Pallai P. V., Burkhardt F., Skoog M., Schreiner K., Bax P., Cohen K. A., Hansen G., Palladino D. E., Harris K. S., Nicklin M. J. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol Chem. 1989 Jun 15;264(17):9738–9741. [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Grakoui A., Galler R., Chambers T. J. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989 Dec;1(3):285–296. [PubMed] [Google Scholar]
- Ruiz-Linares A., Cahour A., Després P., Girard M., Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989 Oct;63(10):4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J., Kent S. B. Enzymatic activity of a synthetic 99 residue protein corresponding to the putative HIV-1 protease. Cell. 1988 Jul 29;54(3):363–368. doi: 10.1016/0092-8674(88)90199-7. [DOI] [PubMed] [Google Scholar]
- Trent D. W., Kinney R. M., Johnson B. J., Vorndam A. V., Grant J. A., Deubel V., Rice C. M., Hahn C. Partial nucleotide sequence of St. Louis encephalitis virus RNA: structural proteins, NS1, ns2a, and ns2b. Virology. 1987 Feb;156(2):293–304. doi: 10.1016/0042-6822(87)90409-0. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Cunningham B. C., Graycar T. P., Estell D. A. Recruitment of substrate-specificity properties from one enzyme into a related one by protein engineering. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R. P., Brown E. L., Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984 Sep;38(2):361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. Changing the binding specificity of a repressor by redesigning an alpha-helix. Nature. 1985 Aug 15;316(6029):601–605. doi: 10.1038/316601a0. [DOI] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Smythers G. W., Oroszlan S. Bovine leukemia virus protease: purification, chemical analysis, and in vitro processing of gag precursor polyproteins. J Virol. 1986 Mar;57(3):826–832. doi: 10.1128/jvi.57.3.826-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Hardy W. R., Shirako Y., Strauss J. H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990 Aug;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]