Abstract
In cells subjected to moderate aminoacyl-tRNA limitation, the peptidyl-tRNA–ribosome complex stalled at the “hungry” codon can slide well beyond it on the messenger RNA and resume translation further downstream. This behavior is proved by unequivocal amino acid sequence data, showing a protein that lacks the bypassed sequence encoded between the hungry codon and specific landing sites. The landing sites are codons cognate to the anticodon of the peptidyl-tRNA. The efficiency of this behavior can be as high as 10–20% but declines with the length of the slide. Interposition of “trap” sites (nonproductive landing sites) in the bypassed region reduces the frequency of successful slides, confirming that the ribosome–peptidyl-tRNA complex passes through the untranslated region of the message. This behavior appears to be quite general: it can occur at the two kinds of hungry codons tested, AUA and AAG; the sliding peptidyl-tRNA can be any of three species tested, phenylalanine, tyrosine, or leucine tRNA; the peptidyl component can be either of two very different peptide sequences; and translation can resume at any of the three codons tested.
Protein synthesis is conventionally pictured as the orderly, sequential addition of amino acids to a growing peptide chain dictated by the sequence of triplets in the messenger RNA. However, alternatives to this linear progression are possible and can occur at surprisingly high frequencies in certain sequence contexts and/or under certain physiological conditions. Limitation for an aminoacyl-tRNA, in particular, raises the probabilities of alternative paths at “hungry” codons cognate to the limiting species. The alternative paths described so far include binding of noncognate aminoacyl-tRNA species (reviewed in refs. 1–3) and frameshifting either to the left (−1) or the right (+1) (reviewed in refs. 4, 5). In addition, peptidyl-tRNA release, normally a low-frequency event, may occur at hungry codons (6–8).
In this communication, we describe another alternative path the ribosome can follow at a hungry codon: sliding over it and moving several codons downstream, resuming translation further along the message. We encountered this phenomenon in the course of studying ribosome behavior at a sequence conducive to leftward frameshifting.
MATERIALS AND METHODS
Bacteria were cultivated at 37°C under forced aeration in M63-glucose medium (9). To retain comparability with ref. 9, the medium was supplemented with leucine, threonine, arginine, and histidine, even though the strain used in the present experiments is prototrophic. The host was strain C92 (relA−, spoT−) carrying the O6 deletion of the early part of lacZ (10). The deletion, provided to us as an F′ plasmid by Jon Beckwith (Harvard Medical School, Boston), was introduced into the C92 chromosome by homogenotization. For unknown reasons, identical plasmids yield twice as much β-galactosidase in this strain as in the CP78/79 lineage we have used previously (9). This proved useful in purifying material for protein sequencing.
All methods of DNA construction and analysis were as described (9). Engineered plasmids (Fig. 1) were transformed into the host strain with selection for the plasmid’s bla gene (1 mg/ml carbenicillin). To measure enzyme synthesis, cultures were grown overnight from single colonies with carbenicillin, diluted, and grown into logarithmic phase. When an OD700 (Beckman spectrophotometer) of 0.2 was reached, the cultures were induced [2 mM isopropyl β-d-thiogalactopyranoside (IPTG), 2.5 mM cAMP], and aliquots were grown in parallel in the absence and presence of 72 μg/ml isoleucine hydroxamate (ILHX) to inhibit isoleucine tRNA charging. Immediately before induction and after about one doubling, samples were withdrawn for measurement of β-galactosidase and protein, as described (9). The plasmid vector containing the engineered forms of the lacZ gene also carries a lacIq regulator gene.
Figure 1.
Oligonucleotides used in the lacZ constructions in this study. Pairs of complementary oligonucleotides with sticky HindIII and BamHI overhangs were ligated into the multiple cloning site in the lacZ gene on pEK-0F which had been simultaneously cut with HindIII and BamHI. The noncoding strands are shown for only the top two oligonucleotides. Symbols representing sequences in L-16 (TTT) shared by the other constructs are ∗, sequences before the slide; −, bypassed regions; and +, sequences after the slide. pEK-0F (pMLB1115EK-0F; ref. 9) is a pBR322 derivative carrying the lacIZ genes. Its lacZ gene contains a modified pUC 9 multiple cloning site, into which we inserted the coding sequence for 18 amino acids ending in the enterokinase cleavage site (Asp-Asp-Asp-Asp-Lys) just upstream of the pUC HindIII site. Enterokinase cleaves the resulting β-galactosidase after the Lys, producing an N terminus of Val-Ser followed by our sequence of interest, with the result that Edman sequence data are greatly improved (9). The left-shift reporter, named LefShf here, has an ATA isoleucine codon following the “shifty” quadruplet T TTC and is identical to that reported in figure 3 of ref. 9. The family of leap reporters have similar sequences, but the TTT triplet two codons before the isoleucine ATA was changed to TAT so that there would be only one phenylalanine codon, to avoid ambiguity in protein sequence. The zero frame control, L-0f, is identical to L-16 except that the blocking TAG terminator in the 0 reading frame has been changed to a sense TAC and a T two nucleotides further downstream has been eliminated, thus continuing the 0 frame into the β-galactosidase coding sequence.
RESULTS
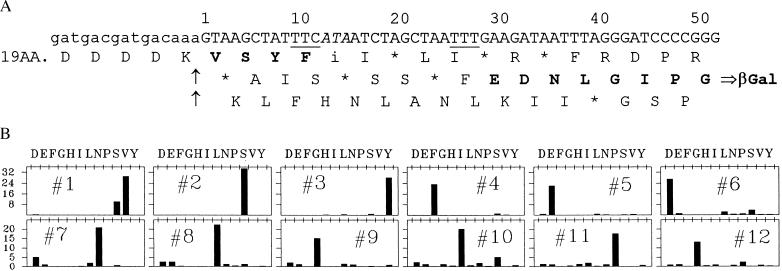
Fig. 2A shows the coding-strand sequence of the relevant region of a lacZ reporter-gene construct we call L-16 (TTT), in which the sequence at positions 9–15 is very prone to leftward frameshifting during limitation for isoleucyl tRNA (9). However, leftward frameshifting at this position, or anywhere else, cannot give rise to β-galactosidase in this construct because the β-galactosidase protein is encoded in the other alternative, or rightward (+1), reading frame, the translation product of which is shown. Moreover, this reading frame cannot be accessed by a rightward shift at any isoleucine codon because it is blocked by two terminator triplets located between the isoleucine codons of the 0 reading frame and position 26, as shown. Nonetheless, partial isoleucine-tRNA starvation elicited a more than 40-fold increase in β-galactosidase synthesis with this construct (Table 1), an increase comparable to that observed with LefShf, a reporter of simple leftward frameshifting at the same sequence (Table 1; ref. 9).
Figure 2.
Nucleotide sequence of the coding strand and amino acid sequence of the protein encoded by construct L-16 (TTT) in the relevant region. (A) The coding sequence is shown numbered from the G in the first position of the codon for the valine on the C-terminal side of the enterokinase cut site, shown by arrows. The 0-frame sequence on the N-terminal side of the cut site is indicated schematically. Predicted translation products in all three reading frames are shown, aligned under their corresponding triplets, with termination at nonsense triplets indicated by an ∗. The shifty sequence T TTC ATA is at positions 9–15. Amino acids in boldface are those found in the protein made in isoleucine-tRNA limited cells. The phenylalanine codons at the takeoff and landing sites are underlined. The hungry isoleucine codon is italicized. (B) Cells carrying L-16 (TTT) (see Table 1) were grown into logarithmic phase, the lac promoter was induced, and isoleucyl-tRNA synthetase was partly inhibited by addition of 72 μg/ml ILHX. After two to three doublings of OD, the cells were harvested and β-glactosidase was immunopurified for amino acid sequence analysis as described (9). The β-galactosidase immunoprecipitate was dispersed in 1× EKMax buffer (50 mM Tris⋅HCl, pH 8.0, 1 mM CaCl2, 0.1% Tween 20) and treated with enterokinase for 2 hr at 37°C (9). The cleavage mixture was run on 6% PAGE gels and electroblotted to poly(vinylidene difluoride) membrane. The results of Edman sequence analysis (performed at Southwest Scientific Resources) are shown for each successive cycle, with lag-corrected pmol on the y axis, and the different amino acids on the x axis. Similar results were obtained with another sample sequenced at the Stanford Protein and Nucleic Acid Facility.
Table 1.
Differential rates of codon-jumping induced by ILHX
| Construct name (Landing site sequence) | Translation behavior | Differential rates EU/mg protein × 103
|
|
|---|---|---|---|
| Control | +ILHX | ||
| L-(0F)* | Wild-type control | 570,000 ± 75,000 (4) | 92,000 ± 23,000 (4) |
| LefShf† | Left shift, TTC (ata) | 595 ± 67 (6) | 12,200 ± 2,280 (6) |
| L-16 (TTT)‡ | TTC ➱ 16 ➱ TTT | 322 ± 26 (5) | 16,900 ± 3,630 (5) |
| Landing site sequence variation | |||
| L-16 (TTC) | TTC ➱ 16 ➱ TTC | 352 ± 16 (3) | 12,855 ± 1,166 (3) |
| L-16 (GTT) | TTC ➱ 16 ➱ GTT | 216 ± 2 (2) | 2,740 ± 250 (2) |
| L-16 (GAT) | TTC ➱ 16 ➱ GAT | 217 ± 45 (3) | 743 ± 352 (3) |
| L-16 (GCG) | TTC ➱ 16 ➱ GCG | 147 ± 1.5 (2) | 783 ± 29 (2) |
| TAC-16 (TAC) | TAC ➱ 16 ➱ TAC | 402 ± 11 (4) | 10,092 ± 526 (4) |
| Effect of distance between takeoff and landing | |||
| L-7 (TTT) | TTC ➱ 7 ➱ TTT | 567 ± 63 (4) | 17,840 ± 2,000 (3) |
| L-15 (TTT) | TTC ➱ 15 ➱ TTT | 495 ± 34 (2) | 21,400 ± 189 (2) |
| L-16 (TTT) | TTC ➱ 16 ➱ TTT | 322 ± 26 (5) | 16,900 ± 3,630 (5) |
| L-22 (TTT) | TTC ➱ 22 ➱ TTT | 308 ± 39 (3) | 8,232 ± 1,548 (3) |
| L-22′ (TTT) | TTC ➱ 22′ ➱ TTT | 200 ± 17 (4) | 9,106 ± 661 (3) |
| L-39 (TTT) | TTC ➱ 39 ➱ TTT | 56 ± 5.8 (2) | 1,017 ± 111 (2) |
| L-40 (TTT) | TTC ➱ 40 ➱ TTT | 60 ± 2.5 (2) | 596 ± 35 (2) |
| Traps in the bypassed sequence | |||
| L-16 (TTT) | TTC ➱ 16 ➱ TTT | 322 ± 26 (5) | 16,900 ± 3,630 (5) |
| E-Trap-16 | L-16, early Trap | 238 ± 4 (2) | 7,740 ± 216 (2) |
| L-Trap-16 | L-16, late Trap | 270 ± 7.8 (4) | 7,091 ± 160 (4) |
| L-22 (TTT) | TTC ➱ 22 ➱ TTT | 308 ± 39 (3) | 8,232 ± 1,548 (3) |
| E-Trap-22 | L-22, early Trap | 120 ± 2.0 (3) | 2,372 ± 256 (3) |
| L-Trap-22 | L-22, late Trap | 151 ± 0.3 (3) | 2,688 ± 446 (3) |
Under “Translation behavior” we indicate the takeoff-site codon for peptidyl-tRNA, followed by an arrow symbolizing the hop, its length, and the landing P-site. The data are reported as average differential rates of enzyme synthesis, in Enzyme Units per mg protein ×1000, ±SEM, with the number of replicate experiments in parentheses. In the case of construct L-16 (TTT), simple isoleucine starvation by means of valine inhibition (11) elicited an increase in the differential rate from 444 to 12,900, closely similar to the effect of ILHX.
*Zero-frame control.
Simple left shifter.
Slide reporter.
How do ribosomes starved for isoleucyl tRNA negotiate this messenger sequence so as to synthesize β-galactosidase? To find out, we purified the β-galactosidase and determined its amino acid sequence in the region of interest (Fig. 2B). The sequence following the enterokinase cut site is as expected (Val-Ser-Tyr-Phe) up to the position of the hungry AUA isoleucine codon at positions 13–15; it then continues (Glu-Asp-Asn-Leu-Glu-Ile-Pro-Gly- and four further amino acids not shown in the figure) with precisely the sequence corresponding to the β-galactosidase reading frame from position 29 (compare Figs. 2 A and B). Evidently, ribosomes stalled at positions 13–15 simply hopped over 16 nucleotides, landing so as to resume translation at the GAA codon at positions 29–31. The ribosomes’ gymnastic feat here is analogous to the ribosome “hopping” described by Weiss et al. (12) and O’Connor et al. (13), although the efficiency with which isoleucyl tRNA limitation promotes the hop is much greater and the distance, as we will see, can also be much greater.
The sequence data (Fig. 2B) are very clean, indicating an essentially homogeneous sequence. This homogeneity must mean that virtually all ribosomes that land in the β-galactosidase reading frame land at the same triplet. The only plausible explanation for this uniformly preferred landing site is that it follows a UUU triplet for phenylalanine, synonymous with the UUC triplet on the 5′ side of the takeoff point. Thus, all ribosomes that take off at positions 13–15 have phenylalanyl-peptidyl-tRNA in the peptidyl (P) site, which could reassociate with the UUU codon bordering the landing site. From this perspective, the landing site should be described as codons 26–31, including the UUU.
To test this hypothesis, we made several constructs with changes in the UUU codon of the landing site (Table 1). When the UUU codon was changed to UUC, which is recognized by the same phenylalanine tRNA, ILHX-induced hopping was equally efficient. When the UUU was changed to GUU, complementary to the phenylalanine tRNA anticodon at only the second and third nucleotides, it was reduced by a factor of 6. Protein sequencing of the β-galactosidase (data not shown) confirmed that it still reflected a hop to the triplet following the GUU codon. When the UUU was changed to GAU or GCG, different at two or three positions, hopping was drastically reduced, by a factor of 20. Finally, when we changed the takeoff site to TAC and likewise changed the landing site to TAC, in construct TAC-16 (TAC), we observed hopping virtually as efficient as the cases in which both sites encode phenylalanyl-tRNA. Thus, hopping induced by a hungry codon depends on the ability of the P-site codons of the takeoff and landing sites to bind the same tRNA or peptidyl-tRNA.
In another set of experiments, we varied the distance between the takeoff and landing sites’ phenylalanine codons. The results (Table 1) show that the frequency of successful hops decreases with distance. When the takeoff and landing sites were separated by 7, 15, or 16 nucleotides, enzyme synthesis was about 20% that of the wild-type control; when separated by 22 nucleotides (bypassing two slightly different sequences), it was 10% of the wild-type control; and when separated by 39 or 40 nucleotides, it was only about 1% of the wild-type control. This effect of the spacing of the takeoff and landing site codons is also consistent with the hypothesis that these codons determine the successful resumption of translation. The results also suggest that this is insensitive to reading frame. Hops that landed in the original reading frame (15 and 39) were not notably more or less efficient than those of similar distance (16 and 40, respectively) that landed in the rightward reading frame. The similar results with the two reporters of 22-nucleotide hops suggest, in fact, that the frequency of success is not much affected by the sequence the ribosome bypasses without translating.
On the other hand, the ribosome–peptidyl-tRNA complex presumably does interact with the region it bypasses so as to detect potential landing sites–that is, codons (evidently in any frame) cognate to the peptidyl-tRNA. If this view is correct, then the number of ribosomes arriving at a productive landing site should be reduced by the interposition of nonproductive landing sites between the takeoff site and the productive landing site. Tests of this proposition are shown in Table 1.
The construct designated E-Trap-16 is identical to L-16 (TTT) except for a 1-base change that introduces a UUU landing trap (followed two codons downstream by a terminator) seven nucleotides beyond the takeoff site. This change reduced β-galactosidase synthesis 54% relative to that of L-16. L-Trap-16 differs from L-16 at just two nucleotides, such that a UUU trap (in the wrong reading frame) is introduced at a later position, 11 nucleotides beyond the takeoff site. This change reduced enzyme synthesis by 58% relative to L-16. In E-Trap-22 and L-Trap-22, we interposed UUU traps (in the wrong reading frame) at early and late positions between the takeoff and landing sites of L-22. These changes reduced β-galactosidase synthesis by 71% and 67%, respectively.
Thus, UUU traps in the bypassed region can eliminate about two-thirds of the ribosomes that would otherwise arrive at a productive landing site and yield β-galactosidase. We conclude that the ribosome–peptidyl-tRNA complex does not in fact hop over the bypassed region, but rather slides through it, scanning for triplets complementary to the anticodon of the peptidyl-tRNA as it proceeds.
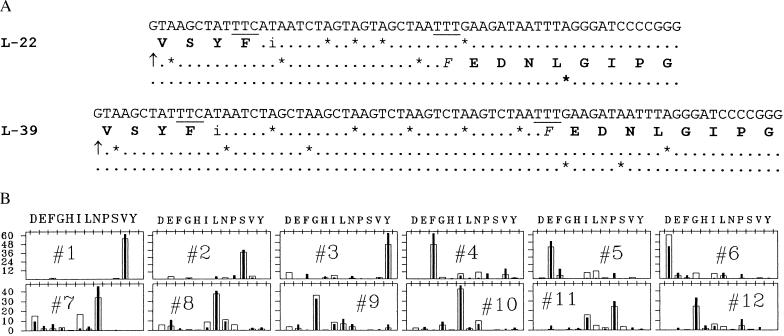
The β-galactosidase produced by constructs that demanded slides of 22 and 39 nucleotides respectively was purified and sequenced as in Fig. 2B. In both cases (Fig. 3) the amino acid sequence confirmed that β-galactosidase was made by the expected movements to the UUU landing site, which is followed by the same downstream sequence in each case.
Figure 3.
Coding strand sequences and protein amino acid sequences of the relevant regions of L22 and L39. (A) The coding sequences are shown as in Fig. 2A for constructs L22 and L39. The predicted amino acid sequences are shown up to the takeoff site and after the predicted landing sites. (B) Cycle bar plots of Edman N-terminal sequence analysis of both proteins, as in Fig. 2B. Filled bars = L22, open bars = L39. Sequence analysis was performed at the Stanford Protein and Nucleic Acid Facility.
In all of the preceding cases, the takeoff site was the 28th codon of a specific and rather peculiar message. Our reporter oligonucleotides all are inserted into a vector that carries an upstream sequence coding for a run of histidines and for the enterokinase cleavage site, which we use to optimize N-terminal sequence analysis (9). To find out whether sliding was specific to this unusual sequence, we inserted the oligonucleotide of L-16 (TTT) into another vector with a completely different upstream sequence, to construct pXA-L16. Fig. 4 shows the two L-16 constructs with different upstream sequences and compares their activities. In pXA-L16, enzyme synthesis was 1/3–1/4 that of L-16, but ILHX stimulated enzyme synthesis by virtually the same factor, 45-fold. Thus, the induction of sliding by a hungry AUA codon is largely independent of upstream sequence, and hence of the nature of the peptide part of the sliding peptidyl-tRNA.
Figure 4.
Sliding in constructs with differing upstream sequences. The nucleotide and predicted amino acid sequences upstream of the slide site are shown for constructs L-16 and pXA-L16. Uppercase letters are used for L-16 and for positions in pXA that are identical, and lowercase letters are used for all positions that are different. Amino acids in boldface are target sites for enterokinase (in L-16) or factor Xa, (in pXA), the cut sites of which are shown by arrows. β-Galactosidase differential rates, in control cells and cells inhibited by ILHX (72 μg/ml), are quoted below each sequence [mean ± SE (no. replicates)] as in Table 1.
In all of the cases described so far, we have induced sliding at a hungry AUA codon by limiting the acylation of isoleucine tRNA. Is there anything special about the hungry AUA codon, or can sliding be initiated at a different hungry codon? The experiments shown in Fig. 5 address this question. Here, we have altered the sequence of L-16 to replace AUA by AAG, coding for lysine tRNA. In addition, we changed the P-site codon to UUA to test still another peptidyl-tRNA for sliding behavior, providing a UUA landing site 16 nucleotides downstream. Finally, we changed the triplet following the landing site to test the influence of that sequence element. Fig. 5A shows the sequence of the coding strand in this construct, which we call L′-16-L′ and indicates its β-galactosidase activity. Lysyl tRNA limitation by lysine hydroxamate promoted a 30-fold increase in β-galactosidase synthesis encoded by this construct. Fig. 5B shows the amino acid sequence of the enzyme made under this condition, confirming the slide from the takeoff sequence UUAAAG to the triplet following the landing site UUA. Note that the first amino acid following the slide in this construct is histidine (CAC), in contrast to the glutamic acid (GAA) in the previous set. We have also confirmed sliding by amino acid sequence analysis in one other case (data not shown), where the first amino acid after the slide is lysine (AAA). Thus, to a first approximation, sliding is independent of the amino acid with which peptide-chain elongation resumes.
Figure 5.
Leaping in construct L′-16-L′ induced by stalling the ribosome at a lysine codon with peptidyl-leucyl-tRNA in the P site. (A) Coding strand sequence of the relevant region of L′-16-L′ as in Fig. 3A, with the predicted amino acid sequence shown up to the takeoff point and after the predicted landing site. Differential rates of β-galactosidase synthesis are reported for one doubling of growth in the control and in 50 μg/ml lysine hydroxamate (each in the presence of 100 μg/ml every amino acid except lysine as in ref. 9), which inhibits growth slightly less than 72 μg/ml ILHX. (B) Cycle bar plot of Edman N-terminal sequence analysis, as in Fig. 2B. Two liters of cells were induced and subjected to lysine hydroxamate (50 μg/ml) inhibition. After about one doubling, the cells were harvested, and β-galactosidase was purified and sequenced as in Figs. 2B and 3B, at the Stanford Protein and Nucleic Acid Facility.
We conclude that ribosome sliding can be promoted when the ribosome is stalled at either AUA or AAG, and perhaps at any hungry codon; that the sliding ribosome can carry peptidyl-tRNA of the phenylalanyl, tyrosyl, or leucyl varieties, and very likely any peptidyl-tRNA, to a cognate landing site downstream; and that translation then resumes with any aminacyl-tRNA encoded in the triplet following the landing site.
DISCUSSION
The constructs under discussion in this paper–including reporters both of frameshifting and of sliding–support about 6 × 10−4 as much enzyme synthesis as the lacZ+ control in growing, uninhibited cells (Table 1). The reporters and the control share the same sequence in the promoter, the ribosome loading region, the first 78 nucleotides of the lacZ coding sequence, and, indeed, nearly all of the lacZ coding sequence. As a consequence, transcription and translation should occur identically in reporters and their controls up to the 0 reading frame terminator triplets of the former (placed by design just downstream of the hungry codon) and beyond that if frameshifting or sliding occurs. Therefore, the 6 × 10−4 ratio of enzyme synthesis almost certainly means that about 1 ribosome in 1,600 performs the acrobatics necessary to generate active β-galactosidase. Similarly, normalizing the rate of enzyme synthesis to that in the 0 reading frame control under conditions of isoleucyl-tRNA limitation should yield the frequency of ribosomes that performs the feat under this condition.
Our isoleucyl-tRNA limitation regime (72 μg/ml ILHX) reduces growth rate ≈80% imposed on relA− cells in which errors of translation of all kinds at hungry codons are exacerbated (1–3). Under these conditions, the average differential rate of enzyme synthesis in the frameshift or shorter slide-reporters increases about 40-fold (Table 1). Moreover, this finding is to be contrasted with an 85% decrease observed in the lacZ+ control because of effects on both transcription of the lacZ gene (14–16) and translation of the other isoleucine codons in the message (2, 3, 17, 18). Normalizing to the lacZ+ control indicates that the frequency of sliding in our L-7, L-15, and L-16 reporters increases from about 6 × 10−4 to between 0.18 and 0.23.
These values are one to two orders of magnitude higher than the frequency of even shorter hops by normal tRNAs reported by Weiss et al. (12) and O’Connor et al. (13), and even somewhat higher than the frequencies observed in certain anticodon loop tRNA mutants that displayed increased hopping (13). Most, but not all of the hops described by these workers were over a nonsense triplet, which is partly consistent with the view that a (presumed) translational pause promotes the hop. Kane et al. (19) reported suggestive evidence for a high-frequency (20%) hop of +6 nucleotides in a mammalian gene overexpressed in Escherichia coli from a takeoff point preceding a very rare AGG codon, so that this event too could be rationalized in terms of ribosome pausing. Our results provide more compelling evidence that ribosome pausing underlies these unusual ribosome movements: limitation for aminoacyl-tRNA converts an ordinary sense codon into a pause site, and this elicits a huge increase in hopping or, as we call it in view of the trap-site experiments, sliding.
Nonetheless, a translational pause may be a necessary but not a sufficient condition for high-frequency sliding. In the case of our experiments, the phenomenon is partly dependent on the relA defect. In our host strain’s relA+ sibling, ILHX still promoted sliding, but about 1/7 as effectively, although the reduction in growth rate was comparable (data not shown). The mechanism by which the relA ppGpp system influences ribosome behavior and accuracy at hungry codons is unclear (2, 3).
There may be sequence contexts around or between the takeoff and landing sites that affect the frequency of sliding, although these preliminary results suggest that these effects are not great. Three different triplets immediately after the landing P site, and three different pairs of takeoff/landing P sites all permitted sliding. No striking secondary-structure sequences are evident between the takeoff and landing sites. Most of the efficient sliders happen to have a UGAA U-turn motif overlapping the landing site, but three (those with TTC, TAC, and TTA takeoff and landing P sites) do not. There is obviously a rich field for much more detailed investigation of sequence constraints affecting this unusual ribosome behavior.
Our results are in some ways analogous to the giant hop of 50 nucleotides discovered by Weiss and colleagues (20, 21) in translation of the gene 60 topoisomerase of phage T4. In this case, the ribosome movement depends on both a terminator codon at the takeoff point and a stem/loop secondary structure just after it (21). Our results demonstrate that starvation-induced pausing at a sense codon can promote analogous ribosome movement very efficiently as well, independent of secondary structure in the untranslated region. The T4 topoisomerase movement also was mysteriously sensitive to the amino acid sequence of the nascent peptide slightly upstream of the takeoff point (21). The two upstream sequences we have tested have virtually nothing in common, as far as we can tell, with each other or with the T4 topoisomerase region in question. Thus, sliding in this study is seemingly not associated with any unique upstream elements or with any obvious consensus-sequence elements.
In each of the cases described above (12, 13, 20, 21) there are matched codons at the take-off and landing sites, and their necessity was demonstrated by mutagenesis in the case of the 50-nucleotide hop in T4 topoisomerase (21). Our results confirm the importance of such matching sites and further demonstrate that they pertain to the movement of peptidyl-tRNA between the P sites at takeoff and landing positions, elicited in the present system by withholding aminoacyl-tRNA cognate to the codon in the takeoff position’s aminoacyl site.
Our results and those just reviewed all differ in regard to the matching takeoff and landing sites from a controversial series of reports by Engelberg-Kulka and colleagues on another translational anomaly in E. coli (22–25). These workers identified an abnormal foreshortened translation product of the trpR gene (22), and later of a trpR-lacZ fusion (23), which they ascribed to a 55-nucleotide hop (23–25). Surprisingly, their analysis shows no matching codons at the putative takeoff and landing sites. Recently, Willis et al. (26) carried out a restudy of the phenomenon and were unable to detect any hopping at all; they concluded instead that the aberrant protein is in fact produced in one case by termination and downstream reinitiation, and in another by (+1) frameshifting without hopping.
Our results, like those of Weiss and colleagues (12, 13, 20, 21), emphasize the importance of matching P sites and therefore seem incompatible with the analysis of Engelberg-Kulka and her colleagues. On the other hand, we did detect some low, residual β-galactosidase activity—about 1% of the lacZ+ control—in constructs without matching sites (Table 1). We have been unable to determine, from amino acid analysis, whether these low values reflect leaping to nonhomologous sites. The most likely nonhomologous site is the TTA triplet in the β-galactosidase reading frame 28 nucleotides downstream of the takeoff site; this would be complementary to the peptidyl-Phe-tRNA’s anticodon at the first and second positions. Amino acid sequence data will be necessary to analyze these and other putative nonhomologous slides. In any case, the hungry codon phenomenon greatly expands the repertoire of slides calling for analysis.
The sequence changes we have examined affect β-galactosidase synthesis in control (uninhibited) cells and in ILHX-inhibited cells in a correlated fashion (r = 0.9 for Table 1 data). This suggests that most of the background enzyme synthesis is caused by the slide events documented with ILHX inhibition. If there is this much sliding in a small region encompassing less than 1% of the lacZ message, then several percent of normal β-galactosidase may include slides somewhere in translation, and an even larger fraction of the ribosomes will have wandered out of reading frame somewhere along the message. These considerations are consistent with earlier studies showing that more than 20% of the ribosomes that initiate translation on the lacZ message fail to stay the course, suggesting a high frequency of processivity errors on a message this long (27–31).
This tendency of ribosomes to wander is greatly stimulated at hungry codons, particularly in relA− cells. High-efficiency translation of heterologous genes in host cells can easily generate hungry codons, with the attendant missense errors, processivity errors, frameshifts (32, 33), and, as the present results show, slides. We think the biotechnology industry would do well to be cognizant of these phenomena and the possible risks they pose.
Acknowledgments
Dr. Jenny Atkinson carried out some preliminary experiments on a different subject which led to those described in this communication. John Miller performed the experiments on leaping during valine inhibition summarized in the legend of Table 1. Bill Hellberg helped with preparation of the manuscript. This work was supported by Grant GM13626-31 from the National Institutes of Health.
ABBREVIATIONS
- ILHX
isoleucine hydroxamate
- P site
peptidyl site
References
- 1.Negre D, Cortay J C, Donini P, Cozzone A J. Biochemistry. 1989;28:1814–1819. doi: 10.1021/bi00430a058. [DOI] [PubMed] [Google Scholar]
- 2.Parker J. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker J. In: Transfer RNA in Protein Synthesis. Hatfield D L, Lee B J, Pirtle R M, editors. Boca Raton, FL: CRC; 1992. pp. pp.191–267. [Google Scholar]
- 4.Farabaugh P. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farabaugh P. Programmed Alternative Reading of the Genetic Code. Austin, TX: R. G. Landes; 1997. [Google Scholar]
- 6.Menninger J R. J Biol Chem. 1978;253:6808–6813. [PubMed] [Google Scholar]
- 7.Menninger J R, Caplan A B, Gingrich P K, Atherly A G. Mol Gen Genet. 1983;190:215–221. doi: 10.1007/BF00330642. [DOI] [PubMed] [Google Scholar]
- 8.Heurgue-Hamard V, Mora L, Guarneros G, Buckingham R H. EMBO J. 1996;15:2826–2833. [PMC free article] [PubMed] [Google Scholar]
- 9.Barak Z, Lindsley D, Gallant J. J Mol Biol. 1996;256:676–684. doi: 10.1006/jmbi.1996.0117. [DOI] [PubMed] [Google Scholar]
- 10.Newton W A, Beckwith J R, Zipser D, Brenner S. J Mol Biol. 1965;14:290–296. doi: 10.1016/s0022-2836(65)80250-9. [DOI] [PubMed] [Google Scholar]
- 11.De Felice M, Levinthal M, Iaccarino M, Guardiola J. Microbiol Rev. 1979;43:42–58. doi: 10.1128/mr.43.1.42-58.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss R B, Dunn D N, Atkins J F, Gesteland R F. Cold Spring Harbor Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 13.O’Conner M O, Gesteland R F, Atkins J F. EMBO J. 1989;8:4315–4323. doi: 10.1002/j.1460-2075.1989.tb08618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Primakoff P. J Bacteriol. 1981;145:410–416. doi: 10.1128/jb.145.1.410-416.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foley D P, Dennis P, Gallant J. J Bacteriol. 1981;145:641–643. doi: 10.1128/jb.145.1.641-643.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel U, Sorensen N, Pedersen S, Jensen K F, Kilstrup N. Mol Microbiol. 1992;6:2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich A K, Li L Y, Parker J. Biochim Biophys Acta. 1991;1089:362–366. doi: 10.1016/0167-4781(91)90177-n. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen M A, Jensen K F, Pedersen S. J Mol Biol. 1994;236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 19.Kane J F, Violand B N, Curran D F, Staten N R, Duffin K L, Bogosian G. Nucleic Acids Res. 1992;20:6707–6712. doi: 10.1093/nar/20.24.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W M, Ao S-Z, Casjens S, Orlandi R, Zeikus R, Weiss R, Winge D, Fang N. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 21.Weiss R B, Huang W M, Dunn D M. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhar I, Miller C, Engelberg-Kulka H. Mol Microbiol. 1992;6:2777–2784. doi: 10.1111/j.1365-2958.1992.tb01457.x. [DOI] [PubMed] [Google Scholar]
- 23.Benhar I, Engelberg-Kulka H. Cell. 1993;72:121–130. doi: 10.1016/0092-8674(93)90056-v. [DOI] [PubMed] [Google Scholar]
- 24.Groisman I, Engelberg-Kulka H. Biochem Cell Biol. 1995;73:1055–1059. doi: 10.1139/o95-113. [DOI] [PubMed] [Google Scholar]
- 25.Engelberg-Kulka H, Schoulaker-Schwarz R. Mol Microbiol. 1994;11:3–8. doi: 10.1111/j.1365-2958.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 26.Wills N M, Ingram J A, Gesteland R F, Atkins J F. J Mol Biol. 1997;272:491–498. doi: 10.1006/jmbi.1997.1187. [DOI] [PubMed] [Google Scholar]
- 27.Manley J L. J Mol Biol. 1978;125:407–432. doi: 10.1016/0022-2836(78)90308-x. [DOI] [PubMed] [Google Scholar]
- 28.Tsunk K, Inouye S, Inouye M. J Biol Chem. 1989;264:4428–4433. [PubMed] [Google Scholar]
- 29.Jorgensen F, Kurland C G. J Mol Biol. 1990;215:511–521. doi: 10.1016/S0022-2836(05)80164-0. [DOI] [PubMed] [Google Scholar]
- 30.Dong H, Kurland C G. J Mol Biol. 1995;248:551–561. doi: 10.1006/jmbi.1995.0242. [DOI] [PubMed] [Google Scholar]
- 31.Kurland C G. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberger R. Dev Biol Stand. 1994;83:21–26. [PubMed] [Google Scholar]
- 33.Kurland C, Gallant J. Curr Opin Biotechnol. 1996;7:489–493. doi: 10.1016/s0958-1669(96)80050-4. [DOI] [PubMed] [Google Scholar]