Abstract
The three yeast A kinase catalytic subunit isoforms are redundant for viability. We demonstrate that they have dramatically different roles in pseudohyphal development: Tpk2 is essential, whereas Tpk3 inhibits. Tpk1 has no discernible effect. Two-hybrid analysis identified the transcription factor Sfl1 as a protein that interacts specifically with Tpk2, but not Tpk1 or Tpk3. Deletion of SFL1 enhances pseudohyphal and invasive growth. Flo11, a cell surface flocculin required for pseudohyphal development, is transcriptionally regulated by Tpk2 and Sfl1. Genetic evidence indicates that Tpk2 acts upstream of Sfl1 in the regulation of Flo11.
Gene duplication with subsequent diversification of function is thought to be a key mechanism in the evolution of genomes (1). Recent evidence suggests that the genome of the yeast Saccharomyces cerevisiae duplicated ≈100 million years ago (2). The consequence of this ancient doubling of the genome is that today many yeast genes are present in multiple copies. This apparent redundancy raises the question of whether the extant copies carry out the same function or whether they have diverged and now perform completely distinct functions.
The yeast A kinases are a good paradigm for studying this problem. cAMP-dependent protein kinase (A kinase) is an inactive heterotetramer consisting of a regulatory dimer and two catalytic subunits. Upon binding cAMP, the regulatory dimer releases the two active catalytic subunits. In yeast there are three strikingly similar catalytic isoforms of the A kinase encoded by three distinct genes TPK1, TPK2, and TPK3. All three proteins consist of an ≈300-residue carboxy-terminal kinase domain that is highly conserved among all A kinases and a short amino-terminal extension. Tpk1 and Tpk3 are ≈88% identical over the kinase domain. In this region, Tpk2 is ≈76% identical to either Tpk1 or Tpk3. Strains deficient in all three A kinase catalytic subunits are inviable, but strains containing any one of the three (e.g., tpk1 tpk2 TPK3) are viable (3). Despite this apparent redundancy, some differences in these double mutants with respect to the response to nutrient deprivation have been observed (4–6).
Although any one of the A kinases suffices for viability, we have found that the three A kinases have different roles in pseudohyphal development. Upon starvation for nitrogen, yeast form cells switch their pattern of growth to make chains of elongated cells called pseudohyphae, which invade the agar surface (7). This dimorphic switch is regulated by components of the mating pheromone response mitogen-activated protein kinase (MAPK) pathway: the p65PAK kinase homolog Ste20, the MEK kinase Ste11, the MAPK kinase (MEK) Ste7, the MAPK Kss1 and the transcription factor Ste12 (8, 9). Ste12 interacts combinatorially with the TEA/ATTS family transcription factor Tec1 to regulate pseudohyphal growth (10). Other studies show that pseudohyphal development also is regulated by cAMP independently of the MAPK pathway (S. Rupp, E. Summers, H.-J. Lo, H. Madhani, and G.R.F., unpublished results). Flo11, a downstream target of Ste12 required for both pseudohyphal development and haploid invasive growth, is an excellent reporter for pseudohyphal development because it responds to signals from both the filamentation MAPK and cAMP pathways (S. Rupp, E. Summers, H.-H. Lo, H. Madhani, and G.R.F., unpublished results, and refs. 12 and 13). Haploid strains do not form pseudohyphae when starved for nitrogen, but they do invade the agar surface when grown in rich medium. Genes known to be required for pseudohyphal growth also are necessary for haploid invasive growth (14).
MATERIALS AND METHODS
Media, Growth Conditions, and Yeast Strains.
Standard yeast genetic techniques and growth media were used (15). Synthetic low ammonia dextrose media (SLAD) was used to assay pseudohyphal growth (7). Pseudohyphal and haploid invasive growth were assayed as described (7, 14). All strains used in this study are congenic to the Σ1278b strain background (7, 16). TPK1 was disrupted with the ptpk1∷URA3 deletion construct (3). The ORFs of TPK2, TPK3, SFL1, and MGA1 were deleted by using the PCR disruption method (17). The flo11∷URA3 plasmid of Lo and Dranginis (12) was used to disrupt FL011. The two-micron plasmids YEpTPK1, YEpTPK2, and YEpTPK3 were used for overexpression of TPK1, TPK2, and TPK3 (3). Standard molecular biology techniques were used (18).
Two-Hybrid Screen.
Two-hybrid analysis tools: the plasmids pGBDU and pGAD, the library Y2HL, and the strain pJ69–4a were kindly provided by Phil James (19). The intact ORFs of TPK1, TPK2, and TPK3 were cloned into pGBDU to give pLR117, pLR115, and pLR118. DPH1 cloned into pGBDU was used as a negative control. pJ69–4α was constructed by switching the mating type of pJ69–4a with Ho. pJ69–4α was transformed with the library Y2HL. A pool of ≈106 independent Leu+ transformants was mated to pJ69–4a transformed with TPK2/pGBDU. The 9 × 107 diploids were plated onto Ade− medium to select for two-hybrid interactions resulting in expression of the GAL2-ADE2 reporter. Of the 1,500 Ade+ colonies that appeared on Ade− medium, ≈1,200 also grew on His− medium. TPK2/pGBDU was lost from these 1,200 colonies by passage over fluoroorotic acid medium. Two hundred of the ≈1,100 clones, which required the presence of the bait TPK2/pGBDU for growth on Ade− medium, were analyzed further. Transformants that grew poorly that did not positively retest for two-hybrid interaction with Tpk2 or that appeared to contain the same insert by crosshybridization were not studied further. The remaining 33 clones were tested for two-hybrid interaction specific to TPK2. The diploid strain pJ69–4a × pJ69–4α transformed with each of these 33 positive clones was additionally transformed with TPK1/pGBDU, TPK2/pGBDU, TPK3/pGBDU, or DPH1/pGBDU to identify proteins that interact specifically with Tpk2 by two-hybrid analysis. These 33 clones represented 11 genes, two of whose products (Mga1 and Sfl1) interacted specifically with Tpk2. SFL1 was isolated four times, MGA1 one time, BCY1 seven times, and GIP2 13 times. All other genes were isolated only once. Inserts from positive clones were sequenced by using the primers TA and 3′ (19).
Northern Analysis.
Haploid strains were grown in yeast extract/peptone dextrose liquid medium at 30°C to an OD600 of ≈1.0. Total RNA was harvested, and 10 μg RNA from each strain was analyzed by Northern blotting (18). A PCR product corresponding to bp 3,502–4,093 of the FLO11 ORF was used to probe for FLO11 message, and an 870-bp PCR product internal to the 3′ exon of ACT1 was used to probe for ACT1 message as a loading control. The amount of FLO11 message in each strain was standardized to the amount of ACT1 message from the same strain and then normalized to the amount of FLO11 message in the wild-type strain.
RESULTS
Null Mutants in the Three A Kinase Genes Have Distinct Pseudohyphal Phenotypes.
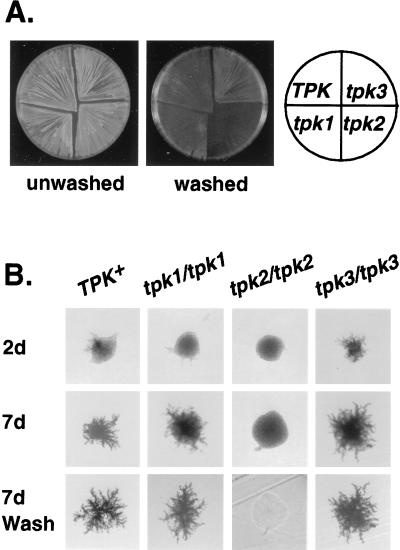
We uncovered the diversity of A kinase function in yeast by observing the pseudohyphal phenotypes of a set of mutants each lacking a single TPK gene (tpk1 TPK2 TPK3, TPK1 tpk2 TPK3, and TPK1 TPK2 tpk3). Diploid tpk2/tpk2 strains grown on SLAD (low nitrogen medium) are completely defective for pseudohyphal development, whereas tpk3/tpk3 strains are greatly enhanced for filamentation (Fig. 1B). Haploid tpk2 mutants are defective for invasive growth on rich medium, and tpk3 mutants are hyperinvasive. tpk1 mutants are indistinguishable from wild-type strains as both haploids and diploids (Fig. 1A). The accentuated filamentation and invasion phenotypes of tpk3 mutants require a functional TPK2 as tpk2 tpk3 double mutants have the same phenotype as tpk2 single mutants, suggesting a key role for Tpk2 in transmitting the pseudohyphal growth signal to downstream targets.
Figure 1.
The tpk null alleles have distinct pseudohyphal and haploid invasion phenotypes. (A) The haploid strains 10560–23C (Tpk+), LRY629 (tpk3), LRY594 (tpk2), and LRY517 (tpk1) were patched onto a yeast extract/peptone/dextrose plate, grown for 3 days at 30°C, and then incubated for 2 days at room temperature. The plates were photographed both before and after gently washing the cells from the surface of the agar with deionized water (14). (B) The homozygous diploid strains LRY841 (Tpk+), LRY838 (tpk1/tpk1), LRY839 (tpk2/tpk2), and LRY840 (tpk3/tpk3) were streaked onto SLAD plates and grown at 30°C. Individual colonies were photographed after 2 days growth and after 7 days growth. The plates were then vigorously washed to remove all noninvasive cells (7).
The level of TPK2 expression is critical to pseudohyphal formation. Strains heterozygous for TPK2 (tpk2/TPK2) have reduced pseudohyphal growth, and strains in which TPK2 is overexpressed show enhanced pseudohyphal growth. No such effects were observed for either Tpk1 or Tpk3.
Sfl1 Interacts Specifically with Tpk2.
As Tpk2 appeared to be uniquely required for pseudohyphal growth, we used a two-hybrid screen to identify proteins that interact specifically with Tpk2. The intact ORF of TPK2 was fused to the Gal4 DNA-binding domain and used to screen a library of yeast genomic fragments fused to the Gal4 transcriptional activation domain (19). Activation of the GAL2-ADE2 reporter, leading to adenine-independent growth of an Ade− strain, was taken as preliminary evidence for an interaction between the Gal4 DNA-binding domain:TPK2 fusion and the TPK2-interacting protein fused to the Gal4 activation domain. Positive clones were retested in the two-hybrid assay with Tpk1, Tpk2, Tpk3, or Dph1 fused to the Gal4-binding domain. DPH1, a gene required for diphthamide biosynthesis, was used to control for interactions that were not specific to the A kinase isoforms.
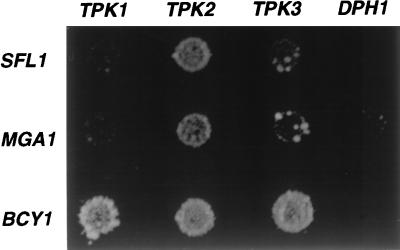
Two classes of proteins interact with Tpk2 in the two-hybrid assay: the first class is indiscriminate and interacts with Tpk1, Tpk2, and Tpk3; the second class is selective and interacts preferentially with Tpk2 (Fig. 2). One of the proteins that interacts with all three catalytic subunits is Bcy1, the negative regulatory subunit of the A kinase, which has been previously shown to bind Tpk1, Tpk2, and Tpk3. The second class consists of Sfl1 and Mga1, both of which interact selectively with Tpk2. Sfl1 and Mga1 are putative transcription factors belonging to a group of five yeast proteins (Mga1, Sfl1, Hsf1, Skn7, and Hms2) containing the helix–turn–helix DNA-binding motif found in the heat shock transcription factor Hsf1. All but Skn7 contain at least one consensus A kinase phosphorylation site ([R/K][R/K] X [S/T]): Sfl1 has five, Mga1 has two, Hsf1 has four, and Hms2 has one (20).
Figure 2.
Sfl1 and Mga1 interact preferentially with Tpk2 by two-hybrid analysis. The strain pJ69–4a × pJ69–4α was transformed with one bait plasmid and one prey plasmid. The two-hybrid bait plasmids used were pLR117 (TPK1 in pGBDU), pLR115 (TPK2 in pGBDU), pLR118 (TPK3 in pGBDU) and as a negative control DPH1 in pGBDU. The two-hybrid prey plasmids SFL1/pGAD, MGA1/pGAD, and BCY1/pGAD were isolated from the Y2HL library (19). Two-hybrid interaction was assayed by growth on Ade− medium.
sfl1 Null Mutants Are Enhanced for Pseudohyphal Growth.
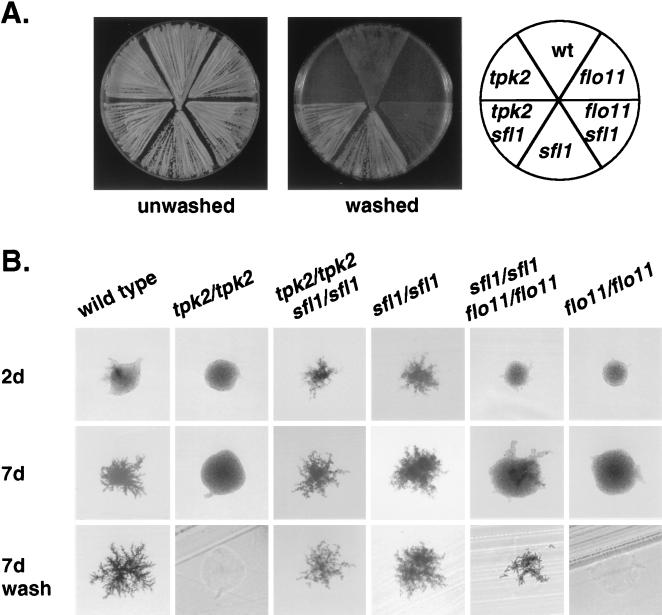
We deleted the ORF of both MGA1 and SFL1. sfl1 loss-of-function mutants have a dramatic phenotype: sfl1 haploid strains are extremely flocculent and hyperinvasive, whereas diploids homozygous for a deletion of sfl1 are greatly enhanced for filamentation (Fig. 3). We constructed an sfl1 tpk2 double mutant to see whether the tpk2 pseudohyphal defect was blocked by loss of function of Sfl1. sfl1 tpk2 double mutants have the same phenotype as sfl1 single mutants, suggesting Sfl1 acts downstream of Tpk2. We could not detect a dramatic defect in pseudohyphal formation for mga1 null mutants.
Figure 3.
Genetic analysis indicates Tpk2 is upstream of Sfl1, which is upstream of Flo11. (A) The haploid strains 10560–23C (wild-type), LRY855 (flo11), LRY857 (sfl1 flo11), LRY801 (sfl1), LRY846 (sfl1 tpk2), and LRY582 (tpk2) were patched onto a yeast extract/peptone/dextrose plate and grown for 3 days at 30°C and then for 2 days at room temperature. The plates were photographed both before and after gently washing the cells from the surface of the agar. (B) The homozygous diploid strains LRY841 (wild-type), LRY839 (tpk2/tpk2), LRY869 (tpk2/tpk2 sfl1/sfl1), LRY829 (sfl1/sfl1), LRY871 (sfl1/sfl1 flo11/flo11), and LRY870 (flo11/flo11) were streaked onto SLAD plates and grown at 30°C. Individual colonies were photographed after 2 days growth and after 7 days growth. The plates then were vigorously washed to remove all noninvasive cells.
FLO11 Is Transcriptionally Regulated by both Tpk2 and Sfl1.
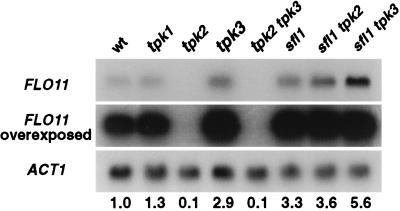
We measured FLO11 mRNA levels in tpk1, tpk2, and tpk3 mutant strains by Northern blotting (Fig. 4). The level of FLO11 mRNA for each Tpk mutant as compared with wild-type corresponds with its colony phenotype: tpk2 mutants show a 10-fold reduction whereas tpk3 mutants have a 3-fold elevation. The levels of FLO11 mRNA also are reduced 10-fold in tpk2 tpk3 double mutants. The level of FLO11 mRNA in tpk1 mutants is indistinguishable from that of wild-type. The dependence of FLO11 expression on Tpk2 suggests that Tpk2 is required for mediating the cAMP signal to FLO11. The proposal that Sfl1 is a downstream target of Tpk2 also is supported by the Northern analysis: FLO11 mRNA levels are increased 3-fold in an sfl1 mutant and are similarly increased in an sfl1 tpk2 double mutant. The levels of FLO11 mRNA in the sfl1 tpk3 double mutant exceeds that of either single mutant.
Figure 4.
FLO11 expression is regulated by TPK2 and SFL1. FLO11 expression levels were determined by Northern analysis. The amount of FLO11 message was standardized to the amount of ACT1 message in each strain and then normalized with respect to wild-type. The results of this quantitation are shown below the Northern analysis. Lane 1: 10560–2B (wild-type), lane 2: LRY520 (tpk1), lane 3: LRY592 (tpk2), lane 4: LRY636 (tpk3), lane 5: LRY645 (tpk2 tpk3), lane 6: LRY738 (sfl1), lane 7: LRY847 (tpk2 sfl1), and lane 8: (tpk3 sfl1).
Phenotypic analysis of sfl1 flo11 double mutants is consistent with the hypothesis that Sfl1 acts upstream of Flo11. sfl1 flo11 haploid mutant strains are defective for invasive growth on rich medium, and sfl1 flo11 homozygous diploid mutants are defective for pseudohyphal development (Fig. 3). Thus, flo11 loss of function blocks the hyperinvasive and hyperfilamentous phenotypes of sfl1 loss of function. However the defect of the sfl1 flo11 double mutant is not as severe as that of the flo11 single mutants, suggesting that Flo11 is not the only downstream target of Sfl1 involved in pseudohyphal development and haploid invasion.
DISCUSSION
We have shown that the three A kinase catalytic subunits of S. cerevisiae, that are redundant for viability, have dramatically distinct roles in pseudohyphal development. Tpk2 is required for pseudohyphal development: loss-of-function mutants are completely defective for both pseudohyphal and invasive growth, and mRNA levels of the target gene FLO11 are reduced 10-fold. In contrast, Tpk3 inhibits pseudohyphal growth; however, this inhibition requires a functional Tpk2. No pseudohyphal effect was observed for Tpk1.
Our data suggest that the Tpk2 signal is mediated by Sfl1. Sfl1 interacts specifically with Tpk2. Loss of function of Sfl1 greatly enhances pseudohyphal growth and increases FLO11 mRNA levels 3-fold. Genetic evidence indicates that Sfl1 acts downstream of Tpk2 and Flo11 acts downstream of Sfl1. Our model, which explains these data, proposes that the specific role of Tpk2 in pseudohyphal growth is due to substrate specificity: Tpk2, not Tpk1 or Tpk3, interacts with Sfl1, thus preventing its transcriptional repression of FLO11 (Fig. 5). FLO11 is the downstream target for several convergent signaling pathways; it is transcriptionally regulated by both the filamentation MAPK pathway and cAMP (S. Rupp, E. Summers, H.-J. Lo, H. Madhani, and G.R.F., unpublished results and ref. 13). We have shown that the cAMP signal is specifically transmitted by Tpk2 to Flo11. Interaction of Tpk2 with Sfl1, a transcriptional repressor of FLO11, positively regulates FLO11 transcription.
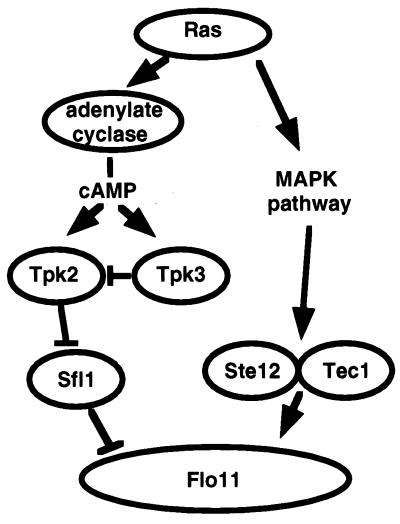
Figure 5.
Both the MAPK pathway and the A kinase pathway converge on FLO11 in the regulation of pseudohyphal growth. The A kinase pseudohyphal signal is mediated specifically by Tpk2.
Sfl1 belongs to a class of five putative transcription factors (Hsf1, Sfl1, Mga1, Hms2, and Skn7) that contain the helix–turn–helix DNA-binding domain of Hsf1. Hsf1 binds to HSE (heat shock element) as a homotrimer (21). Sfl1 is likely to have a more general transcriptional function in addition to its role in pseudohyphal growth because it has been found to interact with components of the RNA polymerase II holoenzyme (22). Mga1 also interacts preferentially with Tpk2, but loss-of-function mutants of Mga1 do not have a dramatic pseudohyphal defect. It is possible that Mga1 has a function in pseudohyphal growth, but that it is functionally redundant, perhaps with Skn7 or Hms2. Another possibility is that Tpk2 has a specific role in cellular processes other than pseudohyphal growth and that the specific interaction between Tpk2 and Mga1 is important for these processes.
Our data show that the three yeast A kinases have distinct roles in pseudohyphal development despite their overlapping function for viability. What differences in the structure of the three proteins could account for the functional differences? The three yeast Tpk proteins have highly divergent amino-terminal extensions. There are differences in length (Tpk1 has 83 residues, Tpk3 has 84, and Tpk2 has 66) and amino acid sequence. The amino-terminal extension of Tpk2 is glutamine-rich: 30% glutamine compared with 11% glutamine in Tpk1 and 2% glutamine in Tpk3. The A kinases of two other fungi in which the cAMP pathway has been implicated in filamentation and pathogenesis, Magnaporthe grisea and Candida albicans, are related more closely to Tpk2 than to either Tpk1 or Tpk3. It is possible that Tpk2 belongs to a class of fungal A kinase catalytic subunits specific to the regulation of dimorphism and polar growth.
In mammalian cells, the multiple A kinase isoforms also may have distinct functions. For example, human T cells express both A kinase type I and A kinase type II; however, data suggest that it is the type I A kinase that inhibits cell proliferation in these cells (11). Whether or not the A kinases can functionally substitute for each other and whether or not the mammalian A kinases have distinct substrate specificities is not yet clear. Our data suggest that the yeast A kinase genes evolved different functions by the acquisition of distinct domains that alter the specificity of their highly conserved catalytic domains. We suggest that one attribute of these novel domains is to promote interactions with other proteins, thus refining the specificity of each A kinase. In the case of Tpk2, the distinct functions observed in pseudohyphal growth result from the specific interaction of the A kinase catalytic subunit Tpk2 with the transcription factor Sfl1.
Acknowledgments
We would like to thank Anne Dranginis, Phil James, and Mike Wigler for plasmids, and Steffen Rupp, Brian Cali, Todd Milne, and Eric Summers for reagents. We are grateful to Steffen Rupp, Todd Milne, and Amir Sherman for critical reading of this manuscript. L.S.R. was supported by National Research Service Award Genome Training Grant HG0039 and G.R.F. is supported by National Institutes of Health Grants GM35101 and GM40266. G.R.F. is an American Cancer Society Professor of Genetics.
ABBREVIATIONS
- MAPK
mitogen-activated protein kinase
- SLAD
synthetic low ammonia dextrose
References
- 1.Cooke J, Nowak M A, Boerlijst M, Maynard-Smith J. Trends Genet. 1997;13:360–364. doi: 10.1016/s0168-9525(97)01233-x. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe K H, Shields D C. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 3.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 4.Cameron S, Levin L, Zoller M, Wigler M. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- 5.Cherry J R, Johnson T R, Dollard C, Shuster J R, Denis C L. Cell. 1989;56:409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- 6.Mbonyi K, van Aelst L, Arguelles J C, Jans A W, Thevelein J M. Mol Cell Biol. 1990;10:4518–4523. doi: 10.1128/mcb.10.9.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 9.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 10.Madhani H D, Fink G R. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 11.Skalhegg B S, Landmark B F, Doskeland S O, Hansson V, Lea T, Jahnsen T. J Biol Chem. 1992;267:15707–15714. [PubMed] [Google Scholar]
- 12.Lo W-S, Dranginis A M. J Bacteriol. 1996;178:7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo W-S, Dranginis A M. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 16.Grenson M, Mousset M, Wiame J M, Bechet J. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- 17.Lorenz M C, Muir R S, Lim E, McElver J, Weber S C, Heitman J. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. In: Current Protocols in Molecular Biology. Chanda V B, editor. New York: Wiley; 1997. [Google Scholar]
- 19.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennelly P J, Krebs E G. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 21.Sorger P K, Nelson H C M. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 22.Song, W. & Carlson, M. (1998) EMBO J. 17, in press. [DOI] [PMC free article] [PubMed]