Abstract
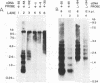
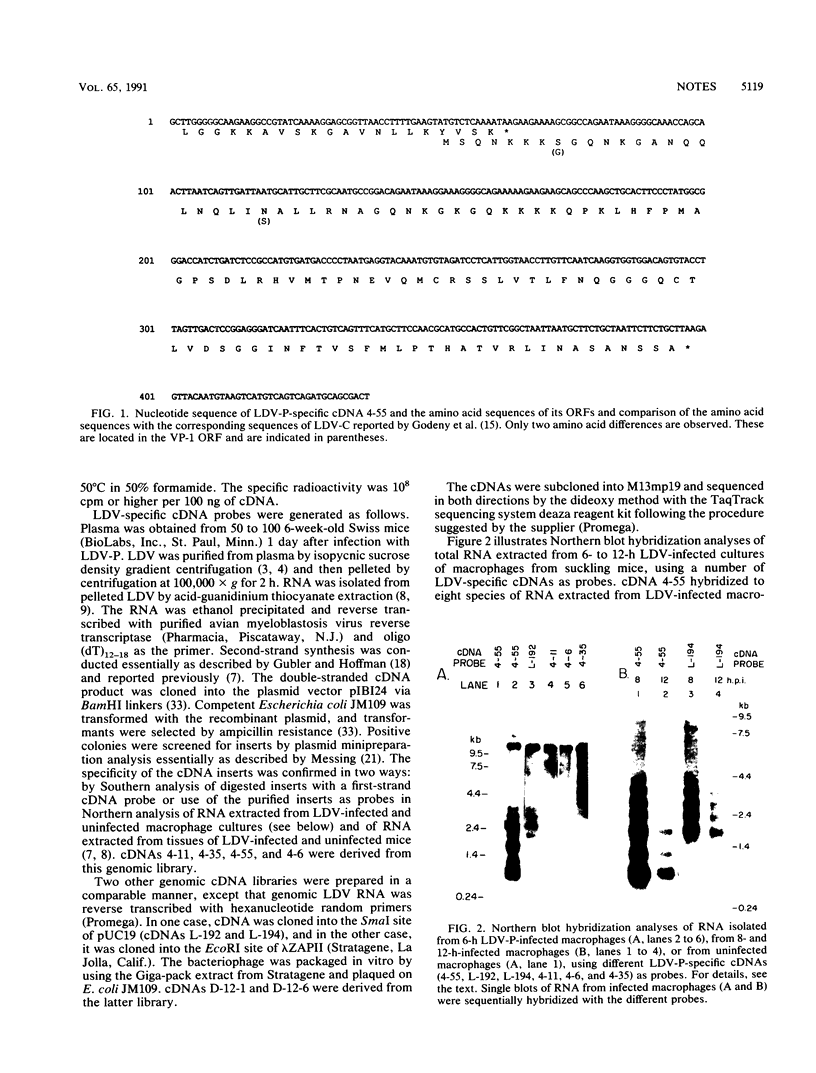
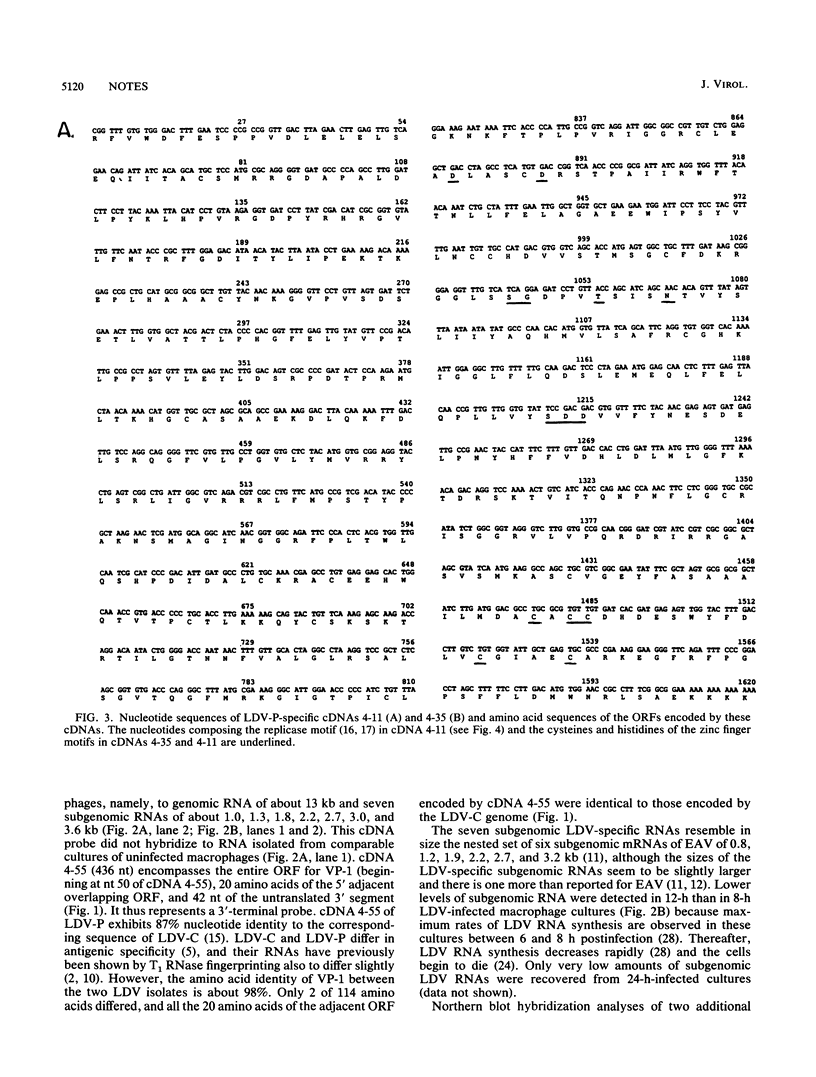
Total RNA was extracted from primary cultures of mouse macrophages isolated from 10-day-old mice 6 to 12 h postinfection with lactate dehydrogenase-elevating virus (LDV). Poly(A)+ RNA was extracted from spleens of 18-h LDV-infected mice. The RNAs were analyzed by Northern (RNA) blot hybridization with a number of LDV-specific cDNAs as probes. A cDNA representing the nucleocapsid protein (VP-1) gene located at the 3' terminus of the viral genome (E. K. Godeny, D. W. Speicher, and M. A. Brinton, Virology 177:768-771, 1990) hybridized to viral genomic RNA of about 13 kb plus seven subgenomic RNAs ranging in size from about 1 to about 3.6 kb. Two other cDNA clones hybridized only to the four or five largest subgenomic RNAs, respectively. In contrast, two cDNAs encoding continuous open reading frames with replicase and zinc finger motifs hybridized only to the genomic RNA. The replicase motif exhibited 75% amino acid identity to that of the 1b protein of equine arteritis virus (EAV) and 44% amino acid identity to those of the 1b proteins of coronaviruses and Berne virus. Combined, the results indicate that LDV replication involves formation of a 3'-coterminal-nested set of mRNAs as observed for coronaviruses and toroviruses as well as for EAV, with which LDV shares many other properties. Overall, LDV, like EAV, possesses a genome organization resembling that of the coronaviruses and toroviruses. However, EAV and LDV differ from the latter in the size of their genomes, virion size and structure, nature of the structural proteins, and symmetry of the nucleocapsids.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton-Darnell M., Plagemann P. G. Structure and chemical-physical characteristics of lactate dehydrogenase-elevating virus and its RNA. J Virol. 1975 Aug;16(2):420–433. doi: 10.1128/jvi.16.2.420-433.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton M. A., Gavin E. I., Fernandez A. V. Genotypic variation among six isolates of lactate dehydrogenase-elevating virus. J Gen Virol. 1986 Dec;67(Pt 12):2673–2684. doi: 10.1099/0022-1317-67-12-2673. [DOI] [PubMed] [Google Scholar]
- Cafruny W. A., Plagemann P. G. Immune response to lactate dehydrogenase-elevating virus: isolation of infectious virus-immunoglobulin G complexes and quantitation of specific antiviral immunoglobulin G response in wild-type and nude mice. Infect Immun. 1982 Sep;37(3):1001–1006. doi: 10.1128/iai.37.3.1001-1006.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafruny W. A., Plagemann P. G. Immune response to lactate dehydrogenase-elevating virus: serologically specific rabbit neutralizing antibody to the virus. Infect Immun. 1982 Sep;37(3):1007–1012. doi: 10.1128/iai.37.3.1007-1012.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Contag C. H., Chan S. P., Wietgrefe S. W., Plagemann P. G. Correlation between presence of lactate dehydrogenase-elevating virus RNA and antigens in motor neurons and paralysis in infected C58 mice. Virus Res. 1986 Dec;6(3):195–209. doi: 10.1016/0168-1702(86)90069-9. [DOI] [PubMed] [Google Scholar]
- Contag C. H., Harty J. T., Plagemann P. G. Dual virus etiology of age-dependent poliomyelitis of mice. A potential model for human motor neuron diseases. Microb Pathog. 1989 Jun;6(6):391–401. doi: 10.1016/0882-4010(89)90081-8. [DOI] [PubMed] [Google Scholar]
- Contag C. H., Plagemann P. G. Age-dependent poliomyelitis of mice: expression of endogenous retrovirus correlates with cytocidal replication of lactate dehydrogenase-elevating virus in motor neurons. J Virol. 1989 Oct;63(10):4362–4369. doi: 10.1128/jvi.63.10.4362-4369.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag C. H., Retzel E. F., Plagemann P. G. Genomic differences between strains of lactate dehydrogenase-elevating virus. Intervirology. 1986;26(4):228–233. doi: 10.1159/000149705. [DOI] [PubMed] [Google Scholar]
- Dominguez G., Wang C. Y., Frey T. K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology. 1990 Jul;177(1):225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Godeny E. K., Speicher D. W., Brinton M. A. Map location of lactate dehydrogenase-elevating virus (LDV) capsid protein (Vp1) gene. Virology. 1990 Aug;177(2):768–771. doi: 10.1016/0042-6822(90)90546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeny E. K., Werner M. R., Brinton M. A. The 3'terminus of lactate dehydrogenase-elevating virus genome RNA does not contain togavirus or flavivirus conserved sequences. Virology. 1989 Oct;172(2):647–650. doi: 10.1016/0042-6822(89)90208-0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989 Jun 26;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Inada T., Mims C. A. Pattern of infection and selective loss of Ia positive cells in suckling and adult mice inoculated with lactic dehydrogenase virus. Arch Virol. 1985;86(3-4):151–165. doi: 10.1007/BF01309821. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Michaelides M. C., Schlesinger S. Structural proteins of lactic dehydrogenase virus. Virology. 1973 Sep;55(1):211–217. doi: 10.1016/s0042-6822(73)81023-2. [DOI] [PubMed] [Google Scholar]
- Onyekaba C. O., Harty J. T., Even C., Hu B. G., Plagemann P. G. Persistent infection of mice by lactate dehydrogenase-elevating virus: effects of immunosuppression on virus replication and antiviral immune responses. Virus Res. 1989 Dec;14(4):297–315. doi: 10.1016/0168-1702(89)90023-3. [DOI] [PubMed] [Google Scholar]
- Onyekaba C. O., Harty J. T., Plagemann P. G. Extensive cytocidal replication of lactate dehydrogenase-elevating virus in cultured peritoneal macrophages from 1-2-week-old mice. Virus Res. 1989 Dec;14(4):327–338. doi: 10.1016/0168-1702(89)90025-7. [DOI] [PubMed] [Google Scholar]
- Ritzi D. M., Holth M., Smith M. S., Swart W. J., Cafruny W. A., Plagemann G. W., Stueckemann J. A. Replication of lactate dehydrogenase-elevating virus in macrophages. 1. Evidence for cytocidal replication. J Gen Virol. 1982 Apr;59(Pt 2):245–262. doi: 10.1099/0022-1317-59-2-245. [DOI] [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Bredenbeek P. J., Horzinek M. C., Rijnbrand R., Spaan W. J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990 Aug 11;18(15):4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Stueckemann J. A., Holth M., Swart W. J., Kowalchyk K., Smith M. S., Wolstenholme A. J., Cafruny W. A., Plagemann P. G. Replication of lactate dehydrogenase-elevating virus in macrophages. 2. Mechanism of persistent infection in mice and cell culture. J Gen Virol. 1982 Apr;59(Pt 2):263–272. doi: 10.1099/0022-1317-59-2-263. [DOI] [PubMed] [Google Scholar]
- Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986 Jul 25;14(14):5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway E. G., Brinton M. A., Gaidamovich SYa, Horzinek M. C., Igarashi A., Käriäinen L., Lvov D. K., Porterfield J. S., Russell P. K., Trent D. W. Togaviridae. Intervirology. 1985;24(3):125–139. doi: 10.1159/000149632. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Vries A. A., Chirnside E. D., Bredenbeek P. J., Gravestein L. A., Horzinek M. C., Spaan W. J. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990 Jun 11;18(11):3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J. A., Snijder E. J., Chirnside E. D., de Vries A. A., Horzinek M. C., Spaan W. J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991 Jun;65(6):2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo M. F., Horzinek M. C., van der Zeijst B. A. Equine arteritis virus-infected cells contain six polyadenylated virus-specific RNAs. Virology. 1982 Apr 30;118(2):345–352. doi: 10.1016/0042-6822(82)90354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]