Abstract
The mouse rump white (Rw) mutation causes a pigmentation defect in heterozygotes and embryonic lethality in homozygotes. At embryonic day (E) 7.5, Rw/Rw embryos are retarded in growth, fail to complete neurulation and die around E 9.5. The Rw mutation is associated with a chromosomal inversion spanning 30 cM of the proximal portion of mouse chromosome 5. The Rw embryonic lethality is complemented by the W19H deletion, which spans the distal boundary of the Rw inversion, suggesting that the Rw lethality is not caused by the disruption of a gene at the distal end of the inversion. Here, we report the molecular characterization of sequences disrupted by both inversion breakpoints. These studies indicate that the distal breakpoint of the inversion is associated with ectopic Kit expression and therefore may be responsible for the dominant pigmentation defect in Rw/+ mice; whereas the recessive lethality of Rw is probably due to the disruption of the gene encoding dipeptidyl aminopeptidase-like protein 6, Dpp6 [Wada, K., Yokotani, N., Hunter, C., Doi, K., Wenthold, R. J. & Shimasaki, S. (1992) Proc. Natl. Acad. Sci. USA 89, 197–201] located at the proximal inversion breakpoint.
Classical genetic studies of the mouse have involved extensive analyses of coat color mutations. Using tester strains that harbor coat color mutations, large scale mutagenesis screens expanded the collection of alleles at loci, such as agouti, brown, albino, dilute, pink-eyed dilution, short ear, piebald-spotting, and others (1). Molecular isolation of genes disrupted in these mutations and characterization of their allelic series revealed new genetic pathways that control developmental processes and basic aspects of cell signaling (2, 3).
A cluster of coat color mutations in the central portion of mouse chromosome 5 includes white spotting (W), patch (Ph), and rump white (Rw) (4). The various alleles of W disrupt the structure or expression of Kit receptor tyrosine kinase (5–9). The role of Kit in the intracellular signal transduction pathways in many distinct cell types is reflected by the pleiotropic affect of W on primordial germ cells, hematopoietic cells, melanocytes, and interstitial cells of Cajal in the small intestine, (9, 10). The Ph mutation is associated with a deletion encompassing another receptor tyrosine kinase gene, Pdgfra (11–14). Homozygous Ph/Ph mice die between embryonic day (E) 9 and E 16 and show severe morphological anomalies (11, 15), with some but not all anomalies observed in embryos with the targeted null-mutation for PDGFαR (16). However, the pigmentation defect apparent in Ph/+ mice cannot be attributed to haploinsufficiency of PDGFRα (16). Expression analysis combined with physical mapping of the Ph deletion and regulatory alleles of W (Wbd and Wsh) (6, 17, 18), suggested the alteration of long range cis-regulatory elements, which cause ectopic expression of Kit in the dermatome of the somites at E 10.5 and E 11.5. Misregulation of Kit, primarily its ectopic expression in neural crest-derived melanoblasts, leads to the abnormal pigment patterns in these mutations, possibly by altering melanocyte dispersal and survival (19, 20).
The least characterized mutation in this cluster, Rw, is associated with depigmentation of the sacrolumbar region in heterozygotes, whereas homozygotes die in utero during early postimplantation development (4). Rw/Rw embryos form three germ layers but arrest after E 7.5 at the end of gastrulation (21). Whereas suppressed recombination between the Rw and the wild-type chromosomes suggested a large chromosomal rearrangement on the Rw chromosome, physical mapping by fluorescense in situ hybridization unequivocally revealed an inversion, named In(5)6H, spanning 30 cM in the proximal third of mouse chromosome 5 (ref. 22 and this publication). The proximal breakpoint of the Rw inversion lies centromeric to the En2 locus, whereas the distal breakpoint maps to the region between the Kit and Pdgfra genes in the central portion of the chromosome (14, 22). These studies suggested that the dominant pigmentation defect in Rw also could be attributed to the long range effect on Kit expression in melanocyte precursors (14, 19). The Rw embryonic lethality is complemented by the W19H deletion, which spans the distal boundary of the Rw inversion, suggesting that the Rw lethality is not caused by the disruption of a gene at the distal end of the inversion (14, 23). Therefore, assuming a simple chromosomal rearrangement, the disrupted gene causing the lethality may map to the proximal inversion breakpoint.
To gain further insight into the molecular basis underlying the developmental defects associated with the Rw mutation, we undertook the molecular cloning and characterization of the Rw inversion breakpoints and sequences surrounding both ends of the Rw inversion. Here, we report that the loss or rearrangement of sequences flanking the distal breakpoint causes misregulation of Kit, expressed from the Rw chromosome. Our analysis found that the proximal breakpoint disrupts the previously described Dpp6 gene, encoding dipeptidyl peptidase-like protein 6 (24). The embryonic form of Dpp6 is expressed in wild-type embryos at early post-implantation stages, suggesting that the disruption of Dpp6 is the underlying cause of the Rw/Rw lethality. This finding suggests the possibility that DPP6 may have a novel function in embryonic development.
MATERIALS AND METHODS
Mice.
Inbred strains of mice (C57BL/6J and C3H/HeJ) and the W19H mutation were obtained from the Jackson Laboratory (Bar Harbor, ME). Rw mice were provided by Colin Beechey and Bruce Cattanach, Medical Research Council, Radiobiology Unit, Harwell, UK.
Pulsed-Field Gel Electrophoresis Analysis.
Methods for pulsed-field gel electrophoresis, including DNA preparation of agarose blocks and restriction analysis have been described previously (25).
Generation and Screening of Cosmid Libraries.
The genomic cosmid library of Rw/+ DNA and subgenomic cosmid library of YAC B20.S3.RA.C6 in SuperCos 1 vector were constructed according to manufacturer’s instructions (Stratagene). Colonies were plated and screened according to published protocols (25).
cDNA Library Screening.
A E 8.5 mouse cDNA library, cloned in λZAP II (Stratagene) and provided by P. Labosky and K. Mahon, was screened by using the radiolabeled 3-kb Dpp6–EcoRI fragment.
Sequencing and Sequence Analysis.
Automated dideoxy terminator cycle sequencing was carried out on cosmid and plasmid DNA by using Dye Terminator sequencing chemistry with Taq FS polymerase from Applied Biosystems. Reaction products were purified by G50 spin columns and analyzed on Applied BioSystems 373A and 377 automated sequencers. Sequence files were edited and assembled into contigs by using the sequencher software (GeneCodes, Ann Arbor, MI). For coding sequence identification, genomic sequences were systemically analyzed with blastn and blastx alogorithms against public databases. Exon predictions were performed with grail1 and grail2, gene finder, and genie programs.
Southern Blot Hybridization.
DNA separated by pulsed field gel electrophoresis analysis (PFGE) or by conventional electrophoresis was transferred to Hybond N+ (Amersham) membrane by capillary blotting in denaturation buffer (25, 26). Filters were UV crosslinked (autocrosslink set up on Stratalinker; Stratagene) and hybridized at 65°C in hybridization buffer. Filters were washed in 0.1× SSC; 0.1% SDS at 65°C.
RNA Analysis.
For Northern blot hybridization, tissue was collected from C57BL/6J and Rw/+ mice. The extraction of total RNA was done by using standard protocols (27). Fifteen micrograms of total RNA was separated in a 1% denaturing formaldehyde gel, transferred to Hybond N (Amersham) membrane, and UV crosslinked (autocrosslink set up on Stratalinker; Stratagene). Hybridization was performed at 65°C in hybridization buffer (25). Filters were washed in 0.1× SSC; 0.1% SDS at 65°C.
In Situ Hybridization.
For in situ hybridization, embryos were fixed in modified Carnoy’s fixative and embedded in paraffin. Embryos were sectioned at 6 μM, and slides were hybridized and washed as described previously (28). Slides were then dipped into XL emmulsion (Eastman Kodak) and developed after several weeks.
RESULTS
Isolation of Sequences Disrupted by the Distal Breakpoint of the Rw Inversion.
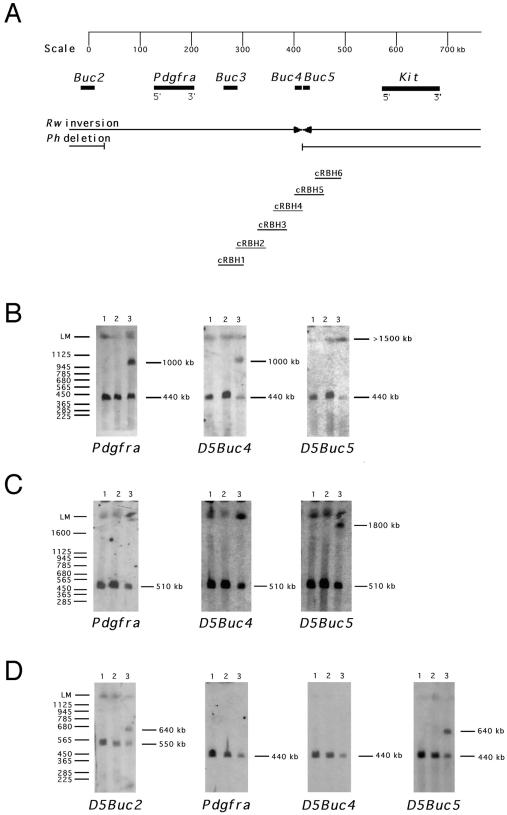
Analysis of the Rw chromosome by fluorescent in situ hybridization and PFGE placed the distal breakpoint of the Rw inversion between Pdgfra and Kit (14) (Fig. 1A). A YAC clone spanning the region around the distal breakpoint was isolated (29), and a subgenomic cosmid library was used to perform a chromosome walk between D5Buc3 and Kit (Fig. 1A). To refine the position of the breakpoint region, unique hybridization probes from the cosmid walk were used in PFGE analysis of Rw/+ and +/+ (C57BL/6J and C3H/HeJ) DNA (Fig. 1 B and C). A Pdgfra probe and several probes isolated from cosmids cRBH1 through cRBH5 displayed distinct hybridization profiles in +/+ and Rw/+ DNA. For example, Pdgfra and probe D5Buc4, detected a 440-kb BssHII DNA fragment on the wild-type chromosome but also hybridized to a 1,000-kb rearranged BssHII DNA fragment in Rw/+ (Fig. 1B). However, the probe D5Buc5, located on the adjacent cosmid cRBH5, hybridized to a Rw-specific fragment of a different size (>1,500 kb). Other probes located distal to cRBH5 also detected this Rw-specific PFGE fragment. These findings show that D5Buc5, located by PFGE analysis 160–220 kb proximal to Kit, maps outside the Rw inversion and that cRBH5 spans the distal inversion breakpoint (Fig. 1).
Figure 1.
PFGE analysis of the chromosomal rearrangements associated with the Rw and Ph mutations. (A) Map positions of the loci Pdgfra, Kit, D5Buc2, D5Buc3, D5Buc4, and D5Buc5 relative to the chromosomal rearrangements associated with Ph and Rw. The probes D5Buc2 and D5Buc3 are described previously (14, 26), and the probes D5Buc4 and D5Buc5 are unique probes from cosmids cRBH4 and cRBH5, respectively, as described in the text. (B) PFGE analysis of the Rw mutation: C57BL/6J (lane 1), C3H/He (lane 2), and Rw/+ (lane 3) DNA digested with BssHII hybridized with Pdgfra, D5Buc4, and D5Buc5. (C) PFGE analysis of the Rw mutation: C57BL/6J (lane 1), C3H/He (lane 2), and Rw/+ (lane 3) DNA digested with NotI hybridized with Pdgfra, D5Buc4, and D5Buc5. (D) PFGE analysis of the Ph mutation. C57BL/6J (lane 1), C3H/He (lane 2), and Ph/+ (lane 3) DNA digested with BssHII hybridized with Pdgfra, D5Buc2, D5Buc4, and D5Buc5. The sizes (in kb) of DNA fragments are shown on the right.
Based on PFGE analysis, the D5Buc3-Kit region also should contain the distal breakpoint for the Ph deletion (14). The Pdgfra locus, D5Buc3, and several loci defined by probes from the cosmids cRBH1–4 (including D5Buc4) were found to be deleted in the Ph mutation (Fig. 1D; refs. 12 and 14 and data not shown). However, D5Buc5 detected the distal breakpoint of the Rw inversion and also detected rearranged fragments in PFGE analysis of Ph/+ DNA (Fig. 1 A and C). Moreover, hybridization of probes D5Buc4 and D5Buc5 to Southern blots containing Ph/+ DNA showed that the same 3.9-kb BamHI DNA fragment disrupted by the distal breakpoint of the Rw inversion also flanks the distal breakpoint of the Ph deletion (Fig. 1 and data not shown). We performed nucleotide sequence analysis of 10 kb of wild-type DNA surrounding the disrupted 3.9-kb BamHI fragment. Analysis of the sequenced region did not reveal homology to any known genes or expressed sequence tags (in GenBank and Swissprot databases), or potential coding sequences, as determined by grail 2 and gene finder. Moreover, cRBH5 did not detect transcripts when hybridized to multiple tissue Northern blots (data not shown), and therefore it is unlikely that the distal breakpoints of Rw and Ph rearrangements disrupt coding sequences of a structural gene. However, several inverted repeat sequences surrounding the closely located breakpoints may represent elements that facilitated chromosome breakage in these two independently isolated mutations.
The Long-Range Effect of the Rw Inversion on Kit Expression.
Discovery that the distal breakpoint of the Rw inversion occurs in the same genomic region as the breakpoints of the Ph deletion and several regulatory alleles of W (Wbd, Wsh, and W57) (6, 17, 18, 26) suggested that the pigmentation defect associated with the Rw mutation might be due to long-range effects on Kit expression. In fact, the examination of Kit expression by in situ hybridization analysis revealed that Kit is ectopically expressed from the Rw chromosome (Fig. 2).
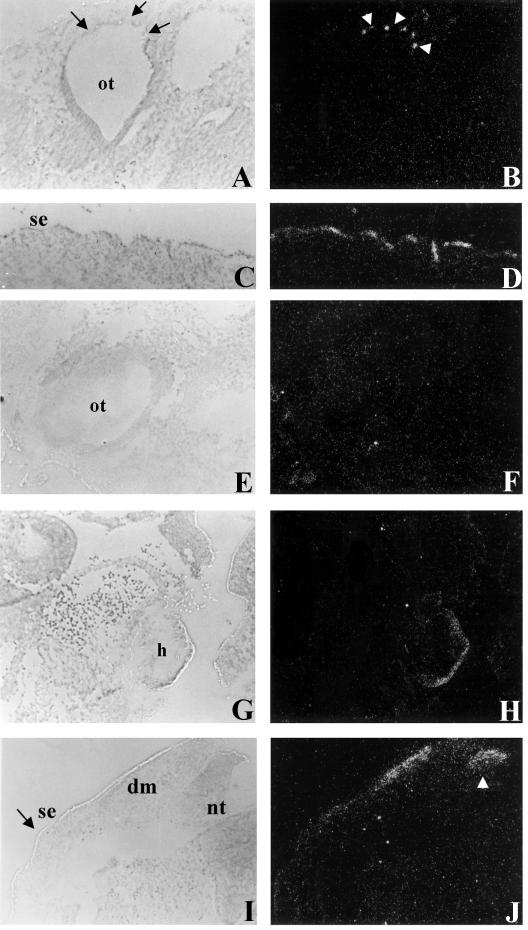
Figure 2.
In situ hybridization of Kit to wild-type and Rw+/+W19H embryos. The expression of Kit in the otic vesicle (ot) in wild-type (A and B) and the absence of Kit expression in Rw+/+W19H (E and F). Ectopic expression of Kit is seen in Rw+/+W19H embryos in the heart (h) (G and H). The expression of Kit in a parasagital section of the surface ectoderm (se) in a wild-type E 10.5 embryo (C and D) and the absence of or reduced expression in transverse sections of surface ectoderm (se, see region denoted by black arrow) from a Rw+/+W19H embryo (I and J). Note that the plane of section in the wild-type (C) is equivalent to the region denoted by the black arrows in the Rw+/+W19H section (I). Ectopic expression is seen in Rw+/+W19H embryos in the dermatome (dm) (I and J) and the dorsal region of the neural tube (nt) (see white arrow head in J).
In situ expression analysis was carried out using Rw+/+W19H compound heterozygous embryos because deletion of Kit on the W19H chromosome allows for the examination of Kit expression specifically from the Rw chromosome. These studies showed that Kit is ectopically expressed in the dorsal neural tube in Rw+/+W19H embryos (Fig. 2 I and J); interestingly, a similar expression pattern is seen in Ph/+ embryos (17, 19). In addition, Kit was observed to be ectopically expressed in the Rw+/+W19H dermatome and heart (Fig. 2G–J), a pattern that is similar to that seen with Wbd and Wsh (6, 18). In contrast, we observed little or no Kit expression in the surface ectoderm and otic vesicle (Fig. 2 E, F, I and J), suggesting that melanoblasts may be reduced in numbers. This possibility is supported by the finding of reduced expression of an early melanoblast marker, tyrosinase-related protein-2 (TRP2) in the otic vesicle (ref. 20 and data not shown) and the fact that adult Rw+/+W19H mice are almost completely depigmented. Overall, our data indicate that the Rw inversion breakpoint located 160–220 kb proximal to Kit has tissue-specific effects on Kit expression during embryogenesis and may be responsible for the dominant pigmentation defect associated with the Rw mutation.
Dpp6 Is Disrupted by the Proximal Breakpoint of the Rw Inversion.
The identification of genomic sequences at the distal breakpoint of the Rw inversion provided the means for isolating sequences juxtaposed to this region on the inverted chromosome. To clone sequences around the proximal breakpoint, a cosmid library of Rw/+ DNA was screened with the D5Buc4 probe (Fig. 1). Restriction analysis showed that an isolated cosmid, cAB1, contains sequences that span the proximal inversion breakpoint. Nucleotide sequence analysis of 29 kb flanking the breakpoint identified four exons corresponding to the gene encoding dipeptidyl aminopeptidase-like protein 6, Dpp6 (24, 30) (Fig. 3B). Chromosomal localization of the Dpp6 gene to the subcentromeric portion of mouse chromosome 5 (31) further confirmed that the isolated sequences span the inversion breakpoint. The DPP6 protein is composed of a short N-terminal cytoplasmic domain, a transmembrane domain, and a long C-terminal extracellular domain (Fig. 3B) and has been shown to lack peptidase activity (24, 32). In the rat, two Dpp6 transcripts, 4.4 and 3.6 kb in length, (DPPX-LV and DPPX-SV), are expressed in the central nervous system whereas a shorter form has a more widespread expression (24, 30). We isolated a mouse cDNA by screening a E 8.5 cDNA library. This embryonic cDNA differs from the reported adult brain isoforms in the first 20 N-terminal amino acids of the hypothetical intracellular domain (Fig. 3B and L. deLecea and G. Sutcliffe, unpublished data).
Figure 3.
Genomic structure of the proximal Rw inversion breakpoints. (A) Wild-type and Rw chromosomal maps give the approximate map positions of genes based on previous studies (14, 22, 31). The Rw inversion is represented by a cross-hatched box. Cosmid cAB1 was isolated by screening a genomic library of Rw/+ DNA with a unique probe from cRBH5 (D5Buc4) as described in the text. Chromosomal distances are not represented on scale. The restriction map of cAB1 is shown. Dashed lines indicate the sequenced region. Solid lines below cAB1 give the approximate positions of Dpp6 exons found by sequencing and grail2 analysis. Arrow shows the positions of the inversion breakpoint. (B) Sequence analysis of the embryonic Dpp6 cDNA (GenBank accession no. AF092507). Shown are the cDNA sequence and the deduced amino acid sequence. The putative intracellular domain is boxed and the transmembrane domain double underlined. The first 20 amino acids are unique in the embryonic DPP6 isoform. The conserved residues, which form the putative catalytic site in nonclassical serine peptidases, are gray shaded, and the aspartic residue, which is replaced by a serine residue shown to be essential for peptidase activity in DPP4, is boxed (32). The location of the proximal Rw inversion breakpoint, which eliminates the C-terminal portion of DPP6, is indicated by an arrowhead.
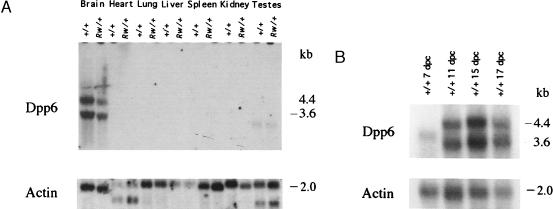
Genomic analysis of the Dpp6 gene on the Rw chromosome places the inversion breakpoint in the coding region between codons corresponding to amino acids 495 and 496 of the predicted protein. These data predict that the inverted chromosome would encode a protein that is missing a significant fraction of the C-terminal region (Fig. 3B). Northern blot analysis confirmed the expression of Dpp6 gene in the adult brain (30); however, truncated transcripts predicted by the genomic rearrangement were not detected in Rw mutant RNA (Fig. 4A). A comparison of steady–state levels of expression of the two Dpp6 transcripts (3.6 and 4.4 kb) in brains of adult Rw/+ and +/+ mice with the level of expression of β actin in the same tissue, shows that Dpp6 is either not expressed from the Rw chromosome or that its truncated message is unstable (Fig. 4A). To investigate whether the disruption of Dpp6 may cause the embryonic lethality of Rw/Rw embryos, we examined the expression of Dpp6 during embryogenesis by Northern blot analysis (Fig. 4B). The two adult mRNA forms of Dpp6 were detected at E 11, E 15, and E 17, whereas at E 7 a transcript of intermediate size, corresponding to the length of the embryonic cDNA clone (3,842 bp), was found to be expressed at a lower level.
Figure 4.
Expression analysis of Dpp6 on the Rw and +/+ chromosome. (A) Northern blot analysis of Dpp6 in +/+ and Rw/+ mice. The Dpp6 cDNA probe detects 4.4 kb and 3.6 kb transcripts in the brain of +/+ and of Rw/+. A transcript of 3.4 kb also was detected in the testes of +/+ and of Rw/+. Total RNA was isolated from adult tissue. RNA loading was assessed by hybridization with β actin and a phosphorimager was used to quantitate and compare levels of expression in +/+ and Rw/+ RNA samples. (B) Analysis of Dpp6 expression in embryonic RNA samples. The probe for Dpp6 detects a 4.4 kb and a 3.6 kb transcripts in 11- to 17-day mouse embryos. A transcript of 3.9 kb was detected in 7-day mouse embryos. The Northern filter (CLONTECH) contains 2 μg of polyadenlyated RNA from E 7, E 11, E 15, and E 17 mouse embryos. RNA loading was assessed by hybridization with β actin.
DISCUSSION
This study describes the molecular characterization of the Rw inversion breakpoints; the distal breakpoint, located 160–220 kb proximal to Kit, probably does not disrupt any transcribed sequences, although it causes ectopic expression of the Kit gene in dermatome, neural tube, and heart at E 10.5. The proximal breakpoint disrupts the gene encoding the dipeptidyl aminopeptidase-like protein 6. This gene of unknown function is expressed in wild-type embryos at an early postimplantation stage when Rw/Rw embryos die and therefore represents a candidate gene for the Rw lethality factor.
The observed overexpression of Kit in Rw+/+W19H embryos and in the previously described Wsh and Wbd mutants, supports the hypothesis of Duttlinger et al. (6, 18) that sequestration of soluble Steel factor, the KIT ligand, by ectopic Kit expression in the dermatome decreases Steel availability to melanoblasts, thereby causing the pigmentation defect seen in adults (6, 17, 19, 33). However, the observation of altered Kit expression in Rw+/+W19H embryos should be interpreted with caution because the analysis was conducted on compound heterozygote embryos, containing only one Kit allele. Therefore, the observed pattern of Kit expression may be due to the synergistic interaction between Rw and W19H. The pigmentation defect in the sacrolumbar region in Rw/+ mice is distinct and different from the “sash-like” depigmentation in Ph, Wsh, Wbd, and W57. Given that different chromosomal rearrangements associated with Ph, Rw, and W alleles display similar or overlapping patterns of Kit expression, it is likely that the loss or juxtaposition of tissue-specific cis-regulatory elements, rather than position-effects on the chromatin structure (34), underlie such long-range effects. However, our data do not permit us to correlate the molecular nature or position of these disrupted sequences with the range of phenotypic anomalies observed in different alleles. Physical mapping has placed chromosomal rearrangements associated with the Ph and Rw mutations and regulatory alleles of W at different positions in the Pdgfra-Kit intergenic region; with the breakpoints of the Ph deletion and the Rw inversion further proximal of the 5′ region of Kit than the breakpoints of the Wsh and Wbd inversions and W57 deletion (6, 17, 18, 26).
A central question is whether the disruption of the Dpp6 gene leads to the developmental arrest and embryonic lethality of Rw/Rw embryos. The biological function of this cell-surface peptidase-like protein is still unresolved. DPP6 protein probably does not possess endopeptidase activity because of an amino acid substitution in the catalytic domain (24, 32). The presence of at least three different forms of the intracellular domain of Dpp6 at different stages of embryonic development and in adult tissues implies that these three DPP6 isoforms may have different functions. Dpp6 is highly expressed in the hippocampus, thalamus, hypothalamus, and striatum and may be involved in neuronal plasticity (30). However, how this predicted role might relate to its role in early postimplantation development remains to be determined. To confirm that the disruption of Dpp6 causes embryonic lethality in Rw, it will be necessary to show that this mutation fails to complement a second loss-of-function mutation in Dpp6 or that Rw/Rw lethality can be rescued by a Dpp6 transgene. Such studies are currently in progress. However, complementation between Rw and loss-of-function allele of Dpp6 would indicate that an independent mutation within the inverted region—a region of suppressed recombination—is responsible for the lethality. The use of the Rw chromosome as a balancer in an ongoing region-specific screen for N-ethyl-N-nitrosourea-induced mutations in the proximal portion of mouse chromosome 5 (35), may facilitate molecular characterization of the gene(s) causing the lethality associated with this mutation.
Acknowledgments
We thank L. de Lecea and G. Suttcliffe for the Dpp6 probe, B. Pavan, K. Steel, and I. Jackson for tyrosinase-related protein-2 (TRP2), K. Steel and A. Bernstein for Kit probes, P. Labosky for the E 8.5 mouse cDNA library, A. Alavizadeh and M. Cohen for help with the expression analysis, M. Santoro of the DNA Sequencing Facility for technical assistance in sequencing and primer walking, and S. Poethig, L. Stubbs, and J. Schimenti for their comments on the manuscript. These studies were supported by National Institutes of Health Grants HD 28410 (to M.B.) and HD 36457 (to C.L.) and National Science Foundation Grant IBN31544 (to C.L.).
ABBREVIATIONS
- E
embryonic day
- PFGE
pulsed field gel electrophoresis analysis
- Rw
Rump white
- Dpp6
dipeptidyl aminopeptidase-like protein 6
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Russell W L. Cold Spring Harbor Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- 2.Jackson I J. Annu Rev Genet. 1994;28:189–217. doi: 10.1146/annurev.ge.28.120194.001201. [DOI] [PubMed] [Google Scholar]
- 3.Barsh G S. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- 4.Searle A G, Truslove G M. Genet Res. 1970;15:227–235. doi: 10.1017/s0016672300001555. [DOI] [PubMed] [Google Scholar]
- 5.Chabot B, Stephenson D A, Chapman V M, Besmer P, Bernstein A. Nature (London) 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 6.Duttlinger R, Manova K, Chu T Y, Gyssler C, Zelenetz A D, Bachvarova R F, Besmer P. Development (Cambridge) 1993;118:705–717. doi: 10.1242/dev.118.3.705. [DOI] [PubMed] [Google Scholar]
- 7.Geissler E N, Ryan M A, Housman D E. Cell. 1988;55:185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 8.Nocka K, Tan J C, Chiu E, Chu T Y, Ray P, Traktman P, Besmer P. EMBO J. 1990;9:1805–1813. doi: 10.1002/j.1460-2075.1990.tb08305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reith A D, Rottapel R, Giddens E, Brady C, Forrester L, Bernstein A. Genes Dev. 1990;4:390–400. doi: 10.1101/gad.4.3.390. [DOI] [PubMed] [Google Scholar]
- 10.Huizinga J D, Thuneberg L, Kluppel M, Malysz J, Mikkelsen H B, Bernstein A. Nature (London) 1995;373:347–349. doi: 10.1038/373347a0. [DOI] [PubMed] [Google Scholar]
- 11.Gruneberg H, Truslove G M. Genet Res. 1960;1:69–90. doi: 10.1017/s0016672300004146. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson D A, Mercola M, Anderson E, Wang C, Stiles C D, Bowen-Pope D F, Chapman V M. Proc Natl Acad Sci USA. 1991;88:6–10. doi: 10.1073/pnas.88.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith E A, Seldin M F, Martinez L, Watson M L, Choudhury G G, Lalley P A, Pierce J, Aaronson S, Barker J, Naylor S L, et al. Proc Natl Acad Sci USA. 1991;88:4811–4815. doi: 10.1073/pnas.88.11.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagle D L, Martin-DeLeon P, Hough R B, Bucan M. Proc Natl Acad Sci USA. 1994;91:7237–7241. doi: 10.1073/pnas.91.15.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schatteman G C, Morrison-Graham K, van Koppen A, Weston J A, Bowen-Pope D F. Development (Cambridge) 1992;115:123–131. doi: 10.1242/dev.115.1.123. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P. Development (Cambridge) 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 17.Duttlinger R, Manova K, Berrozpe G, Chu T-Y, DeLeon V, Timokhina I, Chaganti R S K, Zelenetz A D, Bachvarova R F, Besmer P. Proc Natl Acad Sci USA. 1995;92:3754–3758. doi: 10.1073/pnas.92.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kluppel M, Nagle D L, Bucan M, Bernstein A. Development (Cambridge) 1997;124:65–77. doi: 10.1242/dev.124.1.65. [DOI] [PubMed] [Google Scholar]
- 19.Wehrle-Haller B, Morrison-Graham K, Weston J A. Dev Biol. 1996;177:463–474. doi: 10.1006/dbio.1996.0178. [DOI] [PubMed] [Google Scholar]
- 20.Cable J, Jackson I J, Steel K P. Mech Dev. 1995;50:139–150. doi: 10.1016/0925-4773(94)00331-g. [DOI] [PubMed] [Google Scholar]
- 21.Bucan M, Nagle D L, Hough R B, Chapman V M, Lo C W. Dev Biol. 1995;168:307–318. doi: 10.1006/dbio.1995.1082. [DOI] [PubMed] [Google Scholar]
- 22.Stephenson D A, Lee K-H, Nagle D L, Yen C-H, Morrow A, Miller D, Chapman V M, Bucan M. Mamm Genome. 1994;5:342–348. doi: 10.1007/BF00356552. [DOI] [PubMed] [Google Scholar]
- 23.Lyon M F, Glenister P H, Loutit J F, Evans E P, Peters J. Genet Res. 1984;44:161–168. doi: 10.1017/s0016672300026367. [DOI] [PubMed] [Google Scholar]
- 24.Wada K, Yokotani N, Hunter C, Doi K, Wenthold R J, Shimasaki S. Proc Natl Acad Sci USA. 1992;89:197–201. doi: 10.1073/pnas.89.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann B G, Barlow D P, Lehrach H. Cell. 1987;48:813–825. doi: 10.1016/0092-8674(87)90078-x. [DOI] [PubMed] [Google Scholar]
- 26.Nagle D L, Kozak C A, Mano H, Chapman V M, Bucan M. Hum Mol Genet. 1995;4:2073–2079. doi: 10.1093/hmg/4.11.2073. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Ruangvoravat C P, Lo C W. Dev Dyn. 1992;194:261–281. doi: 10.1002/aja.1001940403. [DOI] [PubMed] [Google Scholar]
- 29.Brunkow M E, Nagle D L, Bernstein A, Bucan M. Genomics. 1995;25:421–432. doi: 10.1016/0888-7543(95)80042-k. [DOI] [PubMed] [Google Scholar]
- 30.de Lecea L, Soriano E, Criado J R, Steffensen S C, Henriksen S J, Sutcliffe J G. Mol Brain Res. 1994;25:286–296. doi: 10.1016/0169-328x(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 31.Wada K, Zimmerman K L, Adamson M C, Yokotani N, Wenthold R J, Kozak C A. Mamm Genome. 1993;4:234–237. doi: 10.1007/BF00417570. [DOI] [PubMed] [Google Scholar]
- 32.Yokotani N, Doi K, Wenthold R J, Wada K. Hum Mol Genet. 1993;2:1037–1039. doi: 10.1093/hmg/2.7.1037. [DOI] [PubMed] [Google Scholar]
- 33.Wehrle-Haller B, Weston J A. Development (Cambridge) 1995;121:731–742. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 34.Hendrich B D, Willard H F. Hum Mol Genet. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- 35.Schimenti J, Bucan M. Genome Res. 1998;8:698–710. doi: 10.1101/gr.8.7.698. [DOI] [PubMed] [Google Scholar]