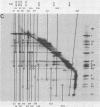
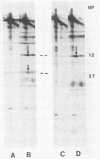
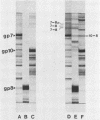
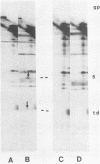
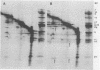
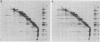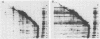Abstract
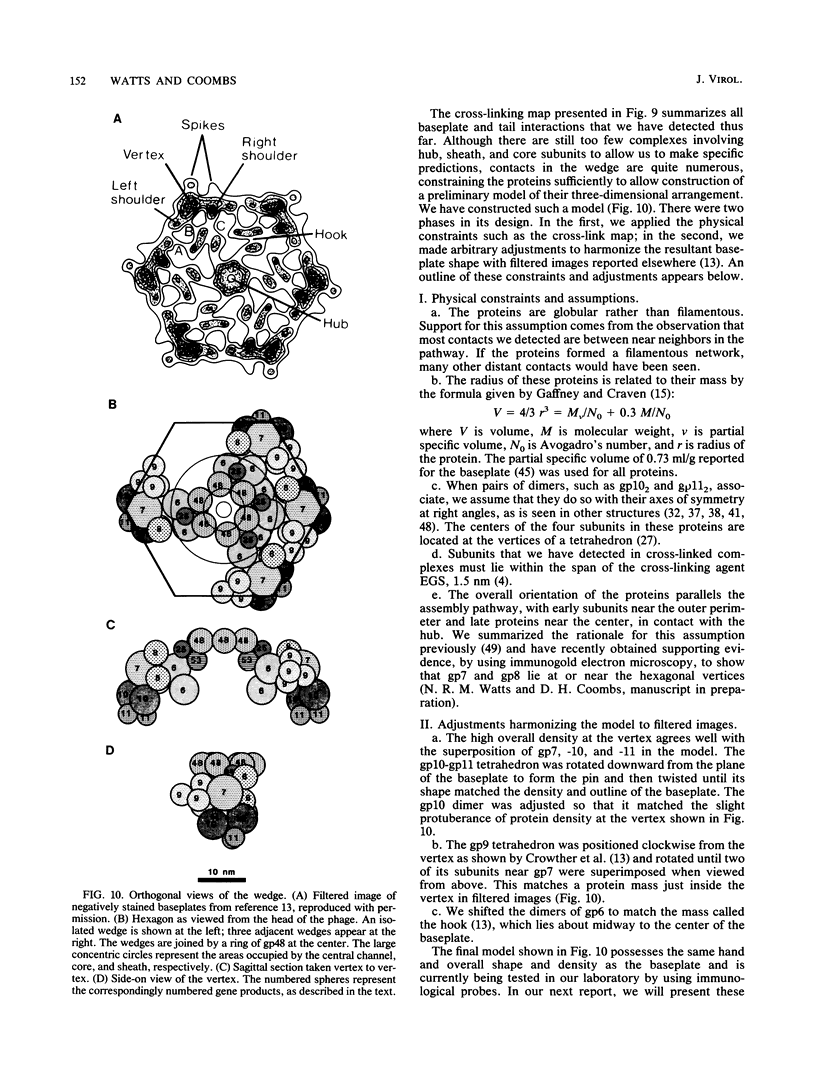
We have carried out a series of reversible chemical cross-linking experiments using the reagent ethylene glycol-bis(succinimidylsuccinate) with the goal of determining the three-dimensional structure of the bacteriophage T4 baseplate. In a previous report, we investigated the near-neighbor contacts in baseplate precursors and substructures (N.R.M. Watts and D.H. Coombs, J. Virol. 63:2427-2436, 1989). Here we report completion of the analysis by examining finished baseplates and tails. Most of the previous contacts were confirmed, and we report several new contacts, including those within the central hub (gp5-gptd2, gp26-gptd), between the hub and the outer wedges (gp6-gp27(2], between baseplate and sheath (gp54-gp18), and between sheath and core (gp19-gp18). On the basis of this and other available information, a partial three-dimensional model of the baseplate is proposed.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdella P. M., Smith P. K., Royer G. P. A new cleavable reagent for cross-linking and reversible immobilization of proteins. Biochem Biophys Res Commun. 1979 Apr 13;87(3):734–742. doi: 10.1016/0006-291x(79)92020-5. [DOI] [PubMed] [Google Scholar]
- Arisaka F., Ishimoto L., Kassavetis G., Kumazaki T., Ishii S. Nucleotide sequence of the tail tube structural gene of bacteriophage T4. J Virol. 1988 Mar;62(3):882–886. doi: 10.1128/jvi.62.3.882-886.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F., Nakako T., Takahashi H., Ishii S. Nucleotide sequence of the tail sheath gene of bacteriophage T4 and amino acid sequence of its product. J Virol. 1988 Apr;62(4):1186–1193. doi: 10.1128/jvi.62.4.1186-1193.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin L. S., Yang C. S. Cross-linking studies of cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase. Biochemistry. 1980 May 13;19(10):2260–2264. doi: 10.1021/bi00551a041. [DOI] [PubMed] [Google Scholar]
- Berget P. B., Warner H. R. Identification of P48 and P54 as components of bacteriophage T4 baseplates. J Virol. 1975 Dec;16(6):1669–1677. doi: 10.1128/jvi.16.6.1669-1677.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Caspar D. L. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980 Oct;32(1):103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley M. P., Wood W. B. Bacteriophage T4 whiskers: a rudimentary environment-sensing device. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3701–3705. doi: 10.1073/pnas.72.9.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs D. H. Density gradient fractionation by piston displacement. Anal Biochem. 1975 Sep;68(1):95–101. doi: 10.1016/0003-2697(75)90683-1. [DOI] [PubMed] [Google Scholar]
- Coombs D. H., Eiserling F. A. Studies on the structure, protein composition and aseembly of the neck of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):375–405. doi: 10.1016/0022-2836(77)90076-6. [DOI] [PubMed] [Google Scholar]
- Coombs D. H., Watts N. R. Generating sucrose gradients in three minutes by tilted tube rotation. Anal Biochem. 1985 Jul;148(1):254–259. doi: 10.1016/0003-2697(85)90654-2. [DOI] [PubMed] [Google Scholar]
- Crowther R. A., Lenk E. V., Kikuchi Y., King J. Molecular reorganization in the hexagon to star transition of the baseplate of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):489–523. doi: 10.1016/0022-2836(77)90081-x. [DOI] [PubMed] [Google Scholar]
- Gaffney P. T., Craven G. R. The use of computerized multidimensional scaling to generate models of the three-dimensional arrangement of ribosomal proteins. Methods Enzymol. 1979;59:602–611. doi: 10.1016/0076-6879(79)59116-2. [DOI] [PubMed] [Google Scholar]
- Gershoni J. M., Palade G. E. Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to a positively charged membrane filter. Anal Biochem. 1982 Aug;124(2):396–405. doi: 10.1016/0003-2697(82)90056-2. [DOI] [PubMed] [Google Scholar]
- KELLENBERGER E., BOLLE A., BOYDELATOUR E., EPSTEIN R. H., FRANKLIN N. C., JERNE N. K., REALE SCAFATI A., SECHAUD J. FUNCTIONS AND PROPERTIES RELATED TO THE TAIL FIBERS OF BACTERIOPHAGE T4. Virology. 1965 Jul;26:419–440. doi: 10.1016/0042-6822(65)90006-1. [DOI] [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol. 1980 Apr;34(1):95–103. doi: 10.1128/jvi.34.1.95-103.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. Roles of bacteriophage T4 gene 5 and gene s products in cell lysis. J Virol. 1980 Apr;34(1):104–107. doi: 10.1128/jvi.34.1.104-107.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975 Dec 25;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol. 1975 Dec 25;99(4):673–694. doi: 10.1016/s0022-2836(75)80179-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975 Dec 25;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971 Jun 28;58(3):693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- Klotz I. M., Langerman N. R., Darnall D. W. Quaternary structure of proteins. Annu Rev Biochem. 1970;39:25–62. doi: 10.1146/annurev.bi.39.070170.000325. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M. Identification of bacteriophage T4D gene products 26 and 51 as baseplate hub structural components. J Virol. 1984 Nov;52(2):344–349. doi: 10.1128/jvi.52.2.344-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lepault J., Leonard K. Three-dimensional structure of unstained, frozen-hydrated extended tails of bacteriophage T4. J Mol Biol. 1985 Apr 5;182(3):431–441. doi: 10.1016/0022-2836(85)90202-5. [DOI] [PubMed] [Google Scholar]
- McFadden B. A., Rao G. R., Cohen A. L., Roche T. E. Isocitrate lyase from Pseudomonas indigofera. V. Subunits and terminal residues and the relation to catalytic activity. Biochemistry. 1968 Oct;7(10):3574–3582. doi: 10.1021/bi00850a035. [DOI] [PubMed] [Google Scholar]
- Meezan E., Wood W. B. The sequence of gene product interaction in bacteriophage T4 tail core assembly. J Mol Biol. 1971 Jun 28;58(3):685–692. doi: 10.1016/0022-2836(71)90033-7. [DOI] [PubMed] [Google Scholar]
- Montag D., Riede I., Eschbach M. L., Degen M., Henning U. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J Mol Biol. 1987 Jul 5;196(1):165–174. doi: 10.1016/0022-2836(87)90519-5. [DOI] [PubMed] [Google Scholar]
- Moody M. F. Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol. 1973 Nov 15;80(4):613–635. doi: 10.1016/0022-2836(73)90200-3. [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Arisaka F., Ishii S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J Virol. 1985 May;54(2):460–466. doi: 10.1128/jvi.54.2.460-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshtrakh M. I., Semenkin V. A. Issledovanie normal'nykh fetal'nogo i vzroslogo gemoglobinov cheloveka metodom messbauérovskoi spektroskopii. Mol Biol (Mosk) 1985 Sep-Oct;19(5):1310–1320. [PubMed] [Google Scholar]
- Penhoet E., Kochman M., Valentine R., Rutter W. J. The subunit structure of mammalian fructose diphosphate aldolase. Biochemistry. 1967 Sep;6(9):2940–2949. doi: 10.1021/bi00861a039. [DOI] [PubMed] [Google Scholar]
- Prilipov A. G., Selivanov N. A., Efimov V. P., Marusich E. I., Mesyanzhinov V. V. Nucleotide sequences of bacteriophage T4 genes 9, 10 and 11. Nucleic Acids Res. 1989 Apr 25;17(8):3303–3303. doi: 10.1093/nar/17.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Association of sindbis virion glycoproteins and their precursors. J Mol Biol. 1982 Jan 15;154(2):325–348. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- Simon L. D., Anderson T. F. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology. 1967 Jun;32(2):279–297. doi: 10.1016/0042-6822(67)90277-2. [DOI] [PubMed] [Google Scholar]
- Simon L. D., Anderson T. F. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology. 1967 Jun;32(2):298–305. doi: 10.1016/0042-6822(67)90278-4. [DOI] [PubMed] [Google Scholar]
- Smith P. R., Aebi U. Studies of the structure of the T4 bacteriophage tail sheath. I. The recovery of three-dimensional structural information from the extended sheath. J Mol Biol. 1976 Sep 15;106(2):243–271. doi: 10.1016/0022-2836(76)90083-8. [DOI] [PubMed] [Google Scholar]
- Sultanova R. A., Chernyak V. Y., Poglazov B. F. Molecular weight of base plates of bacteriophage T4D. Biochim Biophys Acta. 1975 Apr 29;386(2):369–372. doi: 10.1016/0005-2795(75)90280-9. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON H. C., BANASZAK L. J. STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE. STRUCTURE SYMMETRY WITHIN THE MOLECULE. Nature. 1964 Dec 5;204:918–920. doi: 10.1038/204918a0. [DOI] [PubMed] [Google Scholar]
- Watts N. R., Coombs D. H. Analysis of near-neighbor contacts in bacteriophage T4 wedges and hubless baseplates by using a cleavable chemical cross-linker. J Virol. 1989 Jun;63(6):2427–2436. doi: 10.1128/jvi.63.6.2427-2436.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Uchida H. Organization and function of bacteriophage T4 tail. I. Isolation of heat-sensitive T4 tail mutants. Virology. 1973 Mar;52(1):234–245. doi: 10.1016/0042-6822(73)90412-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Uchida H. Organization and function of the tail of bacteriophage T4. II. Structural control of the tail contraction. J Mol Biol. 1975 Feb 25;92(2):207–223. doi: 10.1016/0022-2836(75)90224-7. [DOI] [PubMed] [Google Scholar]