Abstract
An oligoribonucleotide (a 27-mer) that mimics the sarcin/ricin (S/R) domain of Escherichia coli 23S rRNA binds elongation factor EF-G; the Kd is 6.9 μM, whereas for binding to ribosomes it is 0.7 μM. Binding saturates when EF-G and the S/R RNA are equimolar; at saturation 70% of the input RNA is in complexes with EF-G. Binding of EF-G to S/R RNA does not require GTP but is inhibited by GDP; the inhibition by GDP is overcome by GTP. The effects of mutations of the S/R domain nucleotides G2655, A2660, and G2661 suggest that EF-G recognizes the conformation of the RNA rather than the identity of the nucleotides. EF-G also binds to an oligoribonucleotide (an 84-mer) that has the thiostrepton region of 23S rRNA; however, EF-G binds independently to S/R and thiostrepton oligoribonucleotides.
The adoption of a reductionist approach to the studies of the structure and the function of the ribosome, referred to as the ribosome-in-pieces, is a concession that the intact particle is too large and too complex to be amenable to direct analysis. The alternative that we have had recourse to here is to use a small part of the organelle. This strategy requires, for its success, that the ribosome domain of interest retain, in isolation, a relevant structure and a relevant function.
The elongation factor EF-G, in a complex with GTP, binds to the ribosome and catalyzes the translocation of peptidyl-tRNA from the A site to the P site (1, 2). There are three-dimensional structures of EF-G alone (3) and in a complex with GDP (4) from x-ray crystallography; however, little is known of the chemistry of the binding of the factor to ribosomes. There is evidence for the interaction of EF-G with two separate sites in Escherichia coli 23S rRNA: with the thiostrepton region (5–11) and with the sarcin/ricin (S/R) domain (11, 12).
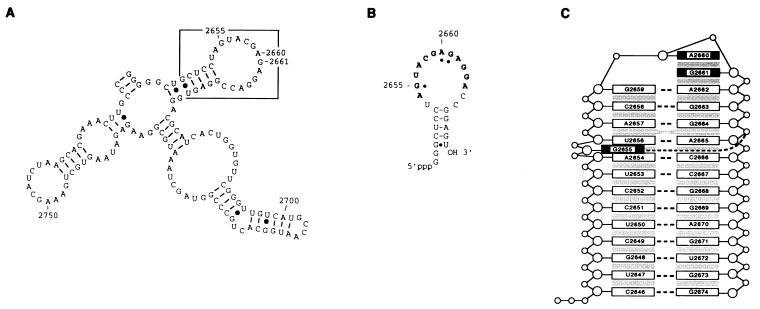
The S/R region of E. coli 23S rRNA (nucleotides 2646–2674), which includes a near universal sequence of 12 nucleotides (2654–2665) (13), is in secondary structure cartoons a stem-loop (Fig. 1A and B); however, the three-dimensional structure, determined by NMR spectroscopy (refs. 14 and 15 and K. Seggerson and P. Moore, personal communication), reveals a more or less continuous helix with a GAGA tetraloop (nucleotides 2659–2662) and a bulged G2655 (Fig. 1C). The domain is the site of action of cytotoxins of two types: sarcin (16), an RNase that cleaves the phosphodiester bond on the 3′ side of G2661 (17); and pokeweed antiviral protein, a ricin-like RNA N-glycosidase that catalyzes the depurination of the 5′ adjacent A2660 (18). The ribotoxins catalyze only these single covalent modifications, which inactivate the ribosome and account entirely for their toxicity.
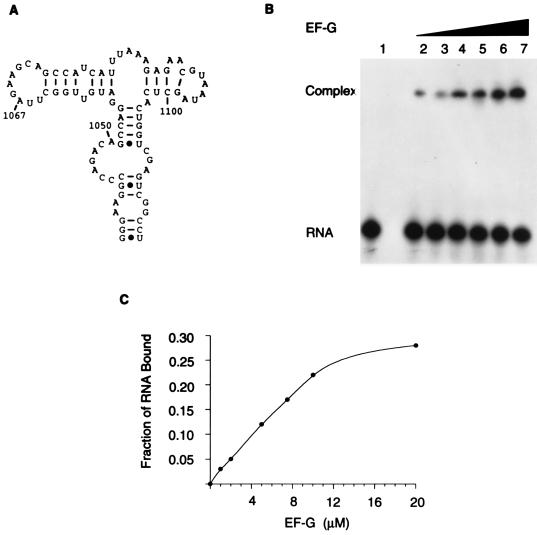
Figure 1.
The S/R domain of 23S rRNA. (A) A portion of E. coli 23S rRNA with the S/R domain boxed. (B) An S/R oligoribonucleotide (a 27-mer) that mimics the sequence of the domain (nucleotides 2648–2672) and has in addition two 5′ guanosine residues that derive from the T7 promoter used to transcribe the RNA. The universal sequence is in boldface letters and the nucleotides protected from chemical modification by EF-G are designated by dots. (C) Representation of the three-dimensional conformation of an E. coli S/R domain oligoribonucleotide determined by NMR spectroscopy (K. Seggerson and P. Moore, personal communication) and based on the NMR determined structure of the closely related eukaryotic S/R domain RNA (14, 15). The numbering is the positions of nucleotides in E. coli 23S rRNA.
The effect of the toxins is de facto evidence that the domain is crucial for ribosome function; crucial, it has been established, because the domain is involved in elongation factor EF-Tu (or EF-1)-dependent binding of aminoacyl-tRNA to ribosomes and elongation factor EF-G (or EF-2)-catalyzed GTP hydrolysis and translocation. The conclusion derives from a series of compelling observations: that in eukaryotes these are the partial reactions of protein synthesis that are most adversely affected by the toxins (19) (peptidyltransferase activity, for example, is not affected); that the S/R domain is the binding site for elongation factors (12); and that EF-Tu and EF-G protect four nucleotides in the S/R domain from chemical modification (11).
MATERIALS AND METHODS
Radioactive oligoribonucleotides were synthesized from synthetic DNA templates with phage T7 RNA polymerase in the presence of the four nucleoside triphosphates (supplemented with [α-32P]ATP) (20, 21). The S/R RNA (a 27-mer) has nucleotides 2648–2672 of the E. coli domain and two additional 5′ guanosines. The thiostrepton RNA (an 84-mer) has nucleotides 1036–1119 of the E. coli domain. The RNA was purified by gel electrophoresis and renatured as described before (20). Physically and enzymatically homogeneous EF-G was prepared from E. coli cells as described by Rohrbach and Bodley (22); the purified protein migrated as a single band when analyzed by electrophoresis in SDS/polyacrylamide gels. The binding reaction contained the following: EF-G (the concentration is in the figure legends); 10 μM oligoribonucleotide; 5 mM Tris⋅HCl (pH 7.6); 2 mM MgCl2; 12.5 mM NaCl; and 10% polyethylene glycol 8000. Incubation was for 10 min at 37°C. Samples were cooled for 10 min at 0°C and binding of EF-G to oligoribonucleotides was assessed by electrophoresis in nondenaturing polyacrylamide gels at 20 V/cm (23). The gels were dried and exposed to x-ray film; quantitation of binding was with imaging plates and a Bio-Imaging Analyzer (Fuji, model BAS 2000) using the program MacBas (Fuji Photofilm). The Kd for binding was determined by gel retardation; the concentration of EF-G in these experiments was 20 μM and the concentration of the oligoribonucleotide was varied from 1 μM to 200 μM. The extent of complex formation (the intensity of the band on the gel) was determined with a Fuji Photoimager. The Kd for binding of EF-G to ribosomes was determined by nitrocellulose filtration (24). The reaction mixture contained 50 mM Tris⋅HCl (pH 7.6), 15 mM MgCl2, 100 mM NH4Cl, 1 mM fusidic acid, 1 mM [3H]GDP (103 cpm/pmol), 0.5 μM E. coli 70S ribosomes, and 0.1–5 μM EF-G. Incubation was for 5 min at 37°C.
RESULTS AND DISCUSSION
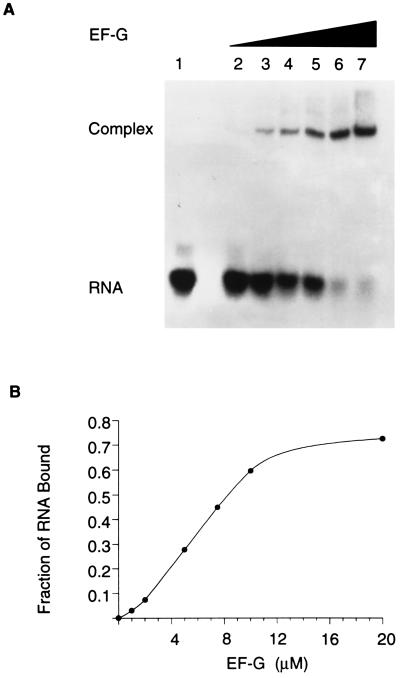
Stable binding of EF-G to 23S rRNA has not heretofore been demonstrated. There is evidence (see above) that the S/R region constitutes at least a portion of the EF-G-binding domain. For this reason an oligoribonucleotide (a 27-mer) that mimics the sequence of nucleotides and the structure of the S/R region of E. coli 23S rRNA was synthesized and the binding of EF-G to the RNA was assessed in a gel-retardation assay (Fig. 2). EF-G binds to the S/R oligoribonucleotide and binding is concentration dependent. At 10 μM EF-G, which approximates the cellular concentration, the binding is near maximal; binding saturates when EF-G and the RNA are equimolar (10 μM here); at saturation, 70% of the RNA is in complexes with EF-G (Fig. 2B). The Kd for the binding of EF-G to the S/R oligoribonucleotide is 6.9 μM; the Kd for binding to ribosomes is 0.7 μM (Table 1). The binding of EF-G to the S/R oligoribonucleotide is specific: there is no binding to poly(U)30, nor to poly(A)30, nor to tRNA, nor to 5S rRNA (results not shown); but more substantial is the observation (see below) that mutations of single nucleotides in the S/R RNA abolish EF-G binding.
Figure 2.
Binding of EF-G to an S/R domain oligoribonucleotide. (A) Gel-retardation assay of the binding of EF-G to radioactive S/R oligoribonucleotide (a 27-mer). The concentrations of EF-G were as follows: lane 1, 0 μM; lane 2, 1 μM; lane 3, 2 μM; lane 4, 5 μM; lane 5, 8 μM; lane 6, 10 μM; and lane 7, 20 μM. The concentration of RNA was 10 μM. (B) Results in A plotted to emphasize that binding saturates and that when the ligands are equimolar the complex contains 70% of the input RNA.
Table 1.
Dissociation constants for binding of EF-G to oligoribonucleotides that mimic the S/R and thiostrepton domains in 23S rRNA
| Oligoribonucleotide | Kd, μM |
|---|---|
| S/R RNA | 6.9 ± 1.9 |
| A2660G | 5.9 ± 1.1 |
| A2660U | 7.5 ± 2.1 |
| A2660C | 8.3 ± 1.6 |
| ΔA2660 | No binding |
| G2661A | 9.2 ± 1.7 |
| G2661U | 8.7 ± 3.9 |
| G2661C | 7.2 ± 3.2 |
| ΔG2661 | 41.8 ± 9.1 |
| G2655A | 40.1 ± 3.2 |
| G2655U | No binding |
| G2655C | No binding |
| ΔG2655 | No binding |
| E73 | 7.8 ± 2.1 |
| Thiostrepton RNA | 17.2 ± 3.6 |
| E. coli ribosome | 0.7 ± 0.13 |
The S/R RNA (a 27-mer) has nucleotides 2648–2672 of E. coli 23S rRNA; mutants of the S/R RNA are identified by specifying the nucleotide that is changed. E73 is an oligoribonucleotide (a 29-mer) that has the eukaryotic S/R region sequence (nucleotides 4311–4337). The thiostrepton oligoribonucleotide (84-mer) has nucleotides 1036–1119 of E. coli 23S rRNA.
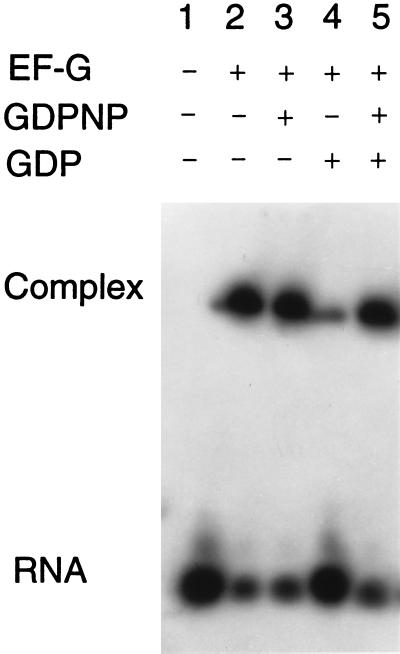
Binding of EF-G to the S/R oligoribonucleotide neither requires nor is increased by guanosine 5′-[β,γ-imido]-triphosphate (GDPNP), a nonhydrolyzable analog of GTP (Fig. 3). It is possible that EF-G is in a conformation favorable for binding to ribosomes even in the absence of GTP or that the S/R RNA induces the favorable conformation. Binding to the S/R RNA is all but abolished by GDP (Fig. 3), a result more in conformity with what occurs physiologically. The antibiotic fusidic acid does not affect binding, neither by itself nor in the presence of either GDPNP or GDP (results not shown). The inhibition of binding by GDP suggests that it induces a conformation of EF-G that is not compatible with stable association of the factor with the S/R RNA and that GTP displaces GDP from EF-G (this G protein has no known nucleotide exchange factor), thereby removing an impediment to binding without itself having a direct affect on EF-G conformation. If this explanation has validity, then GDPNP should overcome the GDP inhibition of the binding of EF-G to S/R RNA. A test gave results that conform to the prediction: GDPNP relieves the GDP inhibition of the binding of EF-G to S/R RNA (Fig. 3).
Figure 3.
Effects of GDPNP and GDP on the binding of EF-G to an S/R domain oligoribonucleotide. The concentrations of the reactants were as follows: EF-G, 20 μM; radioactive S/R oligoribonucleotide, 10 μM; GDPNP, 1 mM; and GDP, 1 mM.
The Kd for the binding of EF-G to an oligoribonucleotide (a 29-mer) that reproduces the sequence of the eukaryotic S/R region (E73 in Table 1) is the same as with the prokaryotic S/R RNA; this suggests conservation of the structure and of the function of the domain. However, EF-G does not sustain protein synthesis by eukaryotic ribosomes; we presume this is because the ribosomal proteins interfere with the binding of the factor. There is a precedent: ricin A-chain does not affect E. coli ribosomes but the toxin catalyzes the specific depurination of A2660 in naked 23S rRNA (25).
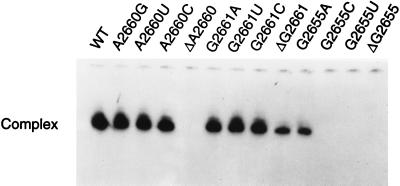
Three of the S/R region nucleotides, G2655, A2660, and G2661 (cf. Fig. 1B), are protected from chemical modification by the binding of EF-G to 70S ribosomes (11) and are presumptive RNA identity elements for the factor, although protection by a ligand from chemical modification does not of necessity connote a direct interaction. Oligoribonucleotides having deletions, transversions, and transitions of these three purines were constructed, and the effects of the mutations on the binding of EF-G were assessed (Fig. 4 and Table 1). The nucleotide most critical for binding of EF-G to the S/R RNA is G2655; deletion, or transversions to C or U, abolish EF-G binding; a transition to A increases the Kd by almost an order of magnitude. The deletion of A2660 also abolishes binding, but neither transitions nor a transversion of the nucleotide affect the interaction. Finally, only the deletion of G2661 significantly affects binding. These results suggest that it is the conformation of the S/R region RNA rather than nucleotide identity that conditions recognition by EF-G.
Figure 4.
Effect of S/R oligoribonucleotide mutations on the binding of EF-G. The concentration of EF-G was 20 μM and that of the radioactive oligoribonucleotides 10 μM. The wild-type (WT) oligoribonucleotide is in Fig. 1B. See Table 1 for the dissociation constants.
There is evidence that the thiostrepton region of 23S rRNA (around A1067; cf. Fig. 5A) is involved in EF-G-dependent functions (5–11). Thiostrepton, an antibiotic that inhibits binding of EF-G to ribosomes (5), binds to a 23S rRNA fragment that has A1067 (26); in addition, methylation of the 2′-hydroxyl of A1067 confers resistance to thiostrepton in strains of Streptomyces aureus (6). EF-G has been cross-linked to 23S rRNA in the vicinity of A1067 (7), and EF-G bound to 70S ribosomes in the presence of fusidic acid or of GDPNP protects A1067 and to a lesser extent A1069 from modification with dimethyl sulfate (11). It is also significant that the ribosomal proteins L11 and L7/L12, which have been implicated in functions of the elongation factors that require the hydrolysis of GTP, bind in (L11; ref. 6), or near (L7/12; ref. 10), the A1067 region.
Figure 5.
Binding of EF-G to a thiostrepton domain oligoribonucleotide. (A) Thiostrepton region of E. coli 23S rRNA. (B) Gel-retardation assay of the binding of EF-G to radioactive thiostrepton oligoribonucleotide (an 84-mer; nucleotides 1036–1119). The concentrations of EF-G were the same as in Fig. 2; the concentration of RNA is 10 μM. (C) Results in B plotted to show the extent of binding.
EF-G binds to a thiostrepton region oligoribonucleotide (an 84-mer; nucleotides 1036–1119 of 23S rRNA), and the extent of the binding is dependent on the concentration of the factor (Fig. 5 B and C) but is less efficient than binding to S/R RNA (compare Fig. 2B and Fig. 5C). When they are equimolar (10 μM), 22% of the thiostrepton RNA is in a complex with EF-G (Fig. 5C). The Kd for the binding reaction with thiostrepton RNA is 17.2 μM, approximately twice that for binding to S/R RNA (Table 1). There was no binding of EF-G to a smaller thiostrepton region RNA (a 30-mer that has nucleotides 1054–1081).
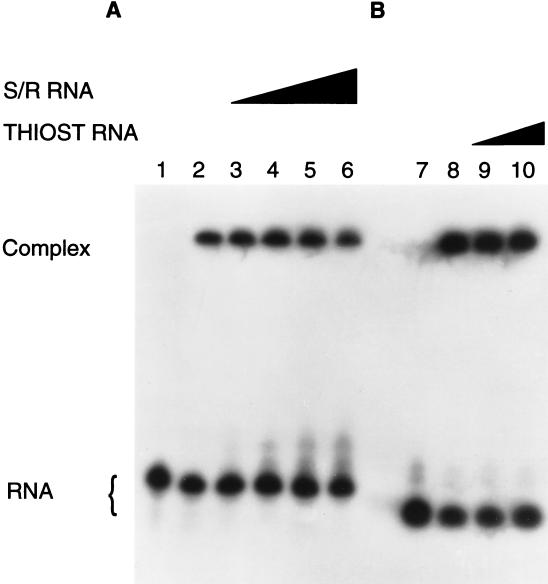
EF-G binds independently to S/R and thiostrepton oligoribonucleotides (Fig. 6); as much as a 10-fold excess of nonradioactive S/R RNA (100 μM) does not affect the binding of EF-G to 10 μM radioactive thiostrepton RNA (Fig. 6A), nor does 50 μM nonradioactive thiostrepton RNA affect binding of EF-G to 10 μM radioactive S/R RNA (Fig. 6B). As expected, nonradioactive S/R and thiostrepton RNAs decreased binding of EF-G to the corresponding radioactive species (results not shown). The results suggest that different EF-G domains associate with the two regions of 23S rRNA. Although the S/R and thiostrepton regions are distant in the primary and secondary structures of 23S rRNA, it is assumed that they are close in the tertiary structure and that together they constitute a single EF-G-binding domain. The preeminent question is whether either or both of these rRNA sites contribute more than factor binding to the biochemistry of protein synthesis; specifically, whether EF-G initiates a conformational change in the rRNA that drives translocation (27).
Figure 6.
Independent binding of EF-G to S/R and thiostrepton oligoribonucleotides. (A) Binding of 10 μM EF-G (except in lane 1, where it was omitted) to radioactive thiostrepton (THIOST) RNA (10 μM) assessed in the presence of increasing concentrations of nonradioactive S/R RNA: lanes 1 and 2, 0 μM; lane 3, 10 μM; lane 4, 20 μM; lane 5, 50 μM; and lane 6, 100 μM. (B) Binding of 10 μM EF-G (except in lane 7, where it was omitted) to radioactive S/R RNA (10 μM) assessed in the presence of increasing concentrations of nonradioactive thiostrepton RNA: lane 8, 0 μM; lane 9, 20 μM; and lane 10, 50 μM.
The results mark the achievement of an important goal: the demonstration of a relevant function of a small ribosome piece. There is significant specific binding of EF-G to relatively small fragments of rRNA; for example, the S/R RNA has 27 nucleotides (only 0.6% of the 4,566 in 70S ribosomes) and lacks ribosomal proteins, yet the Kd for the binding of EF-G is within an order of magnitude of that for the binding to intact ribosomes. It should be possible now to learn more of the chemistry of the interaction of EF-G with its rRNA binding site and, perhaps, something of the functional consequences of the binding.
Acknowledgments
This work was supported by Grant GM 33702 from the National Institutes of Health.
ABBREVIATIONS
- S/R
sarcin/ricin
- GDPNP
guanosine 5′-[β,γ-imido]-triphosphate
References
- 1.Pestka S. J Biol Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- 2.Bodley J W, Zieve F J, Lin L. J Biol Chem. 1970;245:5662–5667. [PubMed] [Google Scholar]
- 3.Ævarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, Al-Karadaghi S, Svensson L A, Liljas A. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czworkowski J, Wang J, Steitz T A, Moore P B. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodley J W, Lin L, Highland J H. Biochem Biophys Res Commun. 1970;41:1406–1411. doi: 10.1016/0006-291x(70)90543-7. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt F J, Thompson J, Lee K, Dijk J, Cundliffe E. J Biol Chem. 1981;256:12301–12305. [PubMed] [Google Scholar]
- 7.Sköld S E. Nucleic Acids Res. 1983;11:4923–4932. doi: 10.1093/nar/11.14.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maassen J A, Möller W. J Biol Chem. 1978;253:2777–2783. [PubMed] [Google Scholar]
- 9.Möller W. In: Ribosomes. Nomura M, Tissières A, Lengyel P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. pp. 711–731. [Google Scholar]
- 10.Beauclerk A A D, Cundliffe E, Dijk J. J Biol Chem. 1984;259:6559–6563. [PubMed] [Google Scholar]
- 11.Moazed D, Robertson J M, Noller H F. Nature (London) 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 12.Hausner T P, Atmadja J, Nierhaus K H. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 13.Noller H F. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- 14.Szewczak A A, Moore P B, Chan Y L, Wool I G. Proc Natl Acad Sci USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szewczak A A, Moore P B. J Mol Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 16.Endo Y, Wool I G. J Biol Chem. 1982;257:9054–9060. [PubMed] [Google Scholar]
- 17.Wool I G. In: Ribonucleases: Structures and Functions. D’Alesio G, Riordan J F, editors. San Diego: Academic; 1997. pp. 131–162. [Google Scholar]
- 18.Marchant A, Hartley M R. J Mol Biol. 1995;254:848–855. doi: 10.1006/jmbi.1995.0660. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Puentes C, Vazquez D. FEBS Lett. 1977;78:143–146. doi: 10.1016/0014-5793(77)80292-5. [DOI] [PubMed] [Google Scholar]
- 20.Endo Y, Chan Y L, Lin A, Tsurugi K, Wool I G. J Biol Chem. 1988;263:7917–7920. [PubMed] [Google Scholar]
- 21.Gurevich V V, Pokrovskaya I D, Obukhova T A, Zozulya S A. Anal Biochem. 1991;195:207–213. doi: 10.1016/0003-2697(91)90318-n. [DOI] [PubMed] [Google Scholar]
- 22.Rohrbach M S, Bodley J W. Methods Enzymol. 1979;60:606–614. doi: 10.1016/s0076-6879(79)60057-5. [DOI] [PubMed] [Google Scholar]
- 23.Konarska M M, Sharp P A. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 24.Bodley J W, Weissbach H, Brot N. Methods Enzymol. 1974;30:235–238. doi: 10.1016/0076-6879(74)30026-2. [DOI] [PubMed] [Google Scholar]
- 25.Endo Y, Tsurugi K. J Biol Chem. 1988;263:8735– 8739. [PubMed] [Google Scholar]
- 26.Thompson J, Cundliffe E. Biochimie. 1991;73:1131–1135. doi: 10.1016/0300-9084(91)90156-u. [DOI] [PubMed] [Google Scholar]
- 27.Wool I G, Glück A, Endo Y. Trends Biochem Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]