Abstract
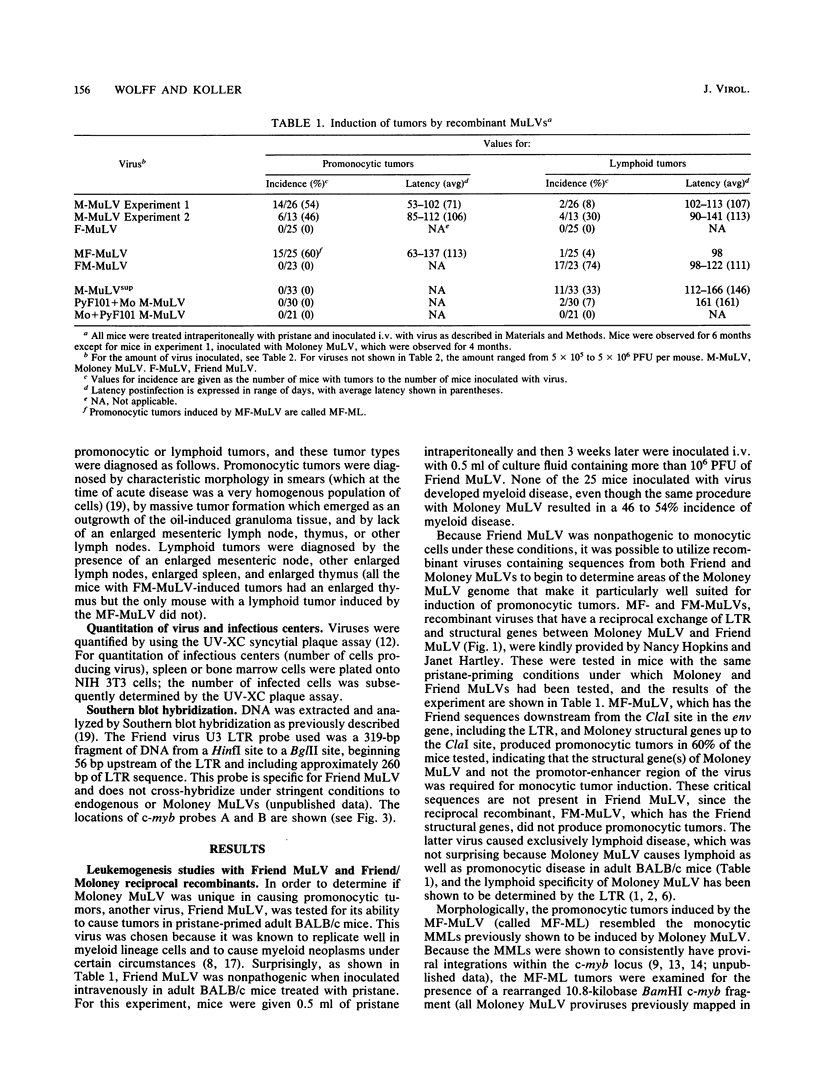
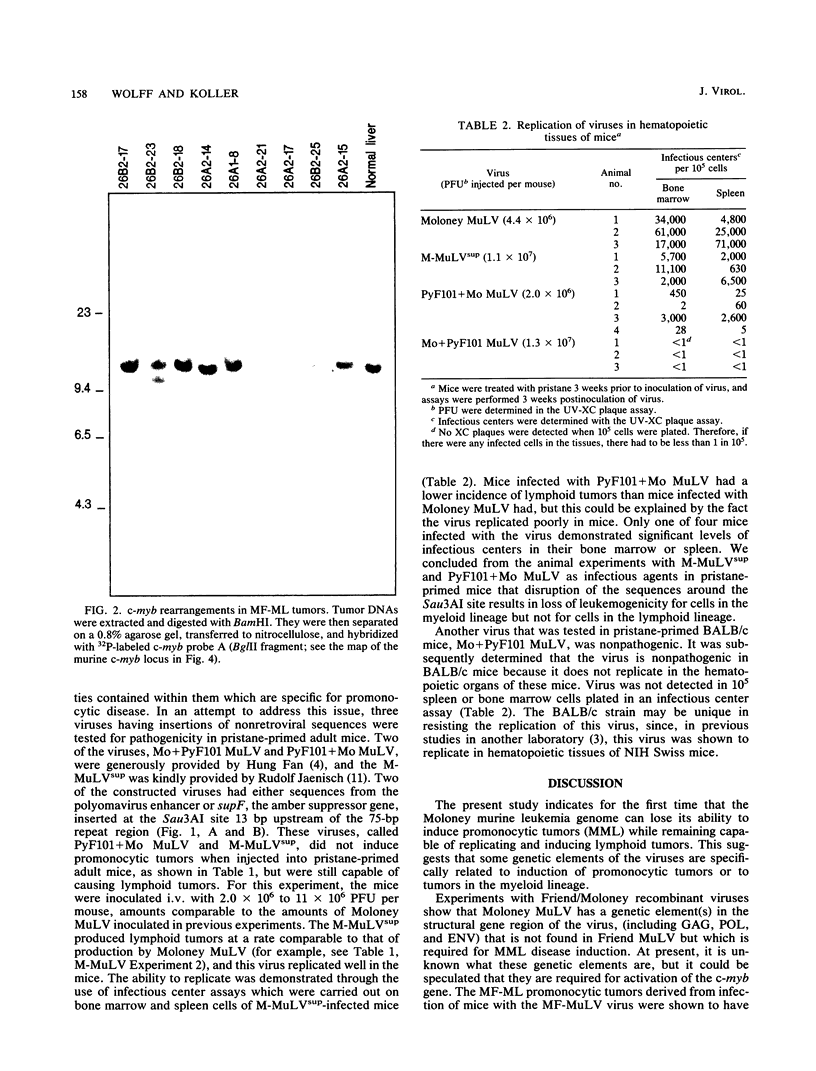
Moloney murine leukemia virus (MuLV) can be a potent inducer of promonocytic leukemias in mice that are undergoing a chronic inflammatory response. The neoplasms are, at least in part, associated with insertional mutagenesis of the c-myb locus. Evidence is presented for the existence of at least two genetic elements of the virus that are crucial to induction of this disease but are not required for viral replication in hematopoietic tissues or induction of lymphoid disease. These genetic elements were detected by testing the pathogenicity of recombinants between Moloney and Friend MuLVs, the latter of which is nonleukemic to myeloid cells under these conditions, and by testing Moloney MuLV-based viruses that have nonretroviral sequences inserted at specific endonuclease sites in their long terminal repeats (LTRs). Analysis of the Moloney/Friend recombinants showed that there are sequences within the structural gene domain of Moloney, but not Friend, MuLV that are necessary for promonocytic leukemia, whereas the LTRs of the MuLVs are equally effective for promonocytic tumor formation and insertional mutagenesis of the c-myb gene. Experiments with viruses which were mutagenized in the LTR by insertions demonstrated that there is a specific genetic element in the U3 region of the LTR of Moloney MuLV, upstream of the 75-base-pair enhancer which, when interrupted, results in loss of leukemogenicity for cells in the monocytic lineage but not cells in the lymphoid lineage. We conclude, therefore, that promonocytic leukemia induction, in Moloney MuLV-infected mice undergoing a chronic inflammatory response, requires specific sequences in the structural gene region of Moloney MuLV as well as other sequences in the regulatory region of the virus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. R., Chandy K. G., Brightman B. K., Gupta S., Fan H. Effects of nonleukemogenic and wild-type Moloney murine leukemia virus on lymphoid cells in vivo: identification of a preleukemic shift in thymocyte subpopulations. J Virol. 1986 Nov;60(2):423–430. doi: 10.1128/jvi.60.2.423-430.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Mittal S., Chute H., Chao E., Pattengale P. K. Rearrangements and insertions in the Moloney murine leukemia virus long terminal repeat alter biological properties in vivo and in vitro. J Virol. 1986 Oct;60(1):204–214. doi: 10.1128/jvi.60.1.204-214.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. R., Krieg A. M., Max E. E., Khan A. S. Negative control region at the 5' end of murine leukemia virus long terminal repeats. Mol Cell Biol. 1989 Feb;9(2):739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda T. J., Cory S., Sobieszczuk P., Holtzman D., Adams J. M. Generation of altered transcripts by retroviral insertion within the c-myb gene in two murine monocytic leukemias. J Virol. 1987 Sep;61(9):2754–2763. doi: 10.1128/jvi.61.9.2754-2763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Anklesaria P., Sakakeeny M. A., Greenberger J. S. Enhancer sequences of a retroviral vector determine expression of a gene in multipotent hematopoietic progenitors and committed erythroid cells. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8662–8666. doi: 10.1073/pnas.84.23.8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavu S., Reddy E. P. Structural organization and nucleotide sequence of mouse c-myb oncogene: activation in ABPL tumors is due to viral integration in an intron which results in the deletion of the 5' coding sequences. Nucleic Acids Res. 1986 Jul 11;14(13):5309–5320. doi: 10.1093/nar/14.13.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Weiher H., Jaenisch R. Replication-competent Moloney murine leukemia virus carrying a bacterial suppressor tRNA gene: selective cloning of proviral and flanking host sequences. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1141–1145. doi: 10.1073/pnas.82.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Shen-Ong G. L., Potter M., Mushinski J. F., Lavu S., Reddy E. P. Activation of the c-myb locus by viral insertional mutagenesis in plasmacytoid lymphosarcomas. Science. 1984 Nov 30;226(4678):1077–1080. doi: 10.1126/science.6093260. [DOI] [PubMed] [Google Scholar]
- Shen-Ong G. L., Wolff L. Moloney murine leukemia virus-induced myeloid tumors in adult BALB/c mice: requirement of c-myb activation but lack of v-abl involvement. J Virol. 1987 Dec;61(12):3721–3725. doi: 10.1128/jvi.61.12.3721-3725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L., Mushinski J. F., Gilboa E., Morse H. C., 3rd Induction of hematopoietic tumors using a viral construct containing c-myc cDNA from normal mouse spleen. Curr Top Microbiol Immunol. 1986;132:33–39. doi: 10.1007/978-3-642-71562-4_5. [DOI] [PubMed] [Google Scholar]
- Wolff L., Mushinski J. F., Shen-Ong G. L., Morse H. C., 3rd A chronic inflammatory response. Its role in supporting the development of c-myb and c-myc related promonocytic and monocytic tumors in BALB/c mice. J Immunol. 1988 Jul 15;141(2):681–689. [PubMed] [Google Scholar]
- Wolff L., Nason-Burchenal K. Retrovirus-induced tumors whose development is facilitated by a chronic immune response: a comparison of two tumors committed to the monocytic lineage. Curr Top Microbiol Immunol. 1989;149:79–87. doi: 10.1007/978-3-642-74623-9_7. [DOI] [PubMed] [Google Scholar]