Abstract
Hybridization-based technologies, such as microarrays, rely on precise probe-target interactions to ensure specific and accurate measurement of RNA expression. Polymorphisms present in the probe–target sequences have been shown to alter probe- hybridization affinities, leading to reduced signal intensity measurements and resulting in false-positive results. Here, we characterize this effect on exon and gene expression estimates derived from the Affymetrix Exon Array. We conducted an association analysis between expression levels of probes, exons and transcripts and the genotypes of neighboring SNPs in 57 CEU HapMap individuals. We quantified the dependence of the effect of genotype on signal intensity with respect to the number of polymorphisms within target sequences, number of affected probes and position of the polymorphism within each probe. The effect of SNPs is quite severe and leads to considerable false-positive rates, particularly when the analysis is performed at the exon level and aimed at detecting alternative splicing events. Finally, we propose simple solutions, based on ‘masking’ probes, which are putatively affected by polymorphisms and show that such strategy results in a large decrease in false-positive rates, with a very modest reduction in coverage of the transcriptome.
INTRODUCTION
Microarray analysis has become an integral part of high-throughput biological research. Microarray-based measurements typically rely on the precise hybridization of a DNA probe to a complementary target DNA or RNA molecule. Advances in technology and miniaturization now allow manufacturers to print up to 10 million probes on a single chip. Such chips are routinely used for truly genome-wide studies of polymorphisms, genomic aberrations (1), gene expression levels (2) and alternative splicing patterns (3). Unfortunately, such massive amounts of data come at the expense of a high potential for false discovery. Common sources of error range from the purely statistical (e.g. multiple testing problems), through experimental techniques, to systematic technical errors (e.g. probe cross-hybridization). As a result, particularly in gene expression analysis, microarray results have often been relegated from the realm of ‘proof’ to the role of a ‘discovery platform’ for further validation. In view of their overall popularity and utility, it is of great importance to minimize systematic errors in microarray experiments. In this study, we focus on one particular source of error: the effect of polymorphisms contained within probe target sequences on hybridization levels. Using expression quantitative trait analysis (eQTA) as an example, we show that this effect can be a major source of error, particularly for the latest generation whole-transcript (WT) arrays.
Association of genetic variants to expression phenotypes is becoming a promising strategy to identify sources of phenotypic diversity among individuals. A large number of genome-wide studies have been conducted in recent years, using various microarray platforms to determine gene expression levels (4–11). This approach usually treats expression data obtained from microarray experiments as a quantitative trait and tests for association with cis-acting polymorphisms. The final goal is to identify regulatory determinants of a particular phenotype, such as a disease state. Once significant associations have been identified, costly and time consuming downstream validations are conducted in order to identify the causative regulatory element. Therefore, it is important to identify candidate cis-acting polymorphisms with a high degree of confidence. Recent studies have shown that mismatches between a microarray probe and its target sequence affect hybridization (12–14) that cause erroneous probe signal estimates. This phenomenon leads to an increase in false-positives, particularly in studies across individuals with different genetic backgrounds (15). Individuals expressing mRNA that perfectly complements the probes on the microarrays hybridize better than individuals with mRNA sequence diversity in the probe–target region. This results in a difference in probe–signal intensity between individuals, even if both groups express the mRNA at the same level (16).
Here, we present a detailed analysis of this phenomenon using Affymetrix Human Exon Array data from our previous study of transcript isoform variation in humans (3) and describe how it affects association results at the probe, exon and gene levels. In addition, to mitigate the effect of polymorphisms, we propose a simple strategy that consists of removing probes that are targeted to annotated polymorphic regions. We show that this approach greatly reduces false-positive rates, particularly for associations at the exon level, with only a small reduction in exon and gene coverage.
METHODS
Microarray data source
In a previous study, we surveyed genetic variation associated with differences in isoform level expression in humans (3). We characterized this effect in a sample of 57 unrelated HapMap individuals of European ancestry (17) for which ∼4 million single nucleotide polymorphism (SNP) genotypes are available. Lymphoblast cells derived from these individuals were grown in triplicates and RNA was extracted from each of these growths and hybridized onto an Affymetrix Human Exon array (n = 171). The resulting probe-fluorescent intensities were used for the present analysis. We restricted our analysis to probes targeting core exons because of their high confidence annotation.
Effect of mismatches on hybridization
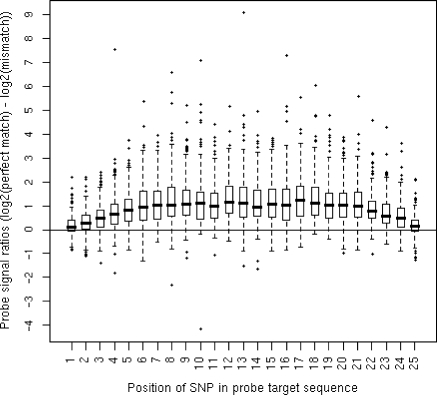
Probe expression signals were quantile-normalized and GC-background corrected using the Affymetrix Power Tools (APT) software package (Affymetrix). To investigate how mismatches affect probe-to-target hybridization on the Affymetrix Human Exon array, we took advantage of the high-resolution genotyping information available from HapMap cell lines and identified 6110 probes that were targeted to a region with only one SNP in at least 1 of the 57 HapMap individuals. These probes were selected because the exon and gene they targeted were considered expressed. Expression of an exon or gene was established using the detected above background (DABG) metric generated by Affymetrix. This metric represents the probability that an exon or gene is expressed below the background. We used false discovery rate (FDR) correction (18) to establish the significance threshold for expression above background at DABG ≤0.02 and DABG ≤0.043 for exons and genes, respectively. Next, we categorized each of these probes in 25 bins, depending on the position of the SNP within the target region (from 5′ to 3′ end). For each of these bins, we determined the fold change between the average probe intensity derived from individuals with a perfect complementary target region and the average probe intensity from individuals with one mismatch (Figure 1).
Figure 1.
Boxplots illustrating the positional effect of SNPs within the probe target region. Probe signal ratios between perfect complementary regions and regions with a single mismatch.
Masking procedure
We have previously shown (3,19) that SNPs located within probe-targets affect their hybridization to Affymetrix Human Exon array probes and consequently cause erroneous expression estimates. To mitigate this effect, we devised a simple procedure that consists of removing all probes from the analysis whose target region contains a known SNP. In total, we found 21 843 core probes target regions out of 1 096 799 probes overlapping at least one polymorphic HapMap II SNP (release 21).
Preprocessing and summarization of hybridization data
To study how probe-to-target hybridization is affected by SNPs, we generated two data sets of exon and gene expression estimates. The APT software package was used to quantile-normalize and GC-background correct each data set at the probe level. The average probe set (representing exons) and meta-probe set (representing genes) expression scores (averaged from triplicates) for each data set were computed using the probe logarithmic error intensity model (Affymetrix). The first data set consisted of probe set and meta-probe set expression estimates produced by summarizing all core probes, regardless of polymorphic probe target regions. The second data set was generated by implementing our masking procedure (see above). Thus, probe set and meta-probe set expression scores, for this last data set, were estimated from probes where no HapMap SNP overlapped their target region.
Association analyses
For each of the two data sets, the first generated from the full core probe list and the second from the masked core probe list, we examined probe, exon, and transcript expression estimates (averaged from triplicate samples for each individual) for association with flanking HapMap SNPs (release 21). One of the objectives of our previous analysis (3) was to identify possible cis-regulatory determinants of differential alternative splicing. The presence of linkage disequilibrium in humans has created haplotype blocks, where SNPs in close proximity to each other escape rearrangements due to recombination. Therefore, assuming physical proximity of a regulatory variant to the target and to limit the cost of multiple testing, we only tested for SNPs within a 50-kb region flanking either side of the gene containing either the probe or probe set. It should be noted that the SNPs associated with a change in microarray hybridization intensity may either be the actual causative SNPs, or simply be in linkage disequilibrium (part of the same haplotype block) with the causative SNP. We measured the level of association between expression scores (probes, probe sets and meta-probe sets) and the genotypes of a given SNP using linear regression analysis, implemented in the Plink software package (20), under a codominant genetic model. This model considers genotypes AA, AB and BB as the independent discrete variable. The genotypes are encoded as 0, 1 and 2, respectively, whereas expression scores were considered a quantitative trait and treated as the dependent variable in the linear regression. Raw P-values were obtained from the linear regression using the standard asymptotic t-statistic. To correct for testing multiple SNPs against each probe set and meta-probe set expression values, we carried out permutation tests (21) followed by 5% FDR correction. Permutation analyses were performed using the ‘label swapping’ and ‘adaptive permutation’ options implemented in Plink. The ‘label swapping’ option is used to preserve the haplotype block structure and the ‘adaptive permutation’ algorithm allows for computationally efficient permutation analyses (20). Subsequently, we performed FDR corrections of 5% on the empirical P-values (from permutations) for association of genotype to the expression at the probe set (P-value <9.73 × 10−9) and meta-probe set levels (P-value <6.07 × 10−7).
Evaluation of SNP mask
To evaluate how SNPs in probe–target regions impacted our association analyses, we estimated the proportion of false-positive and false-negative associations due to polymorphic probe target regions. We treated the association results for the masked data set as the reference (true) data set because they were derived from expression estimates free of influence from known SNPs. This reference data set (see Supplementary Tables 1 and 2) enables us to evaluate the four scenarios described in Table 1. Associations of probe set or meta-probe set, which were significant (P-value below the thresholds) and non-significant (P-value above thresholds) in both masked and unmasked data sets, were classified as true positives and true negatives, respectively. We consider a result a false-positive when a significant association is found in the unmasked data set, but becomes non-significant after masking probes containing SNPs (masked data set). Conversely, associations that were non-significant in the unmasked data set but significant in the masked data set were categorized as false-negatives. The false-positive and -negative rates are computed by: FPR = FP/(FP + TP) and FNR = FN/(FN + TN), respectively. In order to avoid the problem of reduced coverage within the masked data, the above analysis does not include probe sets which were entirely ‘masked’ due to the presence of SNPs.
Table 1.
Comparison of association analyses with and without a SNP mask
| SNP Mask |
||
|---|---|---|
| Positive for association | Negative for association | |
| No Mask | ||
| Positive for association | True positive | False positive |
| Negative for association | False negative | True negative |
RESULTS
Our first objective was to examine the effect of sequence mismatches on probe-to-target hybridization. We selected all probes that contained known SNPs and compared their hybridization intensity between individuals with homozygous match and mismatch genotypes. We illustrated how hybridization intensity changed when a mismatch is present at a given position within a probe in Figure 1. We observed that the position of the polymorphism within the probe's target sequence affects its binding affinity. Probe expression scores show a median ∼2-fold decrease in expression when a polymorphism is present near the middle of the target area i.e. between positions 6 and 21. This effect decreases linearly towards the edges of the target area and the median fold change in the end is near zero i.e. at positions 1 and 25, which supports the theoretical prediction of Lee et al. (22). It should be noted that the variance in the estimate of the effect is very high and that some mismatches decrease hybridization levels by much more than 2-fold; 7.5% of mismatches cause ≥5-fold decrease in signal intensity. Thus, in some cases the effect of SNPs may be very severe. This corroborates suggestions by earlier studies (12–15,23) that mRNA sequence diversity in probe target regions disrupts hybridization and that polymorphisms in the middle of the probe target regions destabilize hybridization more than those closer to the ends.
We next investigated how the association of expression phenotypes to neighboring SNPs, as in our previous analysis (3), are distorted by including probes whose target regions were polymorphic. We characterized this by performing an association analysis between expression levels of probes, exons and transcripts, with the genotypes of neighboring HapMap II SNPs. We compared only the top 1% of significant associations as a way to uniformly correct for multiple testing between the different levels of expression (probe, exon and gene). We observed that probes with polymorphic target regions were highly over-represented in the top 1% of significant association by a factor (odds ratio) of 16.8 (Table 2; χ2 = 33976.74, P-value << 10−16) compared to probes with perfectly complementary probe target regions. We also observed this over-representation at the probe set and meta-probe set levels, although to a lesser degree. In the top 1% of significant associations, we found an enrichment of 6.1-fold (Table 2; χ2 = 1443.88, P-value << 10−16) and 2.5-fold (Table 2; χ2 = 19.45, P-value = 1.03 × 10−5) for probe sets and meta-probe sets, respectively, whose expression estimates included probes that were targeted to polymorphic regions. In addition, this enrichment is also positively correlated with the number of polymorphisms within probe target region at the probe set (Pearson r = 0.956) and meta-probe set (Pearson r = 0.967) levels (Table 2). This further demonstrated that sequence polymorphisms between an Affymetrix Human Exon array probe and its target sequence resulted in changes to hybridization intensity and influenced the apparent association between the SNP genotypes and expression intensities. Given that probe set and meta-probe set expression estimates are derived by summarizing probe signals, erroneous probe signals due to probe target mismatches are a source of error in comparative expression analyses.
Table 2.
Enrichment for probes with polymorphic target region in the top 1% of significant association for probes, probe sets and meta-probe sets
| Number of SNP overlaps | Enrichment (odds ratio) |
||
|---|---|---|---|
| Probe | Probe set | Meta-probe set | |
| All | 16.83 | 4.30 | 2.46 |
| 1 | 16.78 | 1.98 | 1.94 |
| 2 | 19.39 | 5.02 | 2.12 |
| 3 | NA | 10.89 | 2.40 |
| 4 | NA | 15.64 | 3.00 |
| ≥5 | NA | 14.84 | 3.01 |
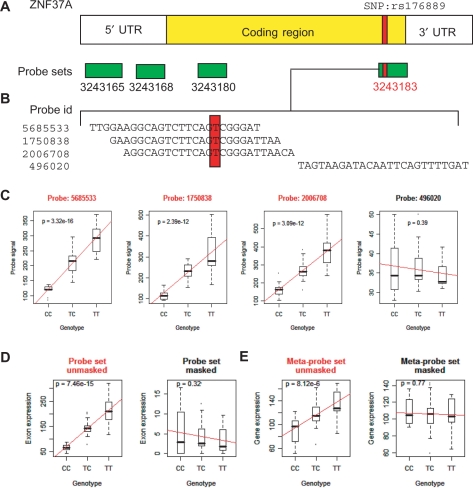
To reduce this source of error, we developed a simple masking procedure where we removed all probes targeted to a known polymorphic region (HapMap phase II SNPs). The remaining probes were used to estimate probe set and meta-probe set expression scores. A detailed example of this procedure and how it reduces the false-positive association caused by polymorphic probe target regions for gene ZNF37A is illustrated in Figure 2. Expression estimates for this gene were derived from four probe sets (Figure 2a), one of which, probe set 3243183, comprised probes targeting a polymorphic region in the 57 HapMap individuals. The first 3 probes from this probe set (Figure 2b) overlapped each other to some degree and targeted a region containing SNP rs176889. Individuals with TT genotypes have higher probe signals than individuals with a TC or CC genotype because the T allele creates a perfectly complementary target to these 3 probes (Figure 2c). The fourth probe, probe 496020, targets a region with no known SNP and shows no significant associations with SNPs rs176889. In addition, we do not find any significant association with neighboring SNPs that could be in linkage disequilibrium with SNP rs176889. Therefore, by using this single probe to estimate the expression of probe set 3243183, we obtain expression estimates that are not affected by erroneous probe signals and in subsequent association analyses (Figure 2d), the same is observed at the gene level (Figure 2e). We only used probe set expressions derived from probes unaffected by SNP to estimate meta-probe set expression scores and find no significant association with neighboring SNPs.
Figure 2.
ZNF37A is an example of a false-positive induced by a SNP (rs176889). (A) The ZNF37A mRNA molecule is illustrated with the coding region in yellow and the 5′ and 3′ UTRs is represented in white. The horizontal green rectangles represent the 4 probe sets that target this transcript. The red bars represent the position of SNP rs176889 in the coding sequence of this transcript. (B) The alignment of the 4 probe sequences that constitute probe set 3 243 183 and SNP rs176889 falls within each of these probes (red box). (C) Plots illustrating the association between each of the 4 probes and the different genotypes for SNP rs176889. Probe 496 020 does not contain any SNP and the association is non-significant. It is the only probe used to estimate probe set 3 243 183 expression scores. (D) Probe set 3 243 183 is no longer a false-positive after our masking procedure. (E) The same is observed at the meta-probe set level, where this gene is not significantly associated with SNP rs176889 or any other neighboring SNPs (results not shown).
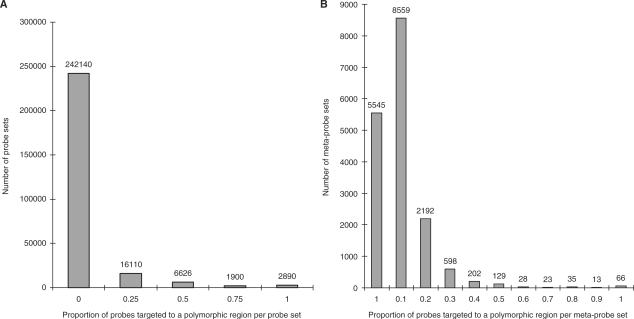
A potential drawback associated with removing problematic probes, is the reduction of probe set and meta-probe set coverage. For this data set, 21 843 (1.99%) probe target sequences overlapped at least one HapMap SNP and the distribution of affected probes per probe set and meta-probe set is illustrated in Figure 3a and b, respectively. We found 1258 (0.47%) probe sets and 99 (0.57%) meta-probe sets where we could not derive any expression estimates because no probes were left after ‘masking’ which is a very modest amount of lost coverage.
Figure 3.
Distribution of probe sets and meta-probe sets containing SNPs. (A) Proportion of affected probes per exon (B) Proportion of probes that contain SNPs per transcript.
Next, we assessed how our masking procedure improved results obtained from our association analyses. For the purpose of the analysis, we assumed that an association is a false-positive when a probe set or meta-probe set is significant in the unmasked data set, and that the same association becomes non-significant after masking probes containing SNPs. This assumption is based on two sources of evidence: (i) the strong over-representation of SNPs in the significant data set and (ii) the fact that in our previous work (3,19) we were unable to experimentally validate an alternative splicing event supported by an SNP-containing probe. We assumed that the expression data set derived by ‘masking’ misbehaving probes represents the best estimates of probe set and meta-probe set expression scores. Using this as the reference (true) data set, we evaluated the four scenarios described in Table 1 by comparing the P-values obtained from the association of the same neighboring SNPs to the same probe sets or meta-probe sets expression score estimated without ‘masking’ problematic probes. It should be noted that the reference set itself may not be free of false-positives (due to sources of errors other than SNPs), but this approach allows us to determine the rates of false-positive results that are induced by the presence of SNPs. We established P-value significance thresholds of 9.73 × 10−9 and 6.07 × 10−7 for probe sets and meta-probe sets, respectively, by permutation testing followed by FDR correction at 5%. We found that the SNP-induced false-positive rate is 86.6 and 8.1% at the probe set and meta-probe set levels, respectively (Table 3). However, false-negative rates do not seem to be influenced by SNPs because, after masking these potentially misbehaving probes, the false-negative rates were reduced by only 0.3 and 0.05% at the probe set and meta-probe sets (Table 3), respectively. This demonstrates that the removal of probe signals impacted by SNPs greatly reduces the rate of false-positives particularly for association conducted at the probe set level (e.g. alternative splicing). We concluded that masking probes targeted to known polymorphic regions does not substantially decrease the coverage of the Human Exon array and effectively reduces the SNP-induced false-positives.
Table 3.
Effect of the masking procedure on results from the association analysis of probe sets and meta-probe sets
| Probe sets | Meta-probe sets | |
|---|---|---|
| False positives | 446 | 9 |
| False negatives | 41 | 4 |
| True positives | 69 | 102 |
| True negatives | 13 359 | 8115 |
| False positive rate | 0.866 | 0.081 |
| False negative rate | 0.003 | 0.0005 |
DISCUSSION
Our analysis suggests that the presence of SNPs within the target sequence of Affymetrix Human Exon array probes causes false-positives when the analysis is conducted at the exon and transcript levels. Exon expression estimates are affected by misbehaving probes at a higher degree then transcript expression estimates because they are summarized from only 4 probe signals, whereas transcript expression estimates rely, on average, on 30 probes. In addition, we demonstrate that ‘masking’ a probe targeted to a known polymorphic region is a simple and effective solution for decreasing the rate of false-positives in an association analysis with individuals of different genetic backgrounds.
Alternative filtering approaches have been suggested. Zhang et al. (24) proposed to remove from the analysis probe sets with 2 or more probes harboring dbSNPs (release 126). This would result in the removal of 1.96% of probe sets—a much more significant reduction than the 0.47% in the approach outlined here. In addition, we do not advocate leaving probe sets containing single SNPs in the analysis, as we show in Table 2, that such probe sets are still ∼2-fold over-represented in the significant data set and are likely to produce false-positive results.
Our analysis takes advantage of the HapMap dataset, which has been genotyped at a high resolution. This constitutes an ideal data set for the purpose of illustration and quantification of the effect of SNPs. However, the results and solutions are applicable to most studies, whenever individuals with diverse genetic backgrounds are being compared. This is typically done in cancer studies and should be taken into consideration, particularly since investigation of alternative splicing and the use of WT arrays are quickly gaining popularity in this field (25,26). Generally, when two large groups of patients and controls are being compared, the effect of SNPs should be minimal in the pooled comparison. However, whenever a single individual or a group of related individuals is being used in a comparison to control samples, the effect of SNPs will be substantial. Similar problems will be encountered in any comparison of alternative splicing across tissues, whenever the tissues do not originate from the same individual. In all such cases, we advocate conservatively masking all probes containing putative SNP sites (from dbSNP). In addition, in our previous study (3) we found a non-trivial effect of still unannotated SNPs. While this problem cannot be corrected for a priori, we advise investigators to carefully monitor the behavior of individual probes before undertaking further costly functional studies—a single significant outlier probe whose behavior is inconsistent with the rest of the probe set may be an indication of a technical problem.
Finally, while we focus our study on the exon array and the analysis of alternative splicing, we would like to point out that other platforms are not immune to this effect. Examples of similar problems have been identified for the Affymetrix 3′ expression arrays (15,16). Other popular expression platforms, such as Agilent and Illumina, use longer probes, which are less sensitive to SNPs, but a slight effect of polymorphisms can be detected in those platforms as well (6,27). Therefore, we advocate preventive measures (such as SNP masking) and vigilance (careful scrutiny of final results), and propose that the next generation of microarray designs avoid, when possible, targeting polymorphic sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank D. Serre, D. Gaffney and E. Harmsen. This work is supported by Genome Canada, Genome Quebec and the Canadian Institutes of Health Research (CIHR). J. M is a recipient of a Canada Research Chair. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, Zhang J, Liu G, Ihara S, Nakamura H, Hurles ME, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 4.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, Parand L, Gagnebin M, Rougemont J, Jongeneel CV, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 2005;14:3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 8.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 9.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 10.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 12.Sliwerska E, Meng F, Speed TP, Jones EG, Bunney WE, Akil H, Watson SJ, Burmeister M. SNPs on chips: the hidden genetic code in expression arrays. Biol. Psychiatry. 2007;61:13–16. doi: 10.1016/j.biopsych.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Vallee M, Robert C, Methot S, Palin MF, Sirard MA. Cross-species hybridizations on a multi-species cDNA microarray to identify evolutionarily conserved genes expressed in oocytes. BMC Genomics. 2006;7:113. doi: 10.1186/1471-2164-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Wu C, Carta R, Zhao H. Free energy of DNA duplex formation on short oligonucleotide microarrays. Nucleic Acids Res. 2007;35:e18. doi: 10.1093/nar/gkl1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter NA, McWeeney SK, Peters ST, Belknap JK, Hitzemann R, Buck KJ. SNPs matter: impact on detection of differential expression. Nat. Methods. 2007;4:679–680. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, Jansen RC. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium IH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 19.Kwan T, Benovoy D, Dias C, Gurd S, Serre D, Zuzan H, Clark TA, Schweitzer A, Staples MK, Wang H, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I, Dombkowski AA, Athey BD. Guidelines for incorporating non-perfectly matched oligonucleotides into target-specific hybridization probes for a DNA microarray. Nucleic Acids Res. 2004;32:681–690. doi: 10.1093/nar/gkh196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, Kruhoffer M, Laurberg S, Borre M, Wang K, et al. Alternative splicing in colon, bladder and prostate cancer identified by exon-array analysis. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M700590-MCP200. (in press). [DOI] [PubMed] [Google Scholar]
- 27.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.