Abstract
It has been long considered that zinc homeostasis in bacteria is maintained by export systems and uptake systems, which are separately controlled by their own regulators and the uptake systems are negatively regulated by Zur which binds to an about 30-bp AT-rich sequence known as Zur-box present in its target promoters to block the entry of RNA polymerase. Here, we demonstrated in vivo and in vitro that in addition to act as a repressor of putative Zn2+-uptake systems, the Zur of the bacterial phytopathogen Xanthomonas campestris pathovar campestris (Xcc) acts as an activator of a Zn2+ efflux pump. The Xcc Zur binds to a similar Zur-box with ∼30-bp AT-rich sequence in the promoters of the genes encoding putative Zn2+-uptake systems but a 59-bp GC-rich sequence with a 20-bp inverted repeat overlapping the promoter's −35 to −10 sequence of the gene encoding a Zn2+-export system. Mutagenesis of the inverted repeat sequence resulted in abolishment of the in vitro binding and the in vivo and in vitro activation of the export gene's promoter by Zur. These results reveal that the Xcc Zur functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters.
INTRODUCTION
Like many other transition metal ions, zinc has paradoxical biological effects. On one hand, zinc is an essential trace element for almost all living organisms and works as a catalytic cofactor for various enzymes and a structural component in numerous proteins, on the other hand, when its intracellular concentration exceeds a threshold, it forms unspecific complex compounds in the cell and then causes toxic effects to the organism (1,2). In consequence, the intracellular Zn2+ level must be precisely regulated. In bacteria, the balance between Zn2+ import and export, known as zinc homeostasis, is maintained mainly through the coordinated expression of import systems and export systems which are separately regulated by their own regulators (3). Many bacteria use the zinc uptake regulator Zur, a Zn2+-sensing metalloregulatory protein belonging to the Fur (Ferric uptake regulator) regulatory protein family, to repress the transcription of Zn2+-uptake systems to maintain a delicate homeostasis (3,4). Zur was first described as a negative transcriptional regulator of genes encoding Zn2+-uptake systems in Escherichia coli (5) and Bacillus subtilis (6). Subsequently, it has been characterized in a number of other bacterial species including Listeria monocytogenes (7), Staphylococcus aureus (8), Salmonella serovar (9), Pasteurella multocida (10), Xanthomonas campestirs (11), Mycobacterium tuberculosis (12) and Streptomyces coelicolor (13). The major role of Zur in these bacteria is also to repress the transcription of genes encoding Zn2+-uptake systems to maintain zinc homeostasis. Recently, it has been demonstrated that the Zur of M. tuberculosis and S. coelicolor also controls intracellular Zn2+ mobilization through regulating some Zn2+-binding ribosomal proteins besides repressing Zn2+ uptake (12,13).
The molecular mechanism by which Zur represses the expression of Zn2+-uptake systems has been studied in some details. In E. coli, the Zur protein binds to an about 30-bp AT-rich sequence known as Zur-box overlapping the −35 to −10 region of the promoter of the Zn2+ uptake operon znuABC to block the entry of the RNA polymerase and thus suppresses the transcription of znuABC (14,15). Similarly, the Zur proteins of B. subtilis, M. tuberculosis and S. coelicolor also bind to a ∼30-bp AT-rich sequence overlapping the −35 to −10 region of their target promoters (12,16,17). The conservation in sequence and location of the Zur-binding sites of E. coli, B. subtilis, M. tuberculosis and S. coelicolor suggests that these Zur proteins may use a similar mechanism to repress the expression of the Zn2+-uptake systems.
To the best of our knowledge, there is not report about a Zur to be involved in the regulation of Zn2+-export systems. Our previous work showed that the zur mutant of the phytopathogenic bacterium Xanthomonas campestris pathovar campestris (Xcc), the causal agent of black rot disease of cruciferous crops (18), is significantly more sensitive to high zinc concentrations and accumulates significantly more zinc than the wild-type strain (11). In this study, we demonstrated that the Zur protein of Xcc not only represses the expression of genes encoding putative Zn2+-uptake systems but also activates the expression of a gene encoding a Zn2+ efflux pump via directly binding to cis-acting elements overlapping the promoters’ −35 to −10 region of these genes. Interestingly, the results displayed that the Zur-binding sequence in the genes encoding the putative Zn2+-uptake systems is distinct to that in the Zn2+-export gene, indicating that the Xcc Zur can recognize at least two distinct targets.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
The bacteria and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown in LB medium (19) at 37°C. Xcc strains were grown in NYG medium (20) at 28°C. Antibiotics were used at the following final concentrations: rifampicin, 50 μg/ml; kanamycin, 25 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 100 μg/ml and tetracycline, 15 μg/ml for E. coli and 5 μg/ml for Xcc.
Table 1.
Bacterial strains and plasmids used in this work
| Strains or plasmids | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| JM109 | RecA1, endA1, gyrA96, thi, supE44, relA1 Δ (lac-proAB)/F’ [traD36, lacIq, lacZ Δ M15] | (32) |
| BL21(DE3)pLysS | F– ompT hsdSB (rB–mB–) gal dcm (DE3) pLysS | Novagen, Germany |
| BL1430 | BL21(DE3)pLysS harboring pET1430, Kanr. | This work |
| Xcc | ||
| 8004 | Wild-type, Rifr | (20) |
| 1430nk | As 8004, but XC1430 (zur)::pK18mob, Rifr, Kanr | (11) |
| 2976nk | As 8004, but XC2976::pK18mob, Rifr, Kanr | This work |
| C2976nk | 2976nk harboring pXC2976, Rifr, Kanr, Tcr | This work |
| wt/pG0267 | 8004 containing plasmid pG0267, Rifr, Tcr | This work |
| zurmt/pG0267 | 1430nk containing plasmid pG0267, Rifr, Kanr, Tcr | This work |
| wt/pG2471-2 | 8004 containing plasmid pG2471-2, Rifr, Tcr | This work |
| zurmt/pG2471-2 | 1430nk containing plasmid pG2471-2, Rifr, Kanr, Tcr | This work |
| wt/pG2976 | 8004 containing plasmid pG2976, Rifr, Tcr | This work |
| zurmt/pG2976 | 1430nk containing plasmid pG2976, Rifr, Kanr, Tcr | This work |
| wt/pG3788 | 8004 containing plasmid pG3788, Rifr, Tcr | This work |
| zurmt/pG3788 | 1430nk containing plasmid pG3788, Rifr, Kanr, Tcr | This work |
| wt/pG2976MT | 8004 containing plasmid pG2976MT, Rifr, Tcr | This work |
| Plasmids | ||
| pET-30a-C(+) | Expression vector, allow the production of fusion proteins containing amino terminal 6xHis-taggted sequences. Kanr | Novagen, Germany |
| pET1430 | pET-30a-C(+)containing the coding region of the Xcc zur gene. Kanr | This work |
| pLAFR6 | Broad host range cloning vector, Tcr | (45) |
| pRK2073 | Helper plasmid,Tra+,Mob+,ColE1, Spcr. | (46) |
| pG0267 | pLAFR6 containing a XC0267 promoter-gusA fusion fragment, Tcr | This work |
| pG2471-2 | pLAFR6 containing a XC2472-XC2471 promoter-gusA fusion fragment, Tcr | This work |
| pG2976 | pLAFR6 containing a XC2976 promoter-gusA fusion fragment, Tcr | This work |
| pG3788 | pLAFR6 containing aXC3788 promoter-gusA fusion fragment, Tcr | This work |
| pG2976MT | pLAFR6 containing a mutant XC2976 promoter-gusA fusion fragment, Tcr | This work |
| pXC2976 | pLAFR6 containing a 1349-bp fragment including XC2976 gene, Tcr | This work |
aRifr, Kanr, Spcr, Tcr = rifampicin-, kanamycin-, spectinomycin-, and tetracycline-resistant, respectively.
DNA manipulation
DNA manipulation was performed following the procedures described by Sambrook et al. (21). The conjugation between the Xcc and E. coli strains was performed as described by Turner et al. (22). Restriction enzymes and DNA ligase were used in accordance with the manufacturer's (Promega, Shanghai, China) instructions.
Construction of the DNA microarray
The genome of the Xcc strain 8004 (GenBank accession numbers CP000050) is composed of 4273 open reading frames (ORFs) (23). A DNA microarray encompassing 4186 ORFs of the strain 8004 has been constructed previously as described by He et al. (24) and used in this study.
RNA isolation and cDNA synthesis
For RNA isolation, 2 ml overnight culture of the wild-type strain 8004 or the zur mutant 1430nk was diluted into 100 ml of NYG medium and grown at 28°C with shaking at 200 r.p.m. When the cells reached an optical density at 600 nm of 1.0, cells were collected by centrifugation for 2 min at 12 000 r.p.m. The cells were resuspended in 10 ml TE (10 mM Tris–HCl, 1mM EDTA) (pH8.0), and frozen in a liquid nitrogen bath for 15 min. Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA), and the contaminated DNA was removed by DNaseI (Promega) treatment. RNA integrity was confirmed by electrophoresis using a 1.3% formaldehyde–agarose gel, and the RNA quality was monitored by reverse transcription polymerase chain reaction (RT–PCR) analysis of two known genes. Three RNA samples were prepared from triplicate cultures of the wild-type strain 8004 and the zur mutant 1430 nk, and used for cDNA synthesis or stored at −80°C until further use.
The RNAs isolated from the wild-type strain 8004 and the zur mutant 1430nk were used to make both Cy3- and Cy5-labeled cDNAs by reverse transcription and Klenow enzyme reaction. Briefly, 20 μg of total RNA and a series of dilutions (from 1 ng to 20 pg) of yeast intergenic spike RNA were utilized in reverse transcription reaction using 3 μg nonamer random primer and 200 units of M-MLV transcriptase (Invitrogen) in 1× first buffer [5 mM Tris–HCl (pH 8.3), 1 mM MgCl2, and 7.5 mM KCl] at 37°C for 1 h. After the completion of the reaction, RNA was hydrolyzed by adding 5 μl of stop buffer (35 mM EDTA, 1.4M NaOH) and neutralized with 1 μl of 10 N acetic acid. cDNA was purified with PCR Clean-up NucleoSpin Extract II kits (Macherey-Nagel, Düren, Germany) and dry vacuumed.
Slide hybridization and data analysis
For expression profiling hybridization, the labeled control and test cDNA samples were quantitatively adjusted according to the efficiency of Cy-dye incorporation, and then mixed with 80 μl hybridization solution (3×SSC, 0.2% SDS, 50% formamide). Prior to loading on the microarray, the mixture was heated to 95°C for 3 min to denature the cDNA in hybridization solution. Hybridization was performed under LifterSlip™ (Erie Company, Portsmouth, NH, USA), which allows for an even dispersal of hybridization solutions between the microarray and coverslip. The hybridization chamber was laid on a Three-phase Tiling Agitator (CapitalBio Corp., Beijing, China) to prompt the microfluidic circulation under the coverslip. The array was hybridized at 42°C overnight and washed with two consecutive washing solutions (0.2% SDS, 2×SSC) at 42°C for 5 min and 0.2% SSC for 5 min at room temperature.
Arrays were scanned with a confocal LuxScan™ scanner (CapitalBio Corp., Beijing, China), and signal intensities were detected and quantified with SpotData software (CapitalBio Corp.) and assembled into EXCEL spreadsheets. Expression ratios were calculated as zur-mutant/wild-type (zur/WT), and the mean, SD and the statistical significance of the expression difference were calculated following the published method (25). Measurements with high variability, those where the standard deviation was equal to or larger than the mean, or those where valid ratios were found for only one of the three replicates, were removed. Three biological replicates of array hybridizations were performed and the data presented were the means of the three replicates.
Construction of an E. coli strain for production of the Xcc His6-Zur protein
A 516-bp PCR product containing the coding region of the Xcc zur gene was amplified using the total DNA of the Xcc wild-type strain 8004 as template and the primer pair 1430PF/1430PR (Table S1) designed according to the sequence of the Zur-encoded ORF XC1430 of the strain 8004 (11,23). After confirmation by sequencing, the amplified DNA fragment was inserted into the BamHI and HindIII sites of the expression vector pET-30a-C(+) (Novagen, Darmstadt, Germany) to generate the recombined plasmid pET1430 (Table 1), and the plasmid pET1430 was introduced into the E. coli strain BL21(DE3)pLysS (Novagen) to create the strain BL1430 (Table 1), which was used for (His)6-Zur protein production.
Xcc His6-Zur protein purification
Ten milliliters of overnight culture of strain BL1430 was used to inoculate a 200 ml LB medium with kanamycin and chloramphenicol and cultured at 37°C with shaking at 200 r.p.m. When the cell concentration reached OD600 = 0.6, IPTG (isopropyl-beta-d-thiogalactopyranoside) was added to the culture at a final concentration of 1 mM, and the culture was further incubated at 37°C for 4 h. Cells were harvested by centrifugation at 4°C for 20 min at 4000 r.p.m. One gram of cells (wet weight) was resuspended in 4 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole) and cells were ultrasonicated for 30 min. The cell lysate was centrifuged at 4000g for 20 min at 4°C, and the clear supernatant was transferred into a 50 ml centrifugation tube and mixed with 2.5 ml of prewashed Ni-NTA resin (Qiagen, Hilden, Germany) in the lysis buffer. After shaking with 200 r.p.m. at 4°C for 2 h, the batch was poured into a plastic Ni-NTA Spin Column (Qiagen) and then washed several times with 50 ml of precooled wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole) until the absorbance at 280 nm was <0.01. Finally, the (His)6-Zur protein was eluted with 500 μl of precooled elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole) and was analyzed by SDS–PAGE or stored at −20°C.
Construction of reporter plasmids
To construct reporter plasmids for the ORFs XC0267, XC2471-2 (XC2471 and XC2472), XC2976 and XC3788, the 150–300 bp region upstream of the start codon (excluding the start codon) of the genes was respectively amplified by PCR using the total DNA of the Xcc wild-type strain 8004 as template and the primer pairs 0267PF/0267PR, 2471-2PF/2471-2PR, 2976PF/2976PR and 3788PF/3788PR (Table S1). To ensure these promoter fragments could be ligated with the promoterless gusA (β-glucuronidase) gene fragment by fusion PCR (26), a 20-nt tag complementary to the first 20 nt of the promoterless gusA fragment was added to the 5′-end of the primers 0267PR, 2471-2PR, 2976PR and 3788PR (Table S1). A 1.8-kb DNA fragment containing the promoterless gusA gene with its RBS (ribosomal-binding site) was amplified by PCR using pLAFR1::Tn5gusA5 as template and the primer pair GusAF/GusAR (Table S1). Each of the amplified promoter fragments and the 1.8-kb gusA fragment were respectively ligated by fusion PCR to generate the promoter-gusA fusion fragments. Then, these fusion fragments were respectively cloned into the broad-host-range vector pLAFR6 to create the reporter plasmids pG0267, pG2471-2, pG2976 and pG3788 (Table 1). These reporter plasmids were respectively introduced into the wild-type strain 8004 to create the strains wt/pG0267, wt/pG2471-2, wt/pG2976 and wt/pG3788, and introduced into the zur mutant 1430nk to create the strains zurmt/pG0267, zurmt/pG2471-2, zurmt/pG2976 and zurmt/pG3788 (Table 1).
Site-directed mutagenesis of the XC2976 promoter
To mutate the XC2976 promoter, ACACCACACC was changed for its left half part CGTGATGTGA of the 20-bp inverted repeat. Firstly, a 130- and 1956-bp DNA fragments were respectively amplified by PCR using the XC2976-promoter-containing reporter plasmid pG2976 (see above) as template and the primer sets 2976PF/2976MTR and 2976MTF/GusAR (Table S1). To ensure the two fragments could be ligated together by fusion PCR, the first 20-nt sequence of the 5′-end of the two primers were designed to complementary each other (Table S1). To obtain the expected nucleotide changes, the sequences of the primers 2976MTR and 2976MTF were designed to contain desired mutant sequence (Table S1). After confirmed by sequencing, the two DNA fragments were ligated by fusion PCR to generate the mutant XC2976 promoter-gusA fusion fragment. Then, the fusion fragment was cloned into the broad-host-range vector pLAFR6 to create the reporter plasmid pG2976MT (Table 1). The plasmid pG2976MT was introduced into the wild-type strain 8004 to create the strain wt/pG2976MT (Table 1).
The mutated XC2976 promoter fragment P2976MT, in which ACACCACACC was changed for the left half part of the 20 bp imperfect inverted repeat CGTGATGTGA of the wild-type XC2976 promoter, was obtained by PCR using pG2976MT DNA as template and the primer set P2976-F/P2976-R (Table S1).
GUS activity assay
The activity of β-glucuronidase (GUS) was determined after the growth of Xcc strains in NYG, NYG supplemented with different levels of ZnSO4 or EGTA for 24 h by measurement of the OD415 using ρ-nitrophenyl β-d-glucuronide as the substrate, as described by Jefferson et al. (27).
Electrophoretic mobility shift assay (EMSA)
EMSA was performed following the procedure described by Gaballa and Helmann (6). Fragments containing the promoter regions of the ORFs XC0267, XC2471-2, XC2976 and XC3788 or the intergenic region between XC2471 and XC2472 (IGR2471-2472, as a negative control) were amplified by PCR using the total DNA of the wild-type strain 8004 as the template and the primer pairs P0267-F/P0267-R, P2471-2-F/P2471-2-R, P2976-F/P2976-R, P3788-F/P3788-R and IGR2471-2-F/IGR2471-2-R (Table S1). After purified by using QIAquick PCR Purification Kit (Qiagen), these PCR products were labeled with [γ-32P]ATP using a 5′-end labeled kit (Takara, Dalian, China). Then the His6-Zur protein was added into a 1.5-ml EP tube containing 20 µl binding buffer [20 mM Tris–HCl (pH 8), 50 mM KCl, 1 mM DTT, 5% glycerol, 0.1 mg of bovine serum albumin per ml, and 5 µg of sheared salmon sperm DNA per ml] to final concentrations of 0, 8, 16, 32, 64, 128, 256 and 508 nM, and 3 pmol labeled DNA was added to each tube and mixed well. The mixtures were incubated at room temperature for 20 min. Samples were then loaded on a 4% polyacrylamide gel prepared and run in 40 mM Tris–acetate buffer (with no EDTA) (pH 8.0). The gel was dried and exposed to a phosphorimager screen (Typhoon 9410, Amersham Biosciences Corp., Piscataway, NJ USA).
To test the effects of Zn2+ and EDTA on protein–DNA binding, the His6-Zur protein was added into 20 µl binding buffer and then ZnSO4 or EDTA was added and mixed well. After incubation for 10 min at room temperature, 1–3 pmol labeled DNA was added and mixed well, and the mixture was incubated at room temperature for 20 min. Samples were then analyzed as described above.
DNaseI footprinting analyses
DNaseI footprinting to map the binding site of Zur in the promoter region of the ORFs XC0267, XC2471-2, XC2976 and XC3788 was performed using fluorescently labeled DNA and the ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA, USA) to resolve the digestion products, as described by Merighi et al. (28). The same primer sets (Table S1) used for amplification of the DNA fragments for EMSA, which contain respectively the promoter region of the ORFs XC0267, XC2471-2, XC2976 and XC3788, were used to amplify the corresponding promoter fragments for DNaseI footprinting analyses. The 5′-ends of the primers P0267-R, P2471-2-R, P3788-R and P2976-F were labeled with 5-carboxytetramethylrhodamine (Tamra) (Sangon, Shangshai, China). DNA fragments containing the promoter region of the ORFs XC0267, XC2471-2, XC2976 and XC3788 were amplified as a 5′-end-labeled PCR product using the total DNA of the wild-type strain 8004 as template. The PCR products were purified by gel electrophoresis and quantified using UV spectrophotometer. The labeled probes (400 ng) were incubated with 120 ng of His6-Zur protein in 20 µl binding buffer [20 mM Tris–HCl (pH 8), 50 mM KCl, 1 mM DTT, 5% glycerol, 0.1 mg of bovine serum albumin per ml, and 5 µg of sheared salmon sperm DNA per ml] at 28°C for 20 min, immediately prior to use, 5 µl RQ1 DNaseI (1 U/µl) was diluted in 100 µl cold Tris–HCl (10 mM, pH 8.0), and then 5 µl DNaseI (Promega) was added to the 20-µl reaction mixture and incubated for 1 min. The reaction was stopped by addition of 5 µl of 0.15M EDTA, 5% SDS and extracted with phenol–chloroform–isoamyl alcohol (25:24:1). The DNA fragments were purified in a Qiagen spin column. Sequencing of the corresponding nonlabeled promoter fragments were performed using ABI BigDye Terminator 2.0 sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Sequencing reactions contained 100 ng of promoter fragment DNA and 3.2 pmol of nonlabeled P0267-R, P2471-2-R, P3788-R and P2976-F primer. A 1-µl aliquot of sequencing product and DNaseI digestion product was loaded onto an ABI 377 DNA sequencer (Applied Biosystems) for a ‘Sequencing running’. Photos of the ‘Gel Image’ were directly taken from the screen of the sequencer. To determine the size of the DNaseI digestion fragments, a 0.1-µl aliquot of Genescan-350 size standard (Applied Biosystems) was combined with either 1 µl sequencing product or 1 µl DNaseI digestion product and loaded onto an ABI 377 DNA sequencer for a ‘Genescan running’. Electropherograms of the Genescan running were analyzed using Genescan 3.1 (Applied Biosystems).
Rapid amplification of cDNA end (5′ RACE)
RNA was extracted from the wild-type strain 8004 grown to the mid-exponential phase (OD600 ≈ 0.6) in the rich medium NYG using the SV Total RNA Isolation System (Promega), and treated with DNaseI (Promega). Five micrograms of the DNA-free RNA and 15 pmol of the gene specific primers GSP1s (Table S1) were incubated at 70°C for 5 min and then 42°C for 1 h in the presence of 1 × M-MuLV-RT buffer, 1 mM dNTPs, 20 U RNase inhibitor (Fermentas, Burlington, Canada) and 200 U M-MuLV Reverse Transcriptase (Fermentas). After treated with 2 U/µl RnaseH (Promega) for 30 min (to remove the remaining RNAs), the reaction product (cDNA) was purified using S.N.A.P.Column (Invitrogen) and finally resolved in 50 µl sterilized dH2O. Then, 10 μl purified cDNA was incubated at 37°C for 10 min in the presence of 0.2 mM dCTP and 20 U of TDT (terminal deoxynucleotidyl transferase) (Invitrogen) to add a poly(C) tail to the 3′-end of the cDNA. Finally, 5 μl poly(C) tailed cDNA was used as template for PCR reaction. To improve assay sensitivity, two rounds of semi-nested PCR were performed. The first semi-nested PCR was performed using 5 μl poly(C) tailed cDNA as template, and AAP and an internal oligonucleotide, GSP2, as primer (Table S1); the second semi-nested PCR was performed using the product of the first semi-nested PCR as template, and AUAP and another internal oligonucleotide, GSP3, as primer (Table S1). After gel purification, the last PCR product was sequenced using GSP3 as primer.
In vitro transcription assays
In vitro transcription assays on the promoter DNA of XC0267, XC2471-2, XC2976 and XC3788 were performed using the experimental procedure described by Friedman et al. (29). The promoter template DNA fragments of XC0267 (P0267), XC2471-2 (P2471-2), XC2976 (P2976) and XC3788 (P3788) were respectively generated by PCR amplification using the total DNA of the wild-type strain 8004 as template and primer sets 0267ivtF/0267ivtR, 2471-2ivtF/2471-2ivtR, 2976ivtF/2976ivtR and 3788ivtF/3788ivtR (Table S1). To generate P2976MT, a template DNA fragment containing a mutated XC2976 promoter in which the 20 bp imperfect inverted repeat sequence is lacking, fragment A and fragment B were respectively amplified using the total DNA of strain 8004 as template and primer sets 2976PF/2976MTR and 2976MTF/2976ivtR (Table S1), and fragments A and B were then linked together to generate the P2976MT fragment by fusion PCR (26). Each promoter template fragment includes the promoter region as well as ∼150-bp coding region downstream of the annotated translation start site of the corresponding gene (23). To remove imidazole, Zur was dialyzed against 200 volumes of Tris·HCl buffer [10 mM Tris·HCl (pH 8.0), 1 mM DTT] at 4°C. For in vitro transcription, Zur was incubated for 30 min at room temperature in transcription buffer [40mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 5 mM DTT, 5% glycerol, 50 mM KCl, 1U RNase inhibitor, 100 µM ZnSO4] containing 4 nM promoter template DNA. Then a NTP mixture [250 µM each of ATP, CTP and GTP, 20 µM UTP, 8 µM (α-32P)UTP (3000 Ci/mmol, 10 mCi/ml)] and 1 U E. coli RNA polymerase Holoenzyme (sigma saturated) (Epicentre, Madison, WI, USA) was added to start the transcription. After incubation at 28°C for 30 min, reactions were terminated by addition of 1 volume of 2× Loading Dye Solution (Fermentas) and chilled on ice. After incubation at 70°C for 10 min, transcription products were run on a 5% denatured polyacrylamide gel containing 7 M urea in 1× Tris borate–EDTA electrophoresis buffer. The transcripts obtained were analyzed by a phosphorimager screen (Typhoon 9410, Amersham Biosciences, USA). Each experiment was repeated three times.
Mutant construction
A mutant of XC2976 was constructed by homologous suicide plasmid integration (30) using pK18mob as the vector (31). A 380-bp internal fragment of XC2976 was amplified using the total DNA of the Xcc wild-type strain 8004 as the template and the primer pair 2976MF/2976MR (Table S1). After confirmation by sequencing, the amplified DNA fragment was cloned into the suicide plasmid pK18mob to create the recombinant plasmid pK2976 (Table 1). The plasmid was transformed from E. coli JM109 (32) into Xcc strain 8004 by triparental conjugation using pRK2073 as the helper plasmid. Transconjugants were screened on NYG supplemented with rifampicin and kanamycin and the obtained transconjugants with a mutation in XC2976 were confirmed by PCR. Confirmation PCR was performed using the total DNA of the transconjugants as the template along with the primer pair P18conF/2976conR (Table S1), and the total DNA of the Xcc wild-type strain 8004 was used as a negative control. The primer P18conF is located in pK18mob, and 2976conR is located downstream of the cloned internal fragment of XC2976. The expected PCR products were further confirmed by sequencing. One of the confirmed mutant transconjugants was designated as 2976nk, and was chosen for further study (Table 1).
Complementation of the mutant 2976nk
For complementation of the XC2976 mutant 2976nk, a 1349-bp DNA fragment containing the entire XC2976 gene (from 200-bp upstream of the start codon to 182-bp downstream of the stop codon) was amplified by PCR using the total DNA of the Xcc wild-type strain 8004 as the template and the primer pair 2976CF/2976CR (Table S1). After confirmation by sequencing, the amplified DNA fragment was cloned into pLAFR6 to generate the recombinant plasmid pXC2976 (Table 1). The plasmid pXC2976 was transferred into the mutant 2976nk by triparental conjugation. The transconjugants carrying pXC2976 were screened on NYG with rifampicin, kanamycin and tetracycline. A confirmed transconjugant representative was named C2976nk (Table 1) and chosen for further study.
Metal ion sensitivity test
For metal ion sensitivity test, 200-µl overnight culture of Xcc strains with optical density at 600 nm of 1.0 (OD600 = 1.0) was inoculated into 200-ml NYG medium supplemented with a certain metal ion to a series of final concentrations, and the cell density for each treatment was measured spectrometrically at 600 nm after incubated at 28°C with shaking at 200 r.p.m. for 24 h. The levels of metal ions used in this study are ZnSO4: 0, 300, 400 and 500 µM; CoCl2: 0, 150, 200, 250, 300 and 350 µM; CdSO4: 0, 40, 50, 60, 70 and 80 µM and NiSO4: 0, 400, 500, 600, 700 and 800 µM.
Measurement of Zn2+ content in cells
Two hundred microliters of overnight culture of Xcc strains were inoculated into 200-ml NYG medium. After incubation at 28°C with shaking at 200 r.p.m. for 12 h, the culture was added with ZnSO4 to a final concentration of 300 μM and incubated for further 4 h. Cells from the culture were harvested and washed twice with 0.1 M LiCl, 0.2 mM EDTA and 0.1 mM EGTA to remove externally bound metal ions. The cell density was adjusted using sterilized ddH2O to an OD600 of 1.0 and the cells were kept on ice. The Zn2+ content in the cells was determined by an atomic absorption spectrophotometer with a HITACHI Model 2000 instrument.
RESULTS
Xcc Zur negatively regulates three genes related to Zn2+-uptake systems and positively regulates a gene related to a Zn2+ efflux pump
Our previous work showed that in Xcc inactivation of zur resulted in hypersensitivity to Zn2+ toxicity (11), indicating that certain important genes involved in Zn2+ homeostasis of Xcc are under the control of Zur. To identify these genes, we screened putative Zur-regulated genes by genome-wide DNA microarray hybridization. RNAs were isolated from the cells of the wild-type Xcc strain 8004 and the Xcc zur mutant 1430nk (Table 1) grown in the Zn2+-replete medium NYG (ca. 10 µM Zn2+) (11), and used to make both Cy3- and Cy5-labeled cDNAs. The labeled cDNAs were hybridized with a previously constructed DNA microarray encompassing 4186 of the 4273 total ORFs of the wild-type strain 8004 (23,24), and the zur mutant/wild-type expression ratios were compared (see Materials and methods section for details). The results showed that 37 and 27 ORFs displayed statistically significant (t-test, P < 0.05) increases and decreases greater than 2-fold in mRNA levels by zur mutation, respectively (Table S2), suggesting that these 64 ORFs may be regulated by Zur.
As shown in Table S2, none of the 64 putative Zur-regulated genes is annotated to have a metal ion homeostasis-related function except XC2976, which was predicted to encode a cobalt–zinc–cadmium resistant protein (23) showing 67% amino acid sequence similarity to the Zn2+ efflux pump CzcD of Ralstonia metallidurans (33). However, when we used the amino acid sequences of the predicted products of these putative Zur-regulated genes to blast (34) the NCBI database (http://www.ncbi.nlm.nih.gov/blast), in addition to XC2976, the predicted products encoded by XC0267, XC2471 and XC3788 displayed amino acid sequence similarity to Zn2+ homeostasis-related proteins identified in other bacteria. XC2471 and XC0267 showed 29 and 54% amino acid sequence similarities to YciA and YciC of B. subtilis, respectively, while XC3788 exhibited 38% similarity to ZnuC of E. coli (Table 2). YciA and YciC are components of the low-affinity Zn2+-uptake system YciABC of B. subtilis (16) and ZnuC is a member of the high-affinity Zn2+-uptake system ZnuABC of E. coli (5). Based on these similarities, we propose that XC2976 is a putative Zn2+ efflux-related gene and XC0267, XC2471 as well as XC3788 are putative Zn2+ uptake-related genes. Interestingly, a survey using the amino acid sequences of the E. coli ZnuA and ZnuB to blast the genome of the Xcc strain 8004 (23) exhibited no homologous sequences (data not shown).
Table 2.
Similarity of the products of genes that showed altered expression in the zur mutant to characterized bacterial metal ion homeostasis-related proteins
| ORFs | Positions in Xcc 8004's genome | Homology [Organism] (accession number) | Amino acid similarity (%) | Characterized function | Reference |
|---|---|---|---|---|---|
| XC0267 | 321454–322779 | YciC [B.subtilis](CAB12130) | 54 | Component of the low-affinity Zn2+ uptake system YciABC | (16) |
| XC2471 | 2989376–2988441 | YciA [B. subtilis] (CAB12128) | 29 | Component of the low-affinity Zn2+ uptake system YciABC | (16) |
| XC2976 | 3563238–3562273 | CzcD [R. metallidurans] (YP_145596) | 67 | Acts as a Zn2+ efflux pump | (33) |
| XC3788 | 4483267–4482527 | ZnuC [E. coli] (P0A9X1) | 38 | Component of the high-affinity Zn2+ uptake system ZnuABC | (5) |
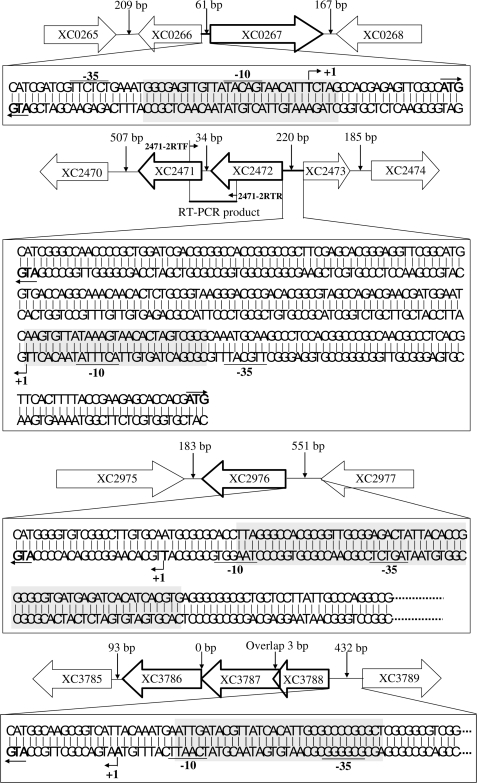
The locations of the putative Zn2+ homeostasis-related genes XC0267, XC2471, XC3788 and XC2976 in the genome of Xcc strain 8004 are shown in Table 2. XC0267 and its adjacent ORF XC0266 are separated by a spacer with only 61 bp and their transcriptions are back to back (Figure 1). The ORFs XC2471 and XC2472 share the same transcriptional direction and are separated by only 34 bp (Figure 1). To determine whether these two ORFs are transcribed together, we performed a RT–PCR using the cDNA converted from the RNA of the wild-type strain 8004 as template and the primers 2471-2RTF/2471-2RTR (Table S1). An amplified DNA fragment with 386 bp in length was obtained (Figure 1; data not shown), indicating that the ORFs XC2471 and XC2472 are together transcribed and they may share the same promoter located in the upstream region of XC2472 (Figure 1). Therefore, we designated the promoter and reporter plasmid of these two ORFs as PXC2471-2 and pG2471-2, respectively (see below). To establish the genetic relationship between zur and XC0267, XC2471, XC2976 as well as XC3788 we constructed plasmid-driven promoter-gusA transcriptional fusion reporters of these genes by fusing the promoterless gusA gene with the promoter of the genes and cloning the fused fragment into the plasmid pLAFR6, respectively. The obtained reporter plasmids were named pG0267, pG2471-2, pG2976 and pG3788, respectively (Table 1; see Materials and methods section for details). These reporter plasmids were introduced into the wild-type strain 8004 (wt) and the zur mutant strain 1430nk (zurmt) to yield reporter strains wt/pG0267, wt/pG2471-2, wt/pG2976, wt/pG3788, zurmt/pG0267, zurmt/pG2471-2, zurmt/pG2976 and zurmt/pG3788, respectively (Table 1). The GUS activities of these strains were determined in Zn2+-rich conditions (see Materials and methods section for details). As shown in Table 3, the GUS activities of the reporter plasmids pG0267, pG2471-2 and pG3788 under the wild-type background are respectively 15-, 5- and 31-fold lower than those under the zur mutation background; in contrast, the GUS activity of pG2976 under the wild-type background is ∼9-fold higher than that under the zur mutation background. The differences are statistically significant (P = 0.01 by t-test). These results demonstrate that the three putative Zn2+-uptake genes XC0267, XC2471, and XC3788 are negatively regulated while the putative Zn2+ efflux gene XC2976 is positively regulated by Zur.
Figure 1.
The genetic organization of the Zur-regulated genes and the detailed genetic elements in their promoters. The genetic organization of XC0267, XC2471, XC2976 and XC3788 loci was based on the genome sequencing data of the Xcc strain 8004 (23). The identified transcriptional start sites (+1) are shown, and the deduced −35 and −10 promoter regions are underlined. Gray boxes denote the position and sequence of the Zur-binding sites in promoter regions of the genes. Value above a vertical narrow denotes the length of the spacer between the nearest two ORFs.
Table 3.
The GUS activity of different reporter strains in zinc-rich and zinc-deficient conditions
| Reporter strain | GUS activity (U)a | ||
|---|---|---|---|
| wt/pG0267 | 0.12 ± 0.02A | 0.33 ± 0.01B | 0.13 ± 0.03A |
| zurmt/pG0267 | 1.78 ± 0.01C | 1.77 ± 0.11C | 1.75 ± 0.13C |
| wt/pG2471-2 | 0.04 ± 0.01A | 0.14 ± 0.02B | 0.06 ± 0.03A |
| zurmt/pG2471-2 | 0.21 ± 0.03C | 0.25 ± 0.02C | 0.22 ± 0.02C |
| wt/pG3788 | 0.13 ± 0.01A | 0.32 ± 0.03B | 0.11 ± 0.02A |
| zurmt/pG3788 | 4.04 ± 0.34C | 4.69 ± 0.17C | 4.57 ± 0.02C |
| wt/pG2976 | 4.46 ± 0.10A | 1.71 ± 0.21B | 5.06 ± 0.23A |
| zurmt/pG2796 | 0.49 ± 0.01C | 0.54 ± 0.02C | 0.51 ± 0.06C |
aβ-Glucuronidase (GUS) activities were respectively determined after the growth of Xcc strains in NYG, NYG supplemented with EGTA to the final concentration of 0.5 mM (NYG + EGTA) or NYG supplemented with EGTA and ZnSO4 to the final concentrations of 0.5 mM and 0.2 mM (NYG + EGTA + Zn2+) for 24 h. Data are the mean ± SD of triplicate measurements. Each experiment was repeated three times and similar results were obtained. The different letters in each horizontal data column indicate significant differences at P = 0.01.
The expression of XC0267, XC2471-2, XC2976 and XC3788 responses to Zn2+ through the mediation of Zur
To determine whether the expression of XC0267, XC2471-2 (XC2471 and XC2472), XC2976 and XC3788 is affected by Zn2+ concentrations, the GUS activities of the reporter strains wt/pG0267, wt/pG2471-2, wt/pG2976 and wt/pG3788 in the Zn2+-rich conditions were compared to those in the Zn2+-deficient conditions. The results showed that when the Zn2+ chelator EGTA was added into the Zn2+-rich NYG medium, the GUS activities of the reporter strains wt/pG0267, wt/pG2471-2 and wt/pG3788 were about 3-, 4- and 2-fold increased, respectively, while the GUS activity of wt/pG2976 was about 2-fold decreased (Table 3). Statistical analysis revealed that these differences are significant (P = 0.01 by t-test). These altered GUS activities were due to the change of the free Zn2+ in the medium, because addition of ZnSO4 into the EGTA-supplemented NYG medium to a final concentration of 0.2 mM could restore the GUS activities of all the reporter strains to the levels in the Zn2+-rich NYG medium (Table 3). The results demonstrate that the expression of XC0267, XC2471-2 and XC3788 are repressed in Zn2+-rich condition and induced in Zn2+-limited condition. On the contrary, XC2976 is induced in Zn2+-rich condition and repressed in Zn2+-limited condition. These further support the possibility that XC0267, XC2471 and XC3788 may play roles in Zn2+ uptake-related function and XC2976 in Zn2+ export-related function.
The GUS activities of the reporter strains wt/pG0267, wt/pG2471-2 and wt/pG3788 in NYG medium with 0.5 mM EGTA are significantly lower than those of zurmt/pG0267, zurmt/pG2471-2 and zurmt/pG3788 in NYG medium without addition of EGTA, respectively (Table 3), indicating that addition of 0.5 mM EGTA could not completely release the repressing effect of Zur. Increasing EGTA concentration did not lead to a further increase of the GUS activities of these reporter strains (data not shown). Furthermore, the GUS activities of the reporter strains zurmt/pG0267, zurmt/pG2471-2, zurmt/pG3788 and zurmt/pG2976 were not affected by the Zn2+ status in the NYG medium (Table 3). These results suggest that the Zn2+ repressing effects on the expression of XC0267, XC2471-2 as well as XC3788 and the inducing effects on the expression of XC2976 are mediated by Zur.
Zur activates the transcription of XC2976 and represses XC0267, XC2471-2 and XC3788 directly
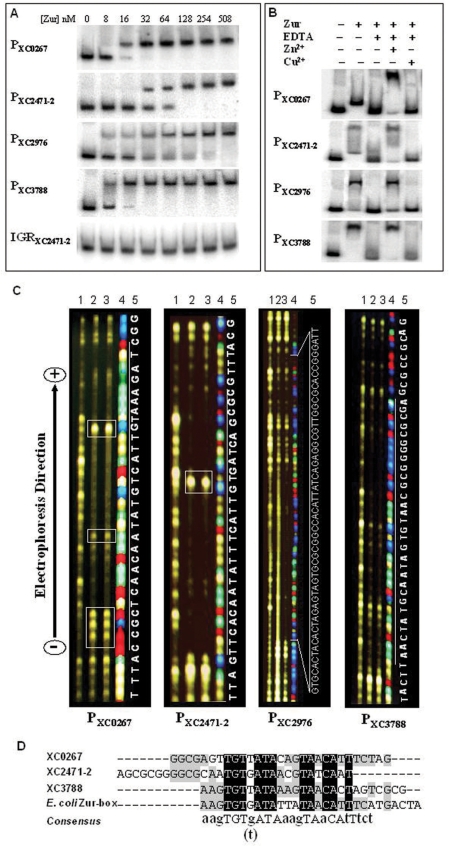
The above studies have demonstrated that the Xcc Zur positively regulates the expression of XC2976 and negatively regulates the expression of XC0267, XC2471-2 and XC3788. To investigate whether Zur directly regulates these genes, we performed EMSA assays to determine if the Xcc Zur binds to the promoter region of these genes. The His6-tagged Zur protein of Xcc was overexpressed and purified from E. coli (see Materials and methods section for details). The DNA fragments of the promoter regions (∼200-bp upstream of the start codon) of the ORFs XC0267, XC2471-2, XC2976 and XC3788 were amplified by PCR using the total DNA of the Xcc wild-type strain 8004 as template and the primer sets P0267-F/P0267-R, P2471-2-F/P2471-2-R, P2976-F/P2976-R and P3788-F/P3788-R (Table S1) and designated as P0267, P2471-2, P2976 and P3788, respectively (see Materials and methods section for details). The binding ability of the His6-Zur to these promoter-containing DNA fragments was evaluated by EMSA (see Materials and methods section). The EMSA results showed that the His6-Zur could bind to P0267, P2471-2, P2976 and P3788 with a high affinity, but not to a DNA fragment containing the intergenic region of the ORFs XC2471 and XC2472 (IGR2471-2472) (Figure 2A). The purified His6-Mip-like protein (35) was unable to bind to the promoter fragments P0267, P2471-2, P2976 and P3786 (data not shown). These indicate that the Zur-DNA binding detected in the EMSA was specific and the His6 tag did not interfere the binding. The results suggest that the Xcc Zur may directly regulate XC0267, XC2471-2, XC2976 and XC3788.
Figure 2.
The interaction of Zur with the promoters of Zur-regulated genes. (A) EMSA of Zur binding to the promoter regions of XC0267 (P0267), XC2471-2 (P2471-2), XC2976 (P2976) and XC3788 (P3788). Zur protein was incubated with 3 pmol of 32P-end-labeled DNA fragment in binding buffer at 28°C for 15 min, and analyzed by 4% polyacrylamide gel electrophoresis. (B) Effect of EDTA, Zn2+ and Cu2+ on the binding of Zur to its target promoters. Zur −, no Zur protein was added to the binding mixture; Zur +, Zur protein was added to the binding mixture to a final concentrations of 254 nM; EDTA −, no EDTA was added to the binding mixture; EDTA +, EDTA was added to the binding mixture to a final concentrations of 2 mM; Zn2+ −, no ZnSO4 was added to the binding mixture; Zn2+ +, ZnSO4 was added to the binding mixture to a final concentrations of 0.25 mM; Cu2+ −, no CuSO4 was added to the binding mixture; Cu2+ +, CuSO4 was added to the binding mixture to a final concentrations of 3 mM. (C) DNaseI footprinting analysis of Zur binding to P0267, P2471-2, P2976 and P3788. Zur protein was incubated with a 5′-TAMRA-labeled DNA fragment in binding buffer at 28°C for 15 min and then DNaseI (Promega) was added to the reaction mixture and incubated for 1 min. The DNaseI digestion products were separated in 4.75% Long Ranger Gel on an ABI 377 DNA sequencer. The sequencing product of the same fragment was included for localization of the binding site. Photos of the ‘Gel Image’ were directly taken from the screen of the sequencer. The concentration of Zur in each reaction was 0 (line 1), 80 (line 2) or 120 (line 3) nM. Line 4, sequencing product; line 5, sequence of the indicated region. The boxed areas indicate the additional DNaseI hypersensitive sites. (D) multiple alignment of the sequences of the Zur-binding sites in the promoters of XC0267, XC2471-2 and XC3788, and E. coli Zur-box. Black boxes denote identical amino acid residues and gray-shaded boxes indicate the place where at least two of the four residues are identical.
To determine the role of Zn2+ in the binding of Zur with its target DNA, we compared the binding ability of Zur to the promoter fragments P0267, P2471-2, P2976 and P3786 in the binding mixtures with and without addition of the Zn2+ chelator EDTA. The EMSA results showed that addition of EDTA blocked Zur-DNA binding and further addition of Zn2+ into the EDTA-containing binding mixture could fully restore the DNA binding ability of Zur (Figure 2B). However, addition of Cu2+ could not return the binding ability (Figure 2B). These reveal that the binding of Zur with its DNA targets requires Zn2+ as a cofactor.
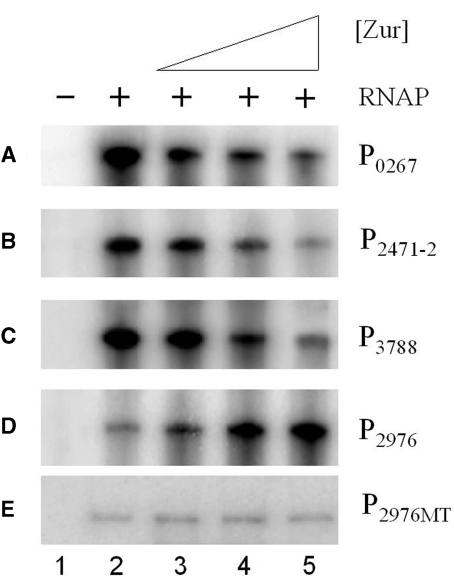
To further clarify whether Zur directly activates the transcription of XC2976 and represses XC0267, XC2471-2 and XC3788, in vitro transcription assays were performed on the promoter DNA of these genes (see Materials and methods section for details). The results showed that addition of Zur to the in vitro reactions decreased the transcription from the promoters of XC0267, XC2471-2 and XC3788, but increased the transcription from the XC2976 promoter (Figure 3A–D). These demonstrate that Zur represses the transcription of XC0267, XC2471-2 and XC3788, and activates the transcription of XC2976, directly.
Figure 3.
(A–E) Zur activates the transcription of the wild-type XC2976 promoter (P2976) but not the mutant XC2976 promoter (P2976MT) and represses XC0267 (P0267), XC2471-2 (P2471-2) and XC3788 (P3788) in vitro. RNA was generated in vitro from a PCR product template containing the promoter as well as 150-bp downstream of the start codon of the corresponding gene. For in vitro transcription, templates were incubated with 0 (lane 2), 7 (lane 3), 36 (lane 4) and 180 ng (lane 5) of Zur protein before the start of transcription on addition of 1 U of RNAP. A negative control (lane 1) was set up without addition of RNAP. Transcription products were then run on a 5% denatured polyacrylamide gel containing 7 M urea in 1× Tris borate–EDTA electrophoresis buffer. Each experiment was repeated three times and similar results were obtained.
Zur recognizes two distinct DNA targets
We employed DNaseI footprinting analysis to ascertain the Zur-binding sequences in its target promoters (see Materials and methods section for details). As shown in Figure 2C, the purified His6-tagged Zur protein bound to a region of 59-bp in length in the promoter of XC2976 and an about 30-bp region in the promoters of XC0267, XC2471-2 and XC3788. Sequencing analysis displayed that the Zur protected regions in the promoters of XC0267, XC2471-2 and XC3788 are about 30-bp AT-rich sequences showing high similarity to the E. coli Zur-binding sequences (Zur-box) (AAGTGTGATATTATAACATTTCATGACTA) (14,15) (Figure 2D). Multiple alignment of the Zur-protected sequences in the promoters of XC0267, XC2471-2 and XC3788 as well as the E. coli Zur box using the Vector NTI Alignment program (Invitrogen) revealed a conserved core sequence aagTGTg(t)ATAaagTAaCAtTtct (Figure 2D). Interestingly, the Zur-protected region in the XC2976 promoter is a 59-bp GC-rich (57.7%) sequence (AATCCCGGTGCGCCAACGCCTCTGATAATGTGGCCGCGCACTACTCTAGTGTAGTGCAC) containing a 20-bp imperfect inverted repeat, GCACTACTCT-AGTGTAGTGC. Sequence comparison exhibited that this inverted repeat sequence displays no significant similarity to the Zur-binding sequences in the promoters of XC0267, XC2471-2 and XC3788. These results, taken together, suggest that the Xcc Zur can recognize two distinct DNA targets.
As shown in Figure 2C, five and one additional DNaseI hypersensitive bands were respectively observed in the Zur-protected regions of XC0267 and XC2471-2 promoters, suggesting that the Xcc Zur might induce local distortions in the target DNAs upon binding. However, no additional hypersensitive band was observed in the Zur-protected regions of XC3788 and XC2976 promoters (Figure 2C), indicating that the interactions of the Xcc Zur with these promoters might be somewhat different. Furthermore, the Zur-protected sequence on the XC2976 promoter was approximately twice as wide as that on the promoters of the XC0266, XC2471-2 and XC3788, suggesting that there might be more Zur molecules bound to the XC2976 promoter. Consistent with this suggestion is the observation that the Zur-P2976 complex migrated more slowly than the Zur-P0266, Zur-P2471-2 and Zur-P3788 complexes (data not shown).
To clarify the Zur-binding site in the above Zur-regulated promoters is located within the −35 to −10 region or outside the region, the transcriptional start sites of XC0267, XC2471-2, XC3788 and XC2976 were determined by 5′ RACE PCR (see Materials and methods section for details). After the transcriptional start site was determined, a potential −35 and −10 sequence in the promoter region were surveyed based on the known features of E. coli promoter sequence (TTGACA [-35]-N17 or N16 [spacer]-TATAAT[-10]-N6 or N7 [spacer] - +1 [transcriptional start site]) (Figure 1). Consistent with the fact that E. coli Zur represses gene expression by binding to the −35 to −10 region of its target promoters to block the entry of RNA polymerase (14,15), the binding site of Xcc Zur in all of the three tested Zur-repressed promoters overlaps the −35 to −10 region (Figure 1). Interestingly, the Zur-binding site in the Zur-activated promoter (PXC2976 of XC2976) also overlaps the −35 to −10 region (Figure 1).
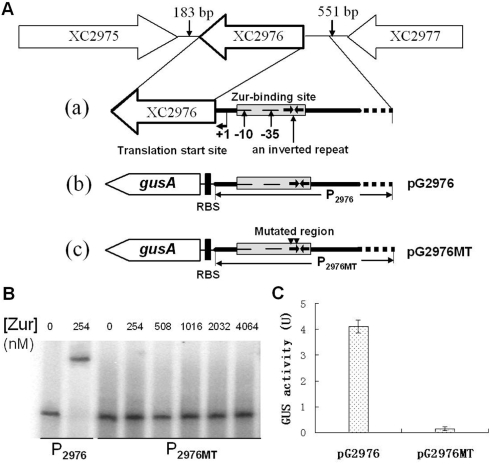
The 20-bp inverted repeat sequence is essential for Zur binding and for activation of XC2976 transcription by Zur
To determine whether the 20-bp imperfect inverted repeat in the XC2976 promoter is indeed a Zur-binding site and the binding of Zur with this element is essential for the transcriptional activation of XC2976, the left half of the 20-bp inverted repeat, CGTGATGTGA, was mutagenized and altered into ACACCACACC (see Materials and methods section for details). This mutagenized XC2976 promoter fragment was designated as P2976MT and used for construction of a gusA transcriptional fusion reporter plasmid named pG2976MT (P2976MT-gusA) (Figure 4A) and for EMSA with Xcc His6-tagged Zur protein. As shown in Figure 4B, the His6-Zur bound specifically to the wild-type XC2976 promoter fragment (P2976) but failed to bind to the mutated XC2976 promoter fragment P2976MT in the same conditions although 20-fold of Zur protein was used, suggesting that the 20-bp imperfect inverted repeat element in the XC2976 promoter is a critical motif for the interaction between Zur and XC2976 promoter. Furthermore, under the Xcc wild-type strain 8004 background (in which the Zur protein is a wild-type), the GUS activity produced by the wild-type XC2976 promoter-gusA fusion reporter plasmid pG2976 was 4.12 U, while the GUS activity produced by the mutagenized XC2976 promoter-gusA fusion reporter plasmid pG2976MT was only 0.14 U (Figure 3C). These results revealed that the Zur was unable to activate the transcription of the mutated XC2976 promoter. To determine whether the 20-bp imperfect inverted repeat is required for in vitro activation of XC2976 promoter, an in vitro transcription assay was performed on the mutated XC2976 promoter in which the 20-bp imperfect inverted repeat element is lacking (see Materials and methods section for details). As expected, although activation was observed at the wild-type XC2976 promoter (P2976), no activation was detected at the mutated XC2976 promoter (P2976MT) (Figure 3C and D). Overall, these findings demonstrate that the 20-bp imperfect inverted repeat is indispensable for Zur binding and activation of XC2976 both in vivo and in vitro.
Figure 4.
Effect of mutation in Zur-binding site on Zur binding and Zur transcriptional activation. (A) The detailed promoter composition of XC2976 (a) and the genetic structure of transcriptional gus fusion with the wild-type (P2976-gusA, pG2976) (b) or mutated Zur-binding site (P2976MT-gusA, pG2976MT) (c). The identified transcriptional start site (+1), the deduced −35 and −10 promoter regions and the Zur-binding site in the XC2976 promoter, as well as the inverted repeat inside the Zur-binding site and the altered region in the Zur-binding site are shown. (B) EMSA of Zur binding to DNA fragments with the wild-type (P2976) or mutated Zur-binding site (P2976MT). Zur protein was incubated with a 32P-end-labeled DNA fragment in binding buffer at 28°C for 20 min, and analyzed by 4% polyacrylamide gel electrophoresis. (C) The GUS activity of reporter plasmids (pG2976, containing a transcriptional gus fusion with the wild-type XC2976 promoter, and pG2976, containing a transcriptional gus fusion with the mutated XC2976 promoter) in the wild-type Xcc strain 8004. Xcc strains wt/pG2976 and wt/pG2976MT were grown in NYG medium for 24 h and the GUS activity was measured as described by Jefferson et al. (27). Data are the mean ± SD of triplicate measurements. Each experiment was repeated three times and similar results were obtained.
XC2976 may be a CDF-type cation efflux pump with broad metal specificity
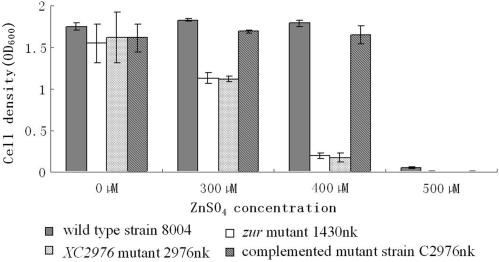
The above data suggest that XC2976 may be involved in Zn2+ efflux. The primary function of the Zn2+ efflux system is to export excessive Zn2+ out of the cytoplasm when the intracellular Zn2+ exceeds a critical level. Disruption of the Zn2+ efflux system will lead to intracellular Zn2+ extreme accumulation that then affects the growth of the bacteria. To verify further if XC2976 encodes a component of the Zn2+ efflux system, we constructed an XC2976 mutant designated 2976nk (Table 1) by homologous suicide plasmid integration (30) and measured its intracellular Zn2+ content as well as its growth rate in an excessive Zn2+ medium (see Materials and methods section for details). The results showed that the Zn2+ content in the mutant cells was about four times higher than that in the wild-type cells when grown in NYG supplemented with 300 µM of ZnSO4 for 4 h (Table 4). The complemented mutant strain contained similar amount of Zn2+ as the wild-type (Table 4), representing that the Zn2+ content in the mutant cells could be restored to the wild-type level by XC2976 in trans. The growth rate of the bacteria was tested in NYG medium supplemented with serial amounts of Zn2+. The results showed that the mutant 2976nk grew poorly in NYG medium supplemented with 400 µM ZnSO4, whereas the wild-type strain 8004 grew essentially normally in the same conditions (Figure 5). The growth of the mutant in the same conditions could be restored to the wild-type level by XC2976 in trans (Figure 5). These results demonstrate that XC2976 may encode a protein involved in Zn2+ efflux system of Xcc.
Table 4.
The intracellular zinc content of Xcc strainsa
| Strains | Intracellular zinc content (µg/1010 cells) |
|---|---|
| 8004 (wild-type) | 1.4 ± 0.11A |
| 1430nk (zur mutant) | 6.2 ± 0.21B |
| 2976nk (XC2976 mutant) | 6.4 ± 0.16B |
| C2976nk (complemented XC2976 mutant) | 1.5 ± 0.08A |
aData are the mean ± SD of triplicate measurements. Each experiment was repeated three times and similar results were obtained. The different letters in each data column indicate significant differences at P = 0.01.
Figure 5.
XC2976 is required for Xcc zinc resistance and the XC2976 and zur mutants display a similar zinc sensitivity level. Two hundred microliters of overnight culture (cell concentration adjusted to OD600 = 1.0) of each Xcc strain were inoculated into 200-ml NYG medium supplemented with ZnSO4 to a final concentration of 300, 400 or 500 µM. Cells were incubated at 28°C with shaking at 200 r.p.m. for 24 h. The cell density was measured spectrometrically at 600 nm. Values are the mean ± SD of triplicate measurements. Each experiment was repeated three times and similar results were obtained.
As shown in Table 2, XC2976 displays 67% similarity at the amino acid level to CzcD, a CDF (cation-diffusion facilitators)-type heavy metal ion efflux system of Ralstonia metallidurans (33). CDFs consist of a protein family involved in the metal ion transport found in archaea, eubacteria and eukarya (36). Ralstonia metallidurans CzcD is a membrane-bound protein with six transmembrane (TM) regions and contributes to the Zn2+ resistance of the bacterium through reducing the intracellular accumulation of the cations (33,37). To test whether XC2976 possesses any structural similarity to R. metallidurans CzcD, we performed a TM domain analysis of XC2976 using the DAS TM prediction server (http://www.biomedi.su.se/-server/DAS) (38). The TM-segment prediction result showed that, like CzcD, XC2976 is also a membrane-bound protein with six TM regions (Data not shown), suggesting further that XC2976 may be a CDF-type Zn2+ efflux pump.
In addition to the effect on Zn2+, the R. metallidurans CzcD also contributes to Co2+ and Cd2+ resistance of the bacterium. To assess whether the czcD homologous XC2976 is responsible for the resistance of Xcc to cobalt and cadmium, the sensitivity of the XC2976-mutant 2976nk to cobalt and cadmium was tested by measuring its growth rate in NYG medium supplemented with serial amounts of Co2+ or Cd2+. The results showed that the mutant failed to grow in NYG medium supplemented with 250 µM CoCl2 or 60 µM CdSO4, whereas the wild-type strain grew almost normally under the same conditions (data not shown). The Co2+ and Cd2+ sensitivity of the mutant could be restored to the wild-type level by introduction of XC2976 in trans (data not shown). We further found that the mutant 2976nk is more sensitive to Ni2+ than the wild-type (data not shown). These demonstrate that XC2976 is required for the resistance of Xcc not only to Zn2+ but also to Co2+, Cd2+ and Ni2+, implying that XC2976 may be a metal efflux pump with broad substrate spectrum.
The above results demonstrate that the Zur of Xcc represses Zn2+ uptake and activates Zn2+ export. In Zn2+-rich medium, the zur mutant accumulated significantly more Zn2+ than the wild-type (11). To determine whether the increased Zn2+ accumulation in the zur mutant cells is due to the disturbance of the effusion or over-uptake of Zn2+, or both, we compared the Zn2+ sensitivity and the intracellular Zn2+ accumulation of the zur mutant 1430nk and the XC2976-mutant 2976nk. The results showed that the Zn2+ sensitivity levels and the intracellular Zn2+ accumulation of the zur mutant are identical to that of the XC2976-mutant (Figure 5; Table 4), indicating that the increased intracellular Zn2+ accumulation is due to the deficiency of Zn2+ effusion. This suggests that the Zn2+ hypersensitivity of the zur mutant is attributed to the suppression of XC2976 expression, not the constitutive expression of Zn2+-uptake systems.
DISCUSSION
Our previous observation that the Xcc zur-mutant accumulates significantly more Zn2+ than the parent strain suggests that Zur may regulate the zinc uptake and/or export systems in Xcc (11). By DNA microarray hybridization, sequence homology comparison, mutagenesis and promoter–reporter analysis, here we have identified three genes coding for putative zinc uptake systems and one gene encoding a zinc export system, which are regulated by Zur in Xcc. Of which, XC2976 encodes a Zn2+ efflux pump with broad metal substrate spectrum and is positively regulated by Zur, and XC0267, XC2471 as well as XC3788 encode putative Zn2+-uptake systems and are negatively regulated by Zur. DNA microarray hybridization result displayed that the expression of more than 60 ORFs was affected by Zur in Xcc. For our interests in this study, we focused on the above four zinc-homeostasis genes and investigated the mechanisms by which Zur regulates their expression.
The results present in this article reveal that the expression of the Zn2+ export gene XC2976 and the putative Zn2+-uptake genes XC0267, XC2471-2 and XC3788 responses to Zn2+ through the mediation of Zur, and Zur binds to the promoters of these genes in a Zn2+-dependent manner. Such influence on the expression of Zn2+-uptake systems has been observed in other bacteria (3,4). DNaseI footprinting analyses showed that the Zur-binding sequence in the three Zur-repressing promoters is an about 30-bp AT-rich sequence, which overlaps the promoters’ −35 to −10 region. This is similar to the findings from previous studies on E. coli and other bacteria. E. coli Zur represses the transcription of the Zn2+-uptake operon znuABC by binding to a ∼30-bp AT-rich sequence overlapping the −35 to −10 region of the znuABC promoter to block the entry of the RNA polymerase (14,15).The Zur of B. subtilis (16), M. tuberculosis (12) and S. coelicolor (17) also binds to a 30-bp AT-rich sequence overlapping the −35 to −10 region of its target promoters. These may suggest that most bacterial Zurs including the Xcc Zur, if not all, regulate negatively the expression of Zn2+-uptake systems by a mechanism similar to that of E. coli Zur.
To our knowledge, this is the first observation that a bacterial Zur positively regulates a Zn2+-export system besides being a repressor for Zn2+-uptake systems. It has been generally considered that the Fur family regulators function as repressors and bacterial metal ion uptake and efflux systems are separately regulated by their own regulators, i.e. members of the MerR and ArsR/SmtB family transcriptional regulators control metal ion efflux systems, while members of the Fur family transcriptional regulators control metal ion uptake systems (3,4,39). In E. coli, for example, the expression of zntA, a gene encoding a P-type ATPase-Zn2+-export system, is regulated by the MerR family transcriptional regulator ZntR (40), while the expression of the high-affinity Zn2+-uptake system zunABC is regulated by the Fur family transcriptional regulator Zur (5). A homology survey displayed that the genome of the Xcc strain 8004 possesses a ZntA [XC3531 (GenBank accession number YP_244594)] and two ArsR/SmtB [XC1489 (GenBank accession number YP_242577) and XC2723 (GenBank accession number YP_243792)] homologues but not ZntR homologue. These facts, taken together, suggest that Xcc may have developed its specific zinc homeostasis machinery, the regulatory mechanism of which is distinct from that of E. coli. A few members of the Fur family have been found to exhibit repressor and activator functions. For instance, the Fur of Neisseria meningitides (41) and Vibrio vulnificus (42), the BosR of Borrelia burgdorferi (43) and the PerR of B. subtilis (44) have been demonstrated to act as an activator and a repressor. These regulators suppress genes’ transcription by binding to their target promoter regions and activate genes’ transcription by binding to an upstream sequence of promoters (41–44). In addition to negative regulation of Zn2+-uptake systems, a few Zur proteins have been found to affect positively the expression of genes with cellular functions beyond the metal ion homeostasis. For example, the Zur of B. subtilis positively regulates the expression of the genes rocD and rocE that are involved in amino acid transport (16), and the Zur of S. typhimurium activates the expression of the virulence-related operon fliAZ (9). Although it has been proposed that the B. substilis Zur may activate the expression of rocE and rocD indirectly (16), the molecular mechanism by which Zur positively regulates gene expression in these bacteria is unknown. The DNA microarray hybridization showed that in addition to the four zinc homeostasis-related genes, the Xcc Zur also regulates other more than 60 genes involved in other cellular processes including hpaB and hrpE, which are involved in pathogenicity and hypersensitive response (Table S2). It is a subject of considerable interest to investigate the mechanisms by which Zur regulates these processes in Xcc.
The gus-promoter transcriptional fusion analysis suggests that the expression of XC2976 responses to Zn2+ through Zur mediation. The EMSA and DNaseI footprinting analyses demonstrate that the Xcc Zur binds to the promoter region of the Zn2+-export gene XC2976. In vitro transcription assay demonstrates that Zur activates the transcription of XC2976 directly. These results reveal that the Xcc Zur directly activates the expression of XC2976. Interestingly, the Xcc Zur binds to a 59-bp GC-rich sequence with a 20-bp imperfect inverted repeat overlapping the −35 to −10 region of the XC2976 promoter, which differs from the ∼30-bp AT-rich sequence bound by Zur in the promoters of the Zn2+-uptake systems. Mutagenesis of the 20-bp imperfect inverted repeat resulted in complete abolishment of the in vitro binding of Zur to the promoter, and the in vivo and in vitro activation of the XC2976 promoter by Zur, indicating that the Xcc Zur positively regulates the transcription of XC2976 by directly binding to a cis-acting element located in the XC2976 promoter. How the Xcc Zur distinguishes these two different target sequences and what is the mechanism by which it activates the transcription of the Zn2+-export gene XC2976 after binding to the promoter remain to be further investigated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to J. Maxwell Dow and Robert P. Ryan for helpful discussions. This work was supported by the National Science Foundation of China (30671142 and 30730004) and the ‘863’ Program of the Ministry of Science and Technology of China (2006AA02Z175). Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nies DH. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999;51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe DK, Morby AP. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 2003;27:291–311. doi: 10.1016/S0168-6445(03)00041-X. [DOI] [PubMed] [Google Scholar]
- 3.Hantke K. Bacterial zinc transporters and regulators. Biometals. 2001;14:239–249. doi: 10.1023/a:1012984713391. [DOI] [PubMed] [Google Scholar]
- 4.Hantke K. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 6.Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalet K, Gouin E, Cenatiempo Y, Cossart P, Hechard Y. Characterisation of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol. Lett. 1999;174:111–116. doi: 10.1111/j.1574-6968.1999.tb13556.x. [DOI] [PubMed] [Google Scholar]
- 8.Lindsay JA, Foster SJ. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology. 2001;147:1259–1266. doi: 10.1099/00221287-147-5-1259. [DOI] [PubMed] [Google Scholar]
- 9.Campoy S, Jara M, Busquets N, Perez De Rozas AM, Badiola I, Barbe J. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect. Immun. 2002;70:4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrido ME, Bosch M, Medina R, Llagostera M, Perez de Rozas AM, Badiola I, Barbe J. The high-affinity zinc-uptake system znuACB is under control of the iron-uptake regulator (fur) gene in the animal pathogen Pasteurella multocida. FEMS Microbiol. Lett. 2003;221:31–37. doi: 10.1016/S0378-1097(03)00131-9. [DOI] [PubMed] [Google Scholar]
- 11.Tang D-J, Li X-J, He Y-Q, Feng J-X, Chen B, Tang J-L. The zinc uptake regulator Zur is essential for the full virulence of Xanthomonas campestris pv. campestris. Mol. Plant Microbe Interac. 2005;18:652–658. doi: 10.1094/MPMI-18-0652. [DOI] [PubMed] [Google Scholar]
- 12.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. Global analysis of Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin JH, Oh SY, Kim SJ, Roe JH. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2) J. Bacteriol. 2007;189:4070–4077. doi: 10.1128/JB.01851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 2000;275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 15.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 16.Gaballa A, Wang T, Ye RW, Helmann JD. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 2002;184:6508–6514. doi: 10.1128/JB.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owen GA, Pascoe B, Kallifidas D, Paget MS. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and sigmaR. J. Bacteriol. 2007;189:4078–4086. doi: 10.1128/JB.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward AC. In: Xanthomonas. Swings JG, Civerolo EL, editors. London: Chapman & Hall; 1993. pp. 51–54. [Google Scholar]
- 19.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbour Laboratory Press: New York; 1972. [Google Scholar]
- 20.Daniels MJ, Barber CE, Turner PC, Sawczyc MK, Byrde RJW, Fielding AH. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad-host-range cosmid pLAFR1. EMBO J. 1984;3:3323–3328. doi: 10.1002/j.1460-2075.1984.tb02298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Turner P, Barber CE, Daniels MJ. Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 1985;199:338–343. [Google Scholar]
- 23.Qian W, Jia Y, Ren S-X, He Y.-Q, Feng J-X, Lu L.-F, Sun Q, Ying G, Tang D.-J, Tang H, et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas. campestris pv. campestris. Genome Res. 2005;15:757–767. doi: 10.1101/gr.3378705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y-Q, Zhang L, Jiang B-L, Zhang Z-C, Xu R-Q, Tang D-J, Qin J, Jiang W, Zhang X, Liao J, et al. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 2007;8:R218. doi: 10.1186/gb-2007-8-10-r218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Haddad H, Tomas C, Alsaker K, Papoutsakis ET. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc. Natl Acad. Sci. USA. 2003;100:1122–1127. doi: 10.1073/pnas.0237337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferson RA, Burges SM, Hirsh D. ß-glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl Acad. Sci. USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merighi M, Majerczak DR, Zianni M, Tessanne K, Coplin DL. Molecular characterization of Pantoea stewartii subsp. stewartii HrpY, a conserved response regulator of the Hrp type III secretion system, and its interaction with the hrpS promoter. J. Bacteriol. 2006;188:5089–5100. doi: 10.1128/JB.01929-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman YE, O’Brian MR. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J. Biol. Chem. 2004;279:32100–32105. doi: 10.1074/jbc.M404924200. [DOI] [PubMed] [Google Scholar]
- 30.Windgassen M, Urban A, Jaeger KE. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbl. Let. 2000;193:201–205. doi: 10.1111/j.1574-6968.2000.tb09424.x. [DOI] [PubMed] [Google Scholar]
- 31.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 33.Anton A, Grosse C, Reissmann J, Pribyl T, Nies DH. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 1999;181:6876–6881. doi: 10.1128/jb.181.22.6876-6881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Zang N, Tang D.-J, Wei M.-L, He Y.-Q, Chen B, Feng J.-X, Xu J, Gan Y.-Q, Jiang B.-L, Tang J.-L. Requirement of a mip-like gene for virulence in the phytopathogenic bacterium Xanthomonas campestris pv. campestris. Mol. Plant Microbe Interac. 2006;20:21–30. doi: 10.1094/MPMI-20-0021. [DOI] [PubMed] [Google Scholar]
- 36.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 37.Munkelt D, Grass G, Nies DH. The chromosomally encoded cation diffusion facilitator proteins DmeF and FieF from Wautersia metallidurans CH34 are transporters of broad metal specificity. J. Bacteriol. 2004;186:8036–8043. doi: 10.1128/JB.186.23.8036-8043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 41.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitides. Mol. Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 42.Lee HJ, Bang SH, Lee KH, Park SJ. Positive regulation of fur gene expression via direct interaction of Fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 2007;189:2629–2636. doi: 10.1128/JB.01791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc. Natl Acad. Sci. USA. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J. Bacteriol. 2005;187:6659–6667. doi: 10.1128/JB.187.19.6659-6667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 46.Leong SA, Ditta GS, Helinski DR. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 1982;257:8724–8730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.