Abstract
In the search of facile and efficient methods for cellular delivery of peptide nucleic acids (PNA), we have synthesized PNAs conjugated to oligophosphonates via phosphonate glutamine and bis-phosphonate lysine amino acid derivatives thereby introducing up to twelve phosphonate moieties into a PNA oligomer. This modification of the PNA does not interfere with the nucleic acid target binding affinity based on thermal stability of the PNA/RNA duplexes. When delivered to cultured HeLa pLuc705 cells by Lipofectamine, the PNAs showed dose-dependent nuclear antisense activity in the nanomolar range as inferred from induced luciferase activity as a consequence of pre-mRNA splicing correction by the antisense-PNA. Antisense activity depended on the number of phosphonate moieties and the most potent hexa-bis-phosphonate-PNA showed at least 20-fold higher activity than that of an optimized PNA/DNA hetero-duplex. These results indicate that conjugation of phosphonate moieties to the PNA can dramatically improve cellular delivery mediated by cationic lipids without affecting on the binding affinity and sequence discrimination ability, exhibiting EC50 values down to one nanomolar. Thus the intracellular efficacy of PNA oligomers rival that of siRNA and the results therefore emphasize that provided sufficient in vivo bioavailability of PNA can be achieved these molecules may be developed into potent gene therapeutic drugs.
INTRODUCTION
Peptide nucleic acids (PNAs) (1) are attractive candidates for the discovery and development of antisense gene therapeutics and antigene drugs (2–4), but rapid progress has been limited by the lack of facile and efficient cellular delivery methods (5,6).
Efficient DNA (and siRNA) transfection reagents, such as cationic lipids and polymers, are positively charged and rely on self-assembly via complexation with the negatively charged DNA. Unmodified aminoethylglycyl PNAs (aegPNAs) are charge neutral and do not interact with cationic lipids or polymers and consequently are not effectively delivered to cells by such reagents. Several modifications of the PNAs have been devised in order to increase the association of PNA oligomers with cationic lipid complexes. These modifications include covalent conjugation to lipophilic ligands [such as fatty acids (7,8) or polyheteroaromates (9)], introduction of negative charges through backbone modification (10–12), or through use of covalent PNA–DNA chimera (13) or PNA/DNA heteroduplexes (14). However, none of these approaches is optimal. Conjugation of PNAs to lipophilic ligands is straightforward, and allows cationic lipoplex delivery of the conjugates (7,8), but unfortunately the conjugates have poor aqueous solubility and in most cases the cellular antisense efficacy is limited. Backbone modifications of the PNAs require synthesis of or access to a number of special monomers and may also affect hybridization efficiency and specificity (12). Synthesis of PNA–DNA chimera likewise requires elaborate synthesis, and results in oligomers with compromised hybridization properties due to the structural incompatibility between the amide PNA and the phosphodiester DNA backbones (13). Finally, PNA/DNA hybrid lipoplex delivery (although being quite efficient) requires additional steps in the delivery protocol and very careful optimization of the complementary DNA sequences for each PNA (14–16). Nonetheless, supplying PNA oligomers with negative charges in order to assure effective interaction with cationic (lipid) cellular delivery agents is indeed an attractive approach provided that uncompromised hybridization characteristics and mismatch discrimination are maintained. We reasoned that simple conjugation of regular aegPNA oligomers to an oligo–phosphate ligand could adequately serve this purpose. In principle phosphorylated peptides containing e.g. phosphoserine or phosphotyrosine could be exploited, but as these are both chemically and biologically unstable—being prone to (enzymatic) hydrolysis—we decided to explore the possibilities of using more stable phosphonate compounds. Thus we have synthesized a glutamine phosphonate and a lysine bis-phosphonate derivative and incorporated these into PNA oligomers by solid phase synthesis.
The PNAs were targeted to the aberrant splice site in the globin intron inserted into a luciferase gene used in the HeLa pLuc705 cell antisense splice correction assay (17). The PNAs were delivered to the cells by lipofectamine, and the results show a dramatically higher antisense activity of the phosphonate conjugated PNAs compared to unmodified PNA with a clear positive correlation to the number of phosphonate groups in the PNA oligomer (up to 12). The bis-phosphonate conjugated PNAs were significantly more active than unmodified PNA delivered by the published PNA/DNA heteroduplex method and showed subnanomolar activity in the HeLa pLuc705 cell assay. These results show that PNA oligomers conjugated to four to six lysine bis-phosphonate units are very efficiently delivered to eukaryotic cells by cationic lipoplexes and therefore hold great promise for a general, straightforward and robust method for delivering gene activity modulating PNAs to eukaryotic cells in culture.
EXPERIMENTAL PROCEDURES
Synthesis of PNA
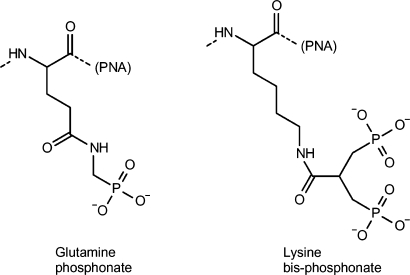
The sequences of the PNAs are listed in Table 1. PNA synthesis was carried out using Boc-chemistry as reported previously (18). Glutamine phosphonate or lysine bis-phosphonate monomers (Figure 1) were covalently linked to PNA at the N-terminal on the solid support via continuous synthesis using Fmoc-chemistry. The resulting PNAs were HPLC purified and characterized by MALDI-TOF mass spectrometry (Table 1). The PNAs were lyophilized and stored at 4°C until use.
Table 1.
PNA oligomers
| No. | Name | Sequence | Mass found (calcd) | (%)a |
|---|---|---|---|---|
| 2389 | Naked-PNA | H-CCTCTTACCTCAGTTACA-NH2 | 4767 (4765) | 95+ |
| 3283 | P3-PNA | H-(Gln-phosphonateb)3-eg1-CCTCTTACCTCAGTTACA-NH2 | 5576 (5576) | 90+ |
| 3242 | P7-PNA | H-(Gln-phosphonate)7-eg1-CCTCTTACCTCAGTTACA-NH2 | 6482f (6464) | 80+ |
| 3284 | P10-PNA | H-(Gln-phosphonate)10-eg1-CCTCTTACCTCAGTTACA-NH2 | 7127f (7128) | # |
| 3323 | bP4-PNA | H-(Lys-bis-phosphonatec)4-CCTCTTACCTCAGTTACA-NH2 | 6181f (6196) | 95+ |
| 3324 | bP5-PNA | H-(Lys-bis-phosphonate)5-CCTCTTACCTCAGTTACA-NH2 | 6545f (6554) | 95+ |
| 3325 | bP6-PNA | H-(Lys-bis-phosphonate)6-CCTCTTACCTCAGTTACA-NH2 | 6915f (6912) | 90+ |
| 3326 | Flk-bP6-PNAd | H-Flk-(Lys-bis-phosphonate)6-CCTCTTACCTCAGTTACA-NH2 | 7427f (7441) | 95+ |
| 2745 | PNA-mismatche | H-eg1-CCTCTGACCTCATTTACA-NH2 | 4911 (4910) | 90+ |
| 3353 | bP6-PNA-mismatche | H-(Lys-bis-phosphonate)6-CCTCTGACCTCATTTACA-NH2 | 6909 (6912) | 95+ |
aPurity (%): 95+ corresponds to single peak with slight foot; 90+ corresponds to single peak with slight foot and/or shoulder peak; 80+ corresponds to single peak with significant foot and/or shoulder peaks; #Broad peak with shoulders, exact purity difficult to estimate.
bStructure is shown in Figure 1.
cStructure is shown in Figure 1.
dFl-bP6-PNA has fluorescein (Fl) attached to the ε-amino group of a lysine residue (39).
eMismatch PNAs of PNA2745 and PNA3353, whereas two mismatches are underlined.
fDue to the polyanionic character pf the phosphonate-conjugated PNAs, the MALDI signal from these were quite weak and broad, and therefore less accurate than for the unmodified PNAs.
Figure 1.
Chemical structures of the glutamine phosphonate and lysine bis-phosphonate amino acids (attached to the N-terminal of PNA).
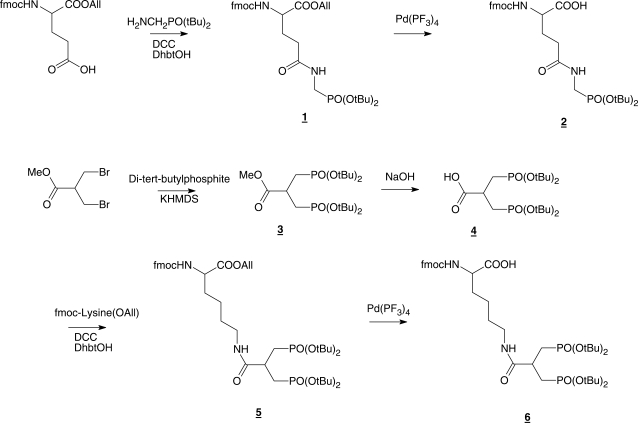
Synthesis of glutamine phosphonate and lysine bis-phosphonate monomers (Scheme 1)
Scheme 1.
4-[N-(Di-tert-butoxy-phosphorylmethyl)-carbamoyl]-2-Fmoc-amino butyric acid allyl ester (1)
Commercially available Fmoc-Glu-OAll (Sigma-Aldrich, Brøndby, Denmark) (3.2 g, 8 mmol) was dissolved in DMF (20 ml) under inert gas. DCC (2.4 g, 12 mmol) and DhbtOH (25% solution in DMF, 6.6 ml, 10 mmol) were added and the mixture was stirred for 20 min. A solution of amino methylphosphonic acid di-tert-butyl ester (19) (1.7 g, 7.2 mmol) in DMF (20 ml) was added and the mixture was stirred overnight under nitrogen. Insoluble solid was removed by filtration and the filtrate was evaporated to dryness under vacuum. The remaining residue was dissolved in EtOAc (100 ml) and extracted with a saturated aqueous solution of Na2CO3 (2 × 50 ml) and brine (1 × 50 ml). The organic phase was dried over Na2SO4 and evaporated to dryness. The crude product was purified by silica gel column chromatography (EtOAc) to provide 1 as a slightly yellow powder (72% yield). 1H NMR (CDCl3) δ 7.8–7.2 (m, 8H), 6.0 (d, J = 8 Hz, 1H), 5.8 (overlapping m, 2H), 5.2 (m, 2H), 4.6 (m, 4H), 4.3 (m, 2H), 4.2 (m, 2H), 3.3 (m, 2H), 2.8 (m, 2H), 1.4, (2s, 18H); 13C NMR (CDCl3) δ 170.72, 169.53, 156.21, 143.90, 143.77, 141.30, 131.62, 127.72, 127.10, 125.24, 125.15, 119.98, 118.63, 83.48, 83.35, 67.28, 66.34, 50.82, 47.13, 39.62, 37.57, 33.98, 30.40 (6C), mass spectrum (FAB), m/z 615 (M+H)+.
4-[N-(Di-tert-butoxy-phosphorylmethyl)-carbamoyl]-2-Fmoc-amino butyric acid (2)
Compund 1 (1.7 g, 2 mmol) was dissolved in THF (4 ml) under inert gas at ambient temperature. N-ethylaniline (1 ml) and a catalytic amount of Pd(PPhe3)4 were added sequentially and completion of the reaction was verified by TLC. Drop-wise addition of the reaction mixture into stirred n-hexane (100 ml) resulted in precipitation of pure 2 (78% yield) as a slightly yellow solid which was filtered off and washed appropriately with n-hexane. 1H NMR (DMSO d6) δ 12.7 (br.s, 1H), 7.8-7.2 (m, 10H), 4.3 (m, 3H), 4.0 (m, 1H), 3.5 (m, 2H), 2.2 (t, J = 8 Hz, 2H), 1.9 (m, 2H), 1.4, (s, 18H); 13C NMR (DMSO d6) δ 175.90, 171.80, 155.06, 136.08, 129.50, 129.33, 128.54, 128.45, 126.98, 126.90, 79.92, 64.78, 54.53, 50.55, 47.58, 45.20, 38.50, 29.69, 28.30 (6C); mass spectrum (FAB), m/z 575 (M+H)+.
3-(Di-tert-butoxy-phosphoryl)-2-(Di-tert-butoxy-phosphorylmethyl)-propionic acid methyl ester (3)
Di-tert-butyl-phosphite (2.56 g, 13.2 mmol) was dissolved in 0.5 M KHMDS solution in toluene (26.4 ml, 13.2 mmol) at 0°C (using an ice bath) under inert gas and stirred for 15 min. A solution of 3-bromo-2-bromomethyl-propionic acid methyl ester (1.54 g, 6 mmol) in dry toluene (5 ml) was added through a syringe. The ice bath was removed and the reaction mixture was stirred overnight at 25°C. Volatiles were removed under reduced pressure and the residue was dissolved in EtOAc (100 ml) and extracted successively with a saturated aqueous solution of Na2CO3 (1 × 50 ml) and KHSO4 (1 × 50 ml) and brine (1 × 50 ml). The organic phase was dried over Na2SO4 and solvents were removed by evaporation to provide 3 as a colorless foam (83% yield). 1H NMR (DMSO-d6) δ 3.6 (s, 3H), 3.3 (overlapping s, 4H), 1.9 (m, 1H), 1.4 (2s, 36H); 13C NMR (DMSO-d6) δ 173.43, 81.96, 52.56, 29.83, 27.46; mass spectrum (FAB), m/z 487 (M+H)+.
6-[3-(Di-tert-butoxy-phosphoryl)-2-(Di-tert-butoxy-phosphorylmethyl)-propionylamino]-2-(9H-floren-9-ylmethylcarboxylamino)-hexanoic acid allyl ester (5)
Compound 3 (2.4 g, 5 mmol) was dissolved in a mixture of methanol and NaOH (2 M) aqueous solution (100 ml, 1 : 1) and stirred for 1 h at 25°C. The solution was concentrated to half of its volume at reduced pressure and 40°C. Remaining solution was washed with diethylether (1 × 50 ml) and acidified to pH 4 with HCl (3 M) at 0°C. This solution was extracted with EtOAc (2 × 100 ml) and the combined organic phases were dried over Na2SO4 and the solvents were removed under reduced pressure to provide 4 as colorless foam (1.5 g, 62%). Freshly made foam was dissolved in DMF (20 ml). DCC (990 mg, 4.8 mmol) and DhbtOH (25% solution in DMF, 2.5 ml, 3.8 mmol) were added and the mixture was stirred for 20 min under inert gas to provide the activated carboxylic acid. In parallel a solution of commercially available Fmoc-lysine-OAll·HCl (Iris Biotech GmbH) (1.5 g, 3.8 mmol) in DMF (20 ml) was prepared and neutralized by adding DIPEA (0.7 ml, 4 mmol) and stirring for 20 min. The neutralized solution was added into activated carboxylic acid solution and the mixture was stirred overnight under inert gas. Insoluble solid was removed by filtration and the filtrate was evaporated under vacuum to dryness. The remaining residue was dissolved in EtOAc (100 ml) and extracted with saturated aqueous solution of Na2CO3 (2 × 50 ml) and brine (1 × 50 ml). The organic phase was dried over Na2SO4 and evaporated to dryness. The crude product was purified by silica gel column chromatography (EtOAc/methanol, 9:1) to provide 5 as a slightly yellow powder (44% yield). 1H NMR (DMSO-d6) δ 7.8-7.3 (m, 10H), 5.8 (m, 1H), 5.3 (m, 2H), 4.6 (m, 3H), 4.3 (m, 2H), 4.0 (m, 1H), 3.3 (m, 2H), 3.0 (m, 2H), 2.7 (m, 1H), 2.0-1.7 (m, 6H), 1.6 (m, 2H), 1.4 (s, 36H); 13C NMR (DMSO-d6) δ 175.30, 172.46, 156.10, 143.70, 142.80, 140.68, 137.40, 132.36, 128.85, 127.57, 127.21, 126.98, 121.30, 120.04, 119.94, 117.57, 81.06 (4C), 65.63, 64.65, 64.27, 53.80, 46.63, 36.73, 33.30, 30.00 (12C), 28.40, 22.68; mass spectrum (FAB), m/z 863 (M+H)+.
6-[3-(Di-tert-butoxy-phosphoryl)-2-(Di-tert-butoxy-phosphorylmethyl)-propionylamino]-2-(9H-floren-9-ylmethylcarboxylamino)-hexanoic acid (6)
The method for preparation of compound 2 was applied to yield compound 6 (68% yield) from 5 as a slightly yellow powder. 1H NMR (DMSO-d6) δ 7.9-7.3 (m, 10H), 4.5 (m, 3H), 3.9 (m, 1H), 3.3 (m, 2H), 3.0 (m, 2H), 2.7 (m, 1H), 2.0-1.7 (m, 6H), 1.6 (m, 2H), 1.4 (s, 36H); 13C NMR (DMSO-d6) δ 177.30, 172.40, 156.30, 143.76, 142.80, 140.66, 137.10, 128.60, 127.50, 127.20, 126.90, 125.22, 121.50, 120.10, 119.90, 81.07 (4C), 65.60, 64.60, 64.20, 53.80, 46.60, 36.70, 33.82, 30.08 (12C), 28.46, 21.00; mass spectrum (FAB), m/z 823 (M+H)+.
Cell culture
HeLa pLuc705 cells were purchased from Gene Tools (Philomath, OR, USA). Cells were grown in RPMI1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma), 1% glutamax (Gibco, Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin (Gibco) and 100 μg/ml streptomycin (Gibco) at 37°C in humidified air with 5% CO2. For studies of HeLa pLuc705 cells in 96-well plate format, cells were trypsinized and seeded (1.2 × 104 cells/well) 16 h before treatment. For studies in 24-well plate, 7.2 × 104 cells/well were used.
Cell delivery of phosphonate conjugated PNA
The cells, replated in 96-well or 24-well plates 16–24 h earlier, were treated with 100 μl/well (for 96-well plate) or 0.3 ml/well (for 24-well plate) of the PNA solution in RPMI1640 (10% FBS, 1% glutamax) for 24 h. Preparation of 0.3 ml delivery solution: 60 μl PNA/lipid complex (formed by incubating 30 μl of PNA solution (in water) with 30 μl of LipofectAMINE2000 (Invitrogen, Carlsbad, CA, USA) (diluted in OPTI-MEM to a final concentration of LipofectAMINE2000 of 8 μl/ml) for 10 min at room temperature) was mixed with 0.24 ml of RPMI1640 (10% FBS, 1% glutamax). Then the cells were incubated with this mixture for 24 h at 37°C and were subjected to further analysis.
PNA/DNA heteroduplex delivery
Formation of PNA/DNA heteroduplexes was performed as described (15), using PNA2389 and complementary DNA (5′-AATATGTAACTGAGGTA-3′). The PNA/DNA heteroduplex was incubated with LipofectAMINE2000 in 50 μl of OPTI-MEM for 15 min and mixed with 250 μl of RPMI1640 (10% FBS, 1% glutamax). The solutions were used for delivery as described above for the phosphonate conjugated PNA.
Luciferase assay
Luciferase activity of the cells was measured by using Bright-Glo Luciferase assay system (Promega, Madison, WI, USA) or Luciferase Assay System (Promega) for 96-well plate format (for screenings) or 24-well plate format, respectively, according to the manufacture's instructions. Luminescent readings from 96-well plate formats (with background subtracted) are presented as relative light units (RLU/well) (as the amount of material does not allow protein determinations). For the cells in 24-well plate formats, cell were lysed with Passive Lysis buffer (Promega) and used for both luciferase analysis and RNA extraction. Luminescent readings from 24-well format were background-subtracted and normalized by protein concentrations and are presented as RLU/μg protein. Protein concentrations were determined by the BCA protein assay (Pierce) according to the manufacturer's instruction.
RT-PCR
Total RNA was extracted from the samples in Passive Lysis buffer using RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. Three ng of total RNA was used for each RT-PCR reaction (20 μl/reaction) by using OneStep RT-PCR kit (Qiagen). Primers for the RT-PCR were: 5′-TTGATATGTGGATTTCGAGTCGTC-3′ (forward primer) and 5′-TGTCAAT-CAGAGTGCTTTTGGCG-3′ (reverse primer). The RT-PCR program was as follows: [(55°C, 35 min) × 1 cycle, (95°C, 15 min) × 1 cycle, (94°C, 0.5 min; 55°C, 0.5 min; 72°C, 0.5 min) × 26–28 cycles]. RT-PCR DNA products were analyzed on a 2% agarose gel with 1 × TBE buffer and visualized by ethidium bromide staining. Gel images were captured by ImageMaster (Pharmacia Biotech, Uppsala, Sweden) and analyzed by UN-SCAN-IT software (Silk Scientific Corporation, Orem, Utah, USA).
Confocal microscopy
Exponentially growing HeLa pLuc705 cells were plated in 8-well Lab-Tek™ chambered coverglass (Nunc, Roskilde, Denmark) at a cell density of 5 × 104 cells/well the day before treatment, and treated with PNA for 24 h as described above. Following treatment, the cells were incubated with nuclear staining dye Syto59 (Molecular probes, 5 μM) for 10 min. Then the cells were washed with Hank's solution and immediately analysed by microscopy using a MultiProbe 2001 Laser Scanning Confocal System equipped with an Argon laser and a red laser diode (excitation wavelength 488 nm and 638 nm, Radiance2000, BioRad, Hercules, CA, USA) connected to a Nicon eclipse TE200 microscope (oil immersion 60X1.4 NA objective, Nikon, Tokyo, Japan). The Lasersharp 2000 software package (BioRad) was used for image acquisition and processing.
Thermal denaturation (Tm)
Tm measurements were performed on a Cary 300 Bio UV-visible spectrophotometer (Varian, Cary, NC, USA) connected to a temperature controller. Thermal melting profiles were obtained in 10 mM sodium phosphate (pH 7.0) containing 0.1 mM EDTA and NaCl at the designated concentration using heating–cooling cycles in the range of 5–95°C at a rate of 0.5°C/min. The melting temperature (Tm) was determined as the maximum of the first derivative of the heating curve. Cuvettes of 1.0 cm path length and 1.0 ml volume were used for all experiments.
RESULTS AND DISCUSSION
Novel efficient and facile methods for cellular (and not least in vivo) delivery of antisense (RNA interference) and antigene (DNA interference) therapeutic drugs in general are highly warranted, and this is particularly pertinent concerning PNA-based technologies. Cationic transfection reagents (such as cationic lipids), which very efficiently deliver negatively charged phosphodiester-derived oligonucleotides to cells in culture, are practically non-effective for charge neutral oligomers like unmodified PNA. However, DNA oligonucleotides may be exploited as hybridization carriers of PNAs, and if properly optimized in terms of PNA–DNA duplex stability, this method appears as the most effective for delivery of PNA to cells in culture (15,16).
Inspired by these observations, we decided to synthesize two readily accessible phosphonate derivatives of glutamine and lysine, respectively (Figure 1, Scheme 1), as a very convenient and general way of introducing a negatively charged domain into a PNA oligomer via continuous solid phase synthesis. The new amino acid monomers, containing either a phosphonate (glutamine) or bis-phosphonate (lysine) moiety (Figure 1), were coupled to the N-terminus end of the PNA as a trimer, heptamer or decamer (mono-phosphonate) or a tetra-, penta- or hexamer (bis-phosphonate) using an antisense PNA sequence which is targeted to an intronic aberrant splice site of the luciferase pre-mRNA in HeLa pLuc705 cells (17).
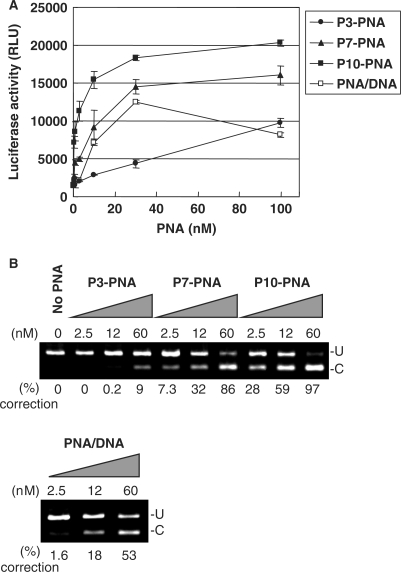
The PNA conjugates were delivered to the HeLa pLuc705 cells in complex with lipofectamine and the antisense activity, which is also a measure of cellular nuclear uptake efficiency, was evaluated quantitatively as induced luciferase activity as a consequence of mRNA splicing correction. The three PNAs, conjugated to 3, 7 or 10 units of glutamine phosphonate monomer, respectively (P3-PNA, P7-PNA, P10-PNA) showed significant dose-dependent antisense activity via lipofectamine delivery (Figure 2A), whereas essentially no activity was found in the absence of the cationic lipid (data not shown). The antisense activity of these PNA conjugates showed a clear dependence on the number of phosphonate monomers, with P10-PNA being most active exhibiting an EC50 [concentration yielding 50% of maximum luciferase activity (and 50% splicing correction)] around 5 nM. Furthermore, the activity found for P7-PNA was comparable to that obtained with unmodified PNA (naked PNA) delivered by the PNA/DNA heteroduplex lipofection method, while P10-PNA was several fold more active. The data obtained in the luciferase assay was quantitatively supported by RT-PCR analysis of mRNA splice correction (Figure 2B). These results clearly show that PNA–phosphonate conjugates are delivered to (the nucleus of) HeLa pLuc705 cells with an efficiency that surpasses that of the PNA/DNA mediated delivery system.
Figure 2.
Comparison of the antisense activity of phosphonate-conjugated PNAs with that of unmodified PNA and a PNA/DNA heteroduplex delivered by lipofection. The phosphonate-conjugated PNAs with 3, 7 and 10 units of phosphonate monomers (respectively P3-PNA, P7-PNA and P10-PNA) or the PNA/DNA heteroduplex were complexed with LipofectAMINE2000 and delivered to the HeLa pLuc705 cells. After 24 h, the cells were subjected to the further analysis. (A) Luciferase activity [using Bright-Gro assay (Promega)]. The values represent the mean ± SD of three experiments. (B) RT-PCR analysis of the mis-splicing correction of luciferase pre-mRNAs by phosphonate-conjugated PNAs and PNA/DNA heteroduplex. U, fragments without correction (268 bp). (C) Fragments with mis-splicing correction (142 bp). The numbers under the figure indicate the relative amount (%) of the corrected form to the sum of corrected form and uncorrected form.
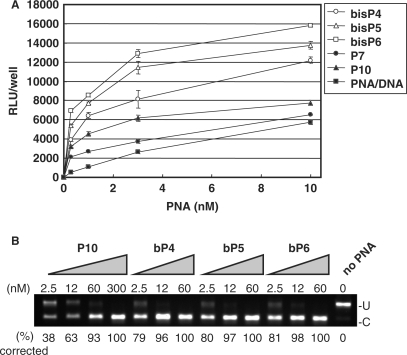
In an attempt to further improve the phosphonate conjugates, we synthesized a bis-phosphonate amino acid, which reduces the number of required coupling steps two-fold for an equivalent number of phosphonate groups. We synthesized PNAs conjugated to 4, 5 or 6 units of the bis-phosphonate lysine monomer, respectively at the N-terminus end (bP4-PNA, bP5-PNA and bP6-PNA) (Table 1) and tested these in the HeLa pLuc705 cell assay. The antisense activity of these PNAs was compared to that of PNAs P7-PNA and P10-PNA as well as to that of the naked PNA delivered as a PNA/DNA heteroduplex (Figure 3A). Also in this series the antisense activity of the bis-phosphonate conjugated PNAs depended on the number of bis-phosphonate monomers—while there were no significant antisense activities in the absence of the cationic lipid (data not shown)—and all of the bis-phosphonate conjugated PNAs showed significantly higher antisense activity than the P10-PNA and thus also than the PNA/DNA heteroduplex. Indeed the activity of bis-P6-PNA was at least 20-fold higher than that of the PNA/DNA duplex, and an EC50 value below 1 nM was indicated.
Figure 3.
Comparison of the antisense activity of bis-phosphonate-conjugated PNAs, phosphonate-conjugated PNAs and PNA/DNA heteroduplex delivered by lipofection. Three PNAs conjugated to 4, 5 and 6 units of bis-phosphonate monomers, respectively (bP4-PNA, bP5-PNA and bP6-PNA) and two phosphonate-conjugated PNAs (P7-PNA, and P10-PNA) and PNA/DNA heteroduplex were complexed with LipofectAMINE2000 and used for delivery to the HeLa pLuc705 cells. After 24 h, the cells were subjected to the further analysis. (A) Luciferase activity [using Bright-Gro assay (Promega)]. The values represent the mean ± SD of three experiments. (B) RT-PCR analysis of the mis-splicing correction of luciferase pre-mRNAs by phosphonate-conjugated PNAs. U, fragments without correction (268 bp). (C) Fragments with mis-splicing correction (142 bp). The numbers under the figure indicate the relative amount (%) of the corrected form to the sum of corrected form and uncorrected form.
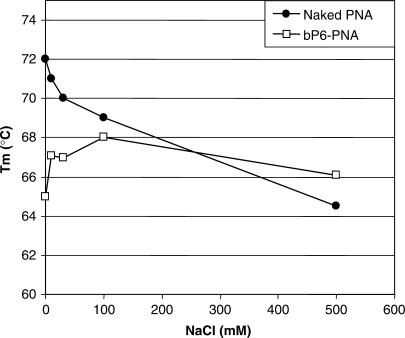
To analyze the effect of PNA–phosphonate conjugation on nucleic acid target binding affinity, thermal stabilities of the PNAs with a complementary DNA or RNA oligonucleotide were investigated (Table 2). Tm values of the PNA–DNA/RNA complexes with phosphonate conjugated PNAs and bis-phosphonate conjugated PNAs were virtually identical to that of the unmodified PNA in 100 mM NaCl (∼69 and ∼78°C for DNA and RNA, respectively). However, as would be expected from electrostatic repulsion considerations, the stability of the phosphonate conjugated PNA–DNA duplex decreases with decreasing ionic strength (Figure 5) as opposed to the behavior of aegPNA–DNA duplexes (20). These results indicate that coupling of the PNA to either phosphonate monomers or bis-phosphonate monomers dramatically improve the cellular delivery of the PNA without affecting the binding affinity of the PNA to the target sequence. Therefore, we further investigated the cellular fate of bP6-PNA, which showed the highest antisense activity among the PNAs tested.
Table 2.
Thermal stability (Tm) of PNA–DNA and –RNA duplexes
| Antisense Oligo# | Name |
Tm (°C)a |
|
|---|---|---|---|
| Complex with sense DNAb | Complex with sense RNAc | ||
| PNA2389 | Naked PNA | 69.0 | 79.0 |
| PNA 3283 | P3-PNA | 70.1 | 78.1 |
| PNA3242 | P7-PNA | 68.0 | 77.2 |
| PNA3284 | P10-PNA | 68.2 | 78.1 |
| PNA3323 | bP4-PNA | 69.0 | 79.0 |
| PNA3324 | bP5-PNA | 69.1 | 78.3 |
| PNA3325 | bP6-PNA | 68.1 | 78.0 |
| PNA2745 | PNA-mismatch | 45.0 | 59.0 |
| PNA3353 | bP6-PNA-mismatch | 46.1 | 59.2 |
| DNA2389d | DNA2389 | 58.0 | 60.0 |
| 2′O-MeRNAe | 2OMe | 56.0 | 75.0 |
aMelting temperatures of duplexes were measured in 10 mM sodium phosphate (pH 7.0) containing 100 mM NaCl and 0.1 mM EDTA.
bSense DNA sequence, 5′-TGTAACTGAGGTAAGAGG-3′.
cSense RNA sequence, 5′-UGUAACUGAGGUAAGAGG-3′.
dDNA2389 sequence, 5′-CCTCTTACCTCAGTTACA-3′.
e2′O-MeRNA sequence, 5′-CCUCUUACCUCAGUUACA-3′.
Figure 5.
Effect of NaCl concentration on the thermal stability of PNA/DNA duplexes. Melting temperature (Tm, °C) of the duplexes formed by PNA (naked PNA (PNA2389) or bP6-PNA (PNA3325) with a complementary DNA (5′-TGTAACTGAGGTAAGAGG-3′) measured in 10 mM sodium phosphate (pH 7.0) containing 0.1 mM EDTA and NaCl at the designated concentration.
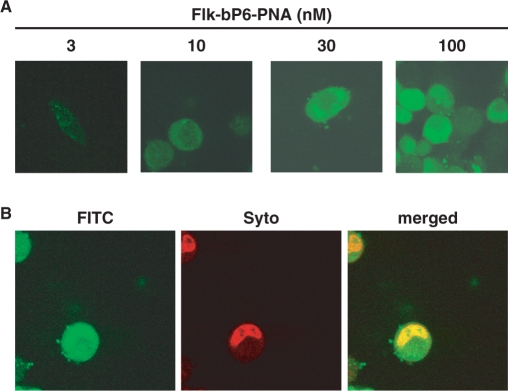
Cellular uptake of bP6-PNA was analyzed by confocal laser scanning microscopy using a fluorescein-labeled PNA (Flk-bP6-PNA). The HeLa pLuc705 cells were treated with a Flk-bP6-PNA/lipofectamine complex for 24 h and subjected to microscopy (Figure 4A and B). While the fluorescence signal was only faint at 10 nM and—almost absent at the lowest PNA concentration (3 nM)—cellular uptake was clearly visible at 30 and 100 nM as indicated by the even intracellular distribution of the green fluorescence appearing both in the cytosol and in the nucleus. To confirm the nuclear localization of the PNA, the cells treated with 30 nM Flk-bP6-PNA were counter-stained in the nucleus by the nuclear staining dye SYTO59 (Figure 4B). Co-localization of PNA (green fluorescent) and nuclear (red fluorescent) staining is very clearly observed as a yellow region on the merged image. These results therefore show that a major fraction of the PNA is indeed present in the nucleus.
Figure 4.
Cellular uptake of bis-phosphonated-PNA (bP6-PNA) in HeLa pLuc705 cells analyzed by confocal microscopy. (A) The cells were treated with 3, 10, 30 or 100 nM of Fl-bP6-PNA (PNA3326). Subsequently, the cells were washed with Hank's solution and subjected to microscopy analysis. (B) The cells were treated with 30 nM of Fl-bP6-PNA and subsequently nuclear-stained with SYTO59 (5 μM, Molecular probes). The merged image was superimposed from the images with green filter (FITC) and red filter (SYTO).
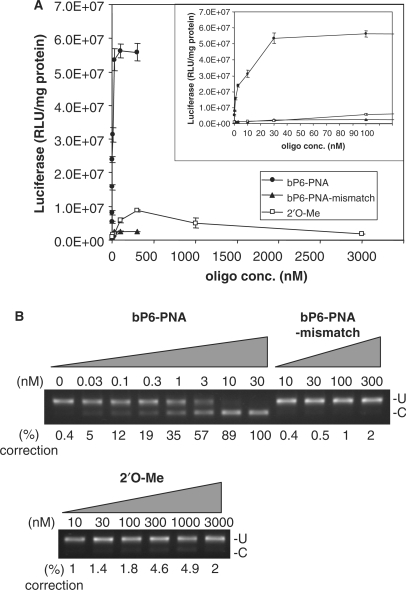
The sequence specificity of the antisense activity was established by using a double mismatch PNA (two base interchange) to the bP6-PNA. This mismatched PNA (PNA 3353) bound to complementary RNA with significantly reduced affinity (ΔTm = −19°C) as compared to the fully matched PNA 3325) (Table 1), and showed <2% antisense activity even at 300 nM (Figure 6A and B). In order to directly compare the efficiency of the bP6-PNA with a compound previously used in the HeLA pLuc705 assay, we used an analogous 2′O-Me-RNA. In a fully parallel experiment (Figure 6) this RNA was only slightly active giving 5% splice correction (10-fold over background) at 300 nM. Therefore, the phosphonate conjugated PNAs, in particular the bP6-PNA, are much more active than the corresponding 2′O-Me-RNA. Surprisingly, the activity of the 2′O-Me-RNA, despite efficient RNA hybridization [Tm = 75°C (Table 2)], did not rise beyond 5% even at higher concentrations (up to 3 μM). However, in other published studies using this RNA (21–23), only luciferase activities and not splice correction data were reported, and judged from this data the luciferase was activated only around 10-fold above background, while maximum activation by the PNA presently described is ≥200-fold. The low activity found for the 2′O-Me-RNA could be caused by rapid degradation due to insufficient nuclease stability (24,25). Finally, it is worth noting that none of these PNAs showed pronounced toxicities above that which can be attributed to the lipofectamine treatment (Figure S1,supplementary).
Figure 6.
Comparison of antisense activity of bP6-PNA with its mismatch control (bP6-PNA-mismatch) and 2′O-Me-RNA. HeLa pLuc705 cells were replated in 24-well plate the day before treatment with bP6-PNA (PNA3325), bP6-PNA mismatch (PNA3353) or 2′O-Me-RNA at the designated concentrations in complex with LipofectAMINE2000 for 24 h and subjected to the further analysis. (A) Luciferase activities were normalized by the protein concentrations. Inserted panel is the same data with the expansion below 100 nM. The values represent the mean ± SD of three experiments. (B) RT-PCR analysis of the mis-splicing correction of luciferase pre-mRNAs by the oligo. U, fragments without mis-splicing correction (268 bp). (C) Fragments with mis-splicing correction (142 bp). The numbers under the figure indicate the relative amount (%) of the corrected form to the sum of corrected form and uncorrected form.
CONCLUSION
The present study shows that conjugation of phosphonate or bis-phosphonate ligands to simple aegPNA oligomers can significantly improve cationic lipoplex-mediated cellular delivery and intracellular gene modulating activity of the PNA oligomer, and most likely cellular activity of other uncharged antisense compounds such as the morpholino oligomers (26,27) should be improved through analogous conjugation. Furthermore, such phosphonate conjugates may be delivered by a large range of systems developed for DNA and siRNA delivery (28–30) including cationic polymers and dendrimers as well as nanoparticles, which all rely on electrostatic interactions between the antisense agent and the delivery vehicle.
In terms of cellular delivery the presently described phosphonate conjugated PNAs may be compared with the PNA–DNA chimera or with the more advanced alternating backbone hydroyprolyl/phosphono PNA (‘HypPNA–pPNA’), which have shown promising properties both in cell culture (11,12) as well as in vivo using a peptide delivery vehicle (31). However, the conjugation strategy described here allows sensitive cellular activity testing and screening of basically unmodified aegPNA (as the phosphonate peptide is essentially a carrier) in order to optimize for gene and sequence target. It will also be straightforward to prepare ‘prodrug’ type of conjugates in which the phosphonate peptide is cleaved off by an esterase (32) or in a reductive environment (via disulfide linkage). In further developments toward medical drugs it may well be advantageous to entirely change delivery strategy for in vivo applications using nanoparticles, carrier peptides or other yet undiscovered modalities, and the phosphonate amino acids may even be included as components in such a strategy. The phosphonate peptide strategy thus increases the flexibility for future in vivo bioavailability optimization and should be considered a complementary strategy to the employment of e.g. cell penetrating peptides (CPPs) (33,34). The latter may eventually be developed for effective in vivo medical use, but are so far severely limited by relatively low potency due to endosomal trapping (35,36) (typically EC50 values around 1 μM is found) as well as significant toxicity. However, novel peptide motifs showing improved cell delivery properties are continuously being discovered (37,38).
Finally, and most importantly the results show that biological activities in the sub-nanomolar range of phosphonate peptide PNA conjugates are obtainable. Thus the intracellular efficacy of aegPNA oligomers rival that of siRNA and the results therefore emphasize that provided sufficient in vivo bioavailability of (derivatives of) aegPNA can be achieved these molecules may be developed into potent gene therapeutic drugs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgements
This work was supported by the Lundbeck Foundation, The Danish Cancer Foundation and the European Commission (6th FP EMIL, NUCAN and TRYLEIDIAG contracts LSHC-CT-2004-503569, NMP4-CT-2004-013775 & INCO-CT-2005-015379). The authors would like to acknowledge Ms Jolanta Ludvigsen for synthesizing the PNAs, Ms Annette W. Jørgensen for performing the Tm measurements and MSc Stanislava Pankratova for help with confocal laser microscopy. Funding to pay the Open Access publication charges for this article was provided by The European Commission (EMIL).
Conflict of interest statement. None declared.
References
- 1.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen PE. Antisense peptide nucleic acids. Curr. Opin. Mol. Ther. 2000;2:282–287. [PubMed] [Google Scholar]
- 3.Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg. Med. Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 4.Good L, Nielsen PE. Progress in developing PNA as a gene-targeted drug. Antisense Nucleic Acid Drug Dev. 1997;7:431–437. doi: 10.1089/oli.1.1997.7.431. [DOI] [PubMed] [Google Scholar]
- 5.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Deliv. Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 6.Kilk K, Langel U. Cellular delivery of peptide nucleic acid by cell-penetrating peptides. Methods Mol. Biol. 2005;98:131–41. doi: 10.1385/1-59259-877-3:131. [DOI] [PubMed] [Google Scholar]
- 7.Ljungstrom T, Knudsen H, Nielsen PE. Cellular uptake of adamantyl conjugated peptide nucleic acids. Bioconjug. Chem. 1999;10:965–972. doi: 10.1021/bc990053+. [DOI] [PubMed] [Google Scholar]
- 8.Mologni L, Marchesi E, Nielsen PE, Gambacorti-Passerini C. Inhibition of promyelocytic leukemia (PML)/retinoic acid receptor-alpha and PML expression in acute promyelocytic leukemia cells by anti-PML peptide nucleic acid. Cancer Res. 2001;61:5468–5473. [PubMed] [Google Scholar]
- 9.Shiraishi T, Bendifallah N, Nielsen PE. Cellular delivery of polyheteroaromate-peptide nucleic acid conjugates mediated by cationic lipids. Bioconjug. Chem. 2006;17:189–194. doi: 10.1021/bc050246z. [DOI] [PubMed] [Google Scholar]
- 10.Efimov VA, Choob MV, Buryakova AA, Kalinkina AL, Chakhmakhcheva OG. Synthesis and evaluation of some properties of chimeric oligomers containing PNA and phosphono-PNA residues. Nucleic Acids Res. 1998;26:566–575. doi: 10.1093/nar/26.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efimov VA, Birikh KR, Staroverov DB, Lukyanov SA, Tereshina MB, Zaraisky AG, Chakhmakhcheva OG. Hydroxyproline-based DNA mimics provide an efficient gene silencing in vitro and in vivo. Nucleic Acids Res. 2006;34:2247–2257. doi: 10.1093/nar/gkl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickstrom E, Choob M, Urtishak KA, Tian X, Sternheim N, Talbot S, Archdeacon J, Efimov VA, Farber SA. Sequence specificity of alternating hydroyprolyl/phosphono peptide nucleic acids against zebrafish embryo mRNAs. J. Drug Target. 2004;12:363–372. doi: 10.1080/10611860412331285242. [DOI] [PubMed] [Google Scholar]
- 13.Uhlmann E. Peptide nucleic acids (PNA) and PNA-DNA chimeras: from high binding affinity towards biological function. Biol. Chem. 1998;379:1045–1052. [PubMed] [Google Scholar]
- 14.Hamilton SE, Simmons CG, Kathiriya IS, Corey DR. Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chem. Biol. 1999;6:343–351. doi: 10.1016/S1074-5521(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 15.Doyle DF, Braasch DA, Simmons CG, Janowski BA, Corey DR. Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry. 2001;40:53–64. doi: 10.1021/bi0020630. [DOI] [PubMed] [Google Scholar]
- 16.Kaihatsu K, Huffman KE, Corey DR. Intracellular uptake and inhibition of gene expression by PNAs and PNA-peptide conjugates. Biochemistry. 2004;43:14340–14347. doi: 10.1021/bi048519l. [DOI] [PubMed] [Google Scholar]
- 17.Kang SH, Cho MJ, Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry. 1998;37:6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- 18.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, et al. Solid-phase synthesis of peptide nucleic acids. J. Pept. Sci. 1995;1:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 19.Genet JP, Uziel J, Port M, Touzin AM, Roland S, Thorimbert S, Tainer S. A practical synthesis of α-aminophosphonic acids. Tetrahedron. Lett. 1992;33:77–80. [Google Scholar]
- 20.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 21.Resina S, Kole R, Travo A, Lebleu B, Thierry AR. Switching on transgene expression by correcting aberrant splicing using multi-targeting steric-blocking oligonucleotides. J. Gene Med. 2007;9:498–510. doi: 10.1002/jgm.1044. [DOI] [PubMed] [Google Scholar]
- 22.Resina S, Abes S, Turner JJ, Prevot P, Travo A, Clair P, Gait MJ, Thierry AR, Lebleu B. Lipoplex and peptide-based strategies for the delivery of steric-block oligonucleotides. Int. J. Pharm. 2007;344:96–102. doi: 10.1016/j.ijpharm.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 23.Sazani P, Kang SH, Maier MA, Wei C, Dillman J, Summerton J, Manoharan M, Kole R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2'-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prater CE, Miller PS. 3′-methylphosphonate-modified oligo-2′-O-methylribonucleotides and their Tat peptide conjugates: uptake and stability in mouse fibroblasts in culture. Bioconjug Chem. 2004;15:498–507. doi: 10.1021/bc049977+. [DOI] [PubMed] [Google Scholar]
- 26.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 27.Wu RP, Youngblood DS, Hassinger JN, Lovejoy CE, Nelson MH, Iversen PL, Moulton HM. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 2007;35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Szoka F.C., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 29.Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv. Drug Deliv. Rev. 2007;59:164–182. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Howard KA, Kjems J. Polycation-based nanoparticle delivery for improved RNA interference therapeutics. Expert Opin. Biol. Ther. 2007;7:1811–1822. doi: 10.1517/14712598.7.12.1811. [DOI] [PubMed] [Google Scholar]
- 31.Morris MC, Gros E, Aldrian-Herrada G, Choob M, Archdeacon J, Heitz F, Divita G. A non-covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucleic Acids Res. 2007;35:e49. doi: 10.1093/nar/gkm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendifallah N, Kristensen E, Dahl O, Koppelhus U, Nielsen PE. Synthesis and properties of ester-linked peptide nucleic acid prodrug conjugates. Bioconjug. Chem. 2003;14:588–592. doi: 10.1021/bc025621r. [DOI] [PubMed] [Google Scholar]
- 33.Herbig ME, Weller KM, Merkle HP. Reviewing biophysical and cell biological methodologies in cell-penetrating peptide (CPP) research. Crit. Rev. Ther. Drug Carrier Syst. 2007;24:203–255. doi: 10.1615/critrevtherdrugcarriersyst.v24.i3.10. [DOI] [PubMed] [Google Scholar]
- 34.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv. Drug Deliv. Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Koppelhus U, Awasthi SK, Zachar V, Holst HU, Ebbesen P, Nielsen PE. Cell-dependent differential cellular uptake of PNA, peptides, and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev. 2002;12:51–63. doi: 10.1089/108729002760070795. [DOI] [PubMed] [Google Scholar]
- 36.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 37.Abes S, Turner JJ, Ivanova GD, Owen D, Williams D, Arzumanov A, Clair P, Gait MJ, Lebleu B. Efficient splicing correction by PNA conjugation to an R6-Penetratin delivery peptide. Nucleic Acids Res. 2007;35:4495–4502. doi: 10.1093/nar/gkm418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Andaloussi S, Johansson HJ, Holm T, Langel U. A novel cell-penetrating peptide, M918, for efficient delivery of proteins and peptide nucleic acids. Mol. Ther. 2007;15:1820–1826. doi: 10.1038/sj.mt.6300255. [DOI] [PubMed] [Google Scholar]
- 39.Lohse J, Nielsen PE, Harrit N, Dahl O. Fluorescein-conjugated lysine monomers for solid phase synthesis of fluorescent peptides and PNA oligomers. Bioconjug. Chem. 1997;8:503–509. doi: 10.1021/bc9700704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.