Abstract
Like its retroviral relatives, the long terminal repeat retrotransposon Ty1 in the yeast Saccharomyces cerevisiae must traverse a permanently intact nuclear membrane for successful transposition and replication. For retrotransposition to occur, at least a subset of Ty1 proteins, including the Ty1 integrase, must enter the nucleus. Nuclear localization of integrase is dependent upon a C-terminal nuclear targeting sequence. However, the nuclear import machinery that recognizes this nuclear targeting signal has not been defined. We investigated the mechanism by which Ty1 integrase gains access to nuclear DNA as a model for how other retroelements, including retroviruses like HIV, may utilize cellular nuclear transport machinery to import their essential nuclear proteins. We show that Ty1 retrotransposition is significantly impaired in yeast mutants that alter the classical nuclear protein import pathway, including the Ran-GTPase, and the dimeric import receptor, importin-α/β. Although Ty1 proteins are made and processed in these mutant cells, our studies reveal that an integrase reporter is not properly targeted to the nucleus in cells carrying mutations in the classical nuclear import machinery. Furthermore, we demonstrate that integrase coimmunoprecipitates with the importin-α transport receptor and directly binds to importin-α. Taken together, these data suggest Ty1 integrase can employ the classical nuclear protein transport machinery to enter the nucleus.
INTRODUCTION
Viruses encounter significant barriers that they must overcome in order to successfully infect cells. Not only must they bind and fuse with the plasma membrane to enter the cell (1), but once within the cell, a subset of viruses must then enter the nucleus and integrate their genome into the host DNA (2). Some retroviruses, such as the gammaretrovirus Murine Leukemia Virus, depend on ongoing mitosis and the breakdown of the nuclear envelope to access genomic DNA (2,3). However, other viruses, such as the lentivirus Human Immunodeficiency Virus Type 1 (HIV-1), have evolved to infect non-dividing, terminally differentiated cells (2,4), and therefore, must have acquired a mechanism to overcome the nuclear barrier and import necessary viral proteins and DNA into the nucleus for integration and replication.
All macromolecular transport between the cytoplasm and the nucleus occurs via a large proteinaceous complex known as the nuclear pore complex (NPC) that is embedded in the nuclear envelope (5–7). Proteins that are destined for the nucleus contain specific signals in their primary sequence termed nuclear localization signals (NLSs) (8). Classical NLS (cNLS) motifs are comprised of one or two clusters of basic amino acid residues termed monopartite or bipartite sequences, respectively (9–11). These cNLS motifs are recognized by the soluble NLS receptor, karyopherin/importin-α, which interacts with another karyopherin receptor, importin-β, to form an import complex (12–14). The directionality of nuclear protein transport is governed by Ran, a Ras-like GTPase that cycles between a GDP- and a GTP-bound state (15,16). The local nucleotide-bound state of Ran is modulated by a number of factors including the cytoplasmic GTPase activating protein (GAP), which facilitates the conversion of Ran-GTP to Ran-GDP in the cytoplasm (17,18), and the nuclear guanine nucleotide exchange factor (GEF), which maintains a nuclear pool of Ran-GTP (19,20). Following translocation of the cargo/receptor import complex through the NPC, Ran-GTP binds importin-β (21) to dissociate the import complex (14,22,23). The importin-β/Ran-GTP complex is then recycled back to the cytoplasm where the RanGAP facilitates the conversion of Ran-GTP to Ran-GDP (17) and the Ran import receptor, Ntf2, binds Ran-GDP to replenish the nuclear pool of Ran (24,25).
The Ty1 retrotransposon undergoes a retrotransposition cycle that is analogous to the ‘lifecycles’ of vertebrate retroviruses. Ty1 contains two open reading frames, TYA1 and TYB1, which are analogous to proteins encoded by the retroviral glycoprotein (gag) and RNA polymerase (pol) genes (26), respectively. In the early steps of the cycle, Ty1 mRNA is synthesized, processed and exported from the nucleus to the cytoplasm. In the cytoplasm, a subset of the mRNA is translated into Gag and Gag-Pol proteins that assemble to form virus-like particles (VLPs) containing two copies of the same Ty1 mRNA genome, Ty1 reverse transcriptase (RT) and integrase (IN) (27). The process of reverse transcription of this mRNA into cDNA occurs within VLPs (27–29). At a minimum, Ty1 cDNA and IN must then reenter the cell nucleus where IN functions to integrate the Ty1 cDNA back into the host genome (27). Since the nuclear membrane does not break down during mitosis in S. cerevisiae (30), these Ty retrotransposons must overcome an intact nuclear envelope in a manner that resembles the barrier posed to retroviruses in non-dividing cells. Thus Ty1 and its yeast host together provide an excellent experimental system to study the nuclear import of retroelement factors.
Ty1 IN contains an atypical NLS at its C-terminus that is required for proper nuclear localization of the IN protein and subsequent replication of the Ty1 retroelement (31,32). This NLS has been described as a non-classical bipartite NLS (31,32) since it consists of two clusters of basic amino acids separated by 29 amino acids. Although this targeting motif has been functionally defined, it is not clear what cellular machinery recognizes this motif and mediates IN import. The 29 amino acid linker is nearly three times longer than previously characterized classical bipartite linker sequences which are typically 10–12 amino acids in length (11,33–35), raising the question of whether this NLS motif mediates binding to the classical import receptor, importin-α. In this study, we investigated the mechanism by which Ty1 IN accesses the nucleus and subsequently the genomic DNA by taking advantage of a number of conditional alleles of essential classical nuclear protein import machinery. Through both in vivo and in vitro analyses, we present evidence that Ty1 IN can utilize the classical protein nuclear import pathway to access the nucleus despite its atypical NLS.
MATERIALS AND METHODS
Strains and plasmids
All DNA manipulations were carried out according to standard methods (36) and all media were prepared by standard procedures (37). Yeast strains and plasmids for this study are listed in Table 1. In all experiments the wildtype strain used was BY4741 (ACY1051). For some analyses, a GFP-fusion protein containing two GFPs fused to the last 54 amino acids of Ty1 integrase was used and will be referred to as GFP2-IN NLS (pAC1804). All chemicals were obtained from Ambion (Austin, TX), Sigma (St Louis, MO), or U.S. Biologicals (Swampscott, MA) unless otherwise specified.
Table 1.
Yeast strains and plasmids
| Strains/plasmids | Description | Reference |
|---|---|---|
| ACY107 | mat a ura3-52, leu2Δ1, trp1Δ63, rna1-1 | (69) |
| ACY110 | mat a ura3-52, leu2Δ1, prp20-1 | (70) |
| ACY114 | mat a ura3-52, leu2Δ1, trp1Δ63, NTF2::HIS3 [+NTF2 URA3] | (24) |
| ACY208 | mat α ade2, ura3-52, leu2Δ1, trp1Δ63, ade2, RSL1::HIS3 [+RSL1 URA3] | (71) |
| ACY212 | mat α ura3-52, leu2Δ1, trp1Δ63, GSP1::HIS3 GSP2::HIS3 [+ GSP1 URA3] | (48) |
| ACY443 | mat α ura3-52, leu2Δ1, trp1Δ63, his3Δ200, SXM1::HIS3 | (52) |
| ACY642 | mat α ura3-52, leu2Δ1, trp1Δ63, his3Δ200, srp1-55::LEU2 [+SRP1 URA3] | (56) |
| BY4741 (ACY1051) | mat a ura3-52, leu2Δ1, trp1Δ63, his3Δ1 | |
| pRS316 (pAC4) | URA3 CEN, AMP | (72) |
| pAC413 | gsp1-1 LEU2 CEN AMP | (48) |
| pAC493 | ΔIBB-SRP1 AMP, pProEX-Htb bacterial expression vector | (45) |
| pSW509 (pAC679) | rsl1-L63A LEU2 CEN AMP | (51) |
| pAC719 | Nab2-GFP URA3 2μ AMP | (42) |
| pAC781 | c-fus-GFP, KAN, pET-28a bacterial expression vector | (44) |
| N-fus GFP (pAC824) | pMET25-N-fus-GFP URA3 CEN AMP | (73) |
| pAC1414 | ntf2-1 TRP1 CEN AMP | (49) |
| pSD600 (pAC1661) | pGAL-Ty1-NEO URA3 CEN AMP | (38) |
| pAR100 (pAC1735) | pGAL-Ty1-HIS3 URA3 CEN AMP | (65) |
| GAL-LacZ (pAC1736) | pGAL-LacZ URA3 CEN AMP | (38) |
| pAC1804 (GFP2-IN NLS) | pMET-GFP-GFP-IN NLS URA3 CEN AMP | This study |
| pAC2326 | SRP1-myc LEU2 CEN AMP | This study |
| DSM1 (pAC2252) | pGAL-LacZ-Ty1 integrase URA3 CEN AMP | (32) |
| pAC2358 | GFP-IN NLS, KAN, pET-28a bacterial expression vector | This study |
| pAC2431 (GFP2-IN NLSmut) | pMET-GFP-GFP-IN NLSmut URA3 CEN AMP | This study |
| pAC2439 (GST-IN NLS) | GST-IN NLS, AMP, pGEX bacterial expression vector | This study |
| pAC2546 (GST-IN NLSmut) | GST-IN NLSmut, AMP, pGEX bacterial expression vector | This study |
Ty1 retrotransposition assay
The Ty1 retrotransposition assay was performed essentially as previously described (38) (Figure 1A). Briefly, wildtype or mutant cells were transformed with either the pAR100 (HIS3) or pSD600 (NEO) test plasmid. Transformants were selected on synthetic complete (SC) medium lacking uracil and including 2% glucose (SC ura− glu). To initiate the transposition assay, nine independent transformants were patched onto SC ura− glu plates. These plates were then replica plated to SC ura− gal and grown for three days at room temperature to induce transposition. Patches were then replica plated sequentially to: (i) yeast peptone dextrose (YPD); (ii) SC medium containing 1.2 g/l 5-fluoroorotic acid (5-FOA) and 2% glucose; and (iii) SC his− glu (pAR100) or YEPD containing G418 (0.2 mg/ml) (pSD600). Growth on the final selection plate was compared to controls: wildtype cells containing the pAR100 or pSD600 plasmid (positive control) or wildtype cells that never contained the test plasmid (negative control).
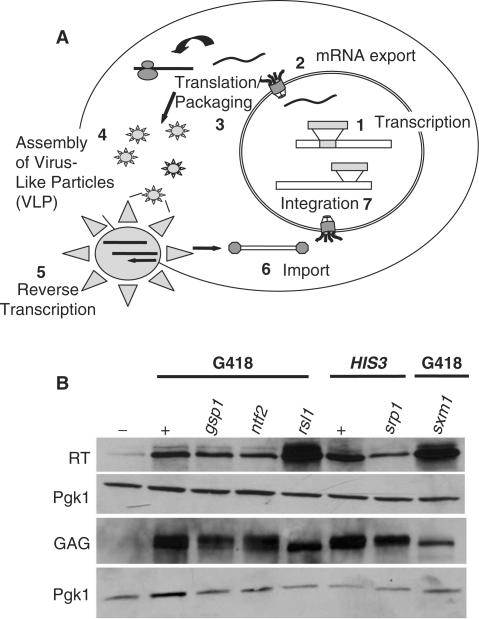
Figure 1.
Analysis of retrotransposition in mutants of classical nuclear transport proteins. (A) A schematic of the Ty1 retrotransposition assay (38). Yeast cells are transformed with the URA3-marked pAR100 (His- cells) or pSD600 (His+ cells) test plasmid and patched onto SC ura− glu plates. Patches are then replica plated to SC ura− gal plates for 3 days at 25°C and sequentially replica plated to (1) SC ura− glu; (2) YPD; (3) 5-FOA; and (4) SC his− glu for pAR100 or YEPD + G418 for pSD600. (B) Transposition levels in nuclear protein import mutants. gsp1-1, ntf2-1, srp1-5, rsl1-L63A, and Δsxm1 mutant cells were transformed with the appropriate test plasmids and replica plated to selection medium. The final selection plates are YEPD + G418 (gsp1-1, ntf2-1, rsl1-L63A, and Δsxm1) and SC his− glu (srp1-55). As controls, each plate also contained wildtype cells either expressing (+) or lacking (−) the appropriate test plasmid. (C) Transport mutants carrying the Ty1 test plasmid grow approximately as well as wildtype cells on selective media. gsp1-1, ntf2-1, rsl1-L63A and Δsxm1 cells were transformed with the pSD600 (G418) test plasmid while srp1-55 cells were transformed with the pAR100 (His) test plasmid. Cell numbers were equalized, cultures were serially diluted 10-fold, and spotted onto either YEPD + G418 plates (top) or SC-his (bottom) glu plates. Plates were then grown at 25°C for 3 days. Wildtype cells either carrying the appropriate Ty1 test plasmid (+) or an empty vector (−) served as positive and negative controls, respectively.
To eliminate any mutants that affect the transposition assay rather than retrotransposition itself, all mutants that showed defects in transposition were retested in two secondary assays. First, because this is a growth-based assay, it is possible that the mutants examined affect cell growth at room temperature or the ability of these cells to convey a His+ or G418R phenotype. To address this point, mutant cells were transformed with the appropriate Ty1 test plasmid (pAR100 or pSD600) and transformants were selected on SC ura− glu. Cells were then grown to saturation in 2 ml cultures at 25°C and were diluted to the same OD600 to standardize the cell number. Cells were serially diluted 10-fold and 3 μl of each dilution was spotted on either synthetic complete medium lacking histidine (SC-his) or SC + G418 (0.2 mg/ml) and incubated at 25°C for 2–4 days. Any mutant population that did not grow as well as the wildtype control or yield a His+ or G418R phenotype was excluded from further analysis.
The second test sought to eliminate mutants that affect induction from the GAL1 promoter, which is used to drive transposition from the Ty1 test plasmids (38). Thus mutants that cannot induce the GAL1 promoter would not display transposition from the Ty1 test plasmid. To eliminate this possibility, mutant cells were transformed with a pGAL1-LacZ reporter plasmid and assayed for LacZ induction. Each mutant containing the LacZ test plasmid was patched onto SC ura− plates and grown at 25°C. After patches had grown, they were replica plated to SC ura− + X-gal (80 μg/ml) plates containing either glucose (negative control) or galactose (positive control). Wildtype cells that were transformed with the pGAL1-LacZ reporter plasmid were used as positive control and the same cells lacking the reporter plasmid served as negative controls. Each mutant was compared in triplicate to the positive and negative controls and mutants failing to show β-galactosidase activity comparable to wildtype cells were excluded from further analysis.
Immunoblot analysis
We used standard methods for immunoblot analysis (39). Briefly, cultures were grown in 5 ml cultures to late log/early stationary phase and then collected by centrifugation at 3000 r.p.m. (735 r.c.f.) for 3 min. Cell pellets were washed once in 1 ml of PBSMT (phosphate-buffered saline, 5 mM MgCl2, 0.5% Triton X-100) for 1 min. Pellets were then resuspended in 500 μl of PBSMT supplemented with protease inhibitors (1 mM PMSF and 3 ng/ml each of aprotinin, leupeptin, chymostatin and pepstatin). Glass beads were added to saturation and cells were lysed with a 2 min pulse in a mini bead beater (Biospec Products). The lysate was cleared by centrifugation at 13 000 r.p.m. (13 800 r.c.f.) for 15 min. Lysate concentrations were determined using a Bradford protein assay (Bio-Rad). For quantitative protein analysis, 3 μg or 15 μg of total lysate was analyzed for the presence of Ty1 Gag or Reverse Transcriptase (RT). Ty1 Gag protein was detected using a 1:40 000 dilution of the Rf1 anti-Gag rabbit polyclonal antibody (26). Ty1 reverse transcriptase (RT) protein was detected using a 1:5000 dilution of an anti-RT rabbit polyclonal antibody (40). PGK-1 protein was detected using a 1:10 000 dilution of 125 μg/ml anti-PGK1 mouse monoclonal antibody (Molecular Probes). For coimmunoprecipitation experiments, GFP proteins were detected using a 1:10 000 dilution of an anti-GFP rabbit polyclonal antibody (41). Myc-tagged proteins were detected using a 1:5000 dilution of 200 μg/ml 9E10 mouse monoclonal c-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA). GST-tagged proteins were detected using a 1:5000 dilution of 200 μg/ml sc-138 mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Direct fluorescence microscopy
Wildtype and mutant cells (gsp1-1, rna1-1, prp20-1, srp1-55 and rsl1 L63A) (refer to Table 1) were transformed with plasmids expressing GFP-fusion proteins, GFP-IN (DSM1), GFP2-IN NLS (pAC1804), GFP2-IN NLSmut (pAC2431), or a control, Nab2-GFP (pAC719) (42). Cultures (2 ml) were grown to log phase in SC ura− glu media. Cells were then collected by centrifugation at 3000 r.p.m. (735 r.c.f.), washed one time in 1 ml of dH2O, and resuspended in 5 ml of SC ura− gal (DSM1) or SC ura−met− glu (pAC1804) medium to induce expression of GFP-fusion proteins overnight at 25°C. Cultures were then split in the morning and half was shifted to 37°C for 2 h (or 18°C overnight for srp1-55 cells) while the other half remained at 25°C. Cells were then incubated with 4.5 nM Hoechst dye (Sigma) to visualize chromatin, indicating the location of the nucleus. GFP was visualized through a GFP-optimized filter (Chroma Technology) and images were captured using IP Lab Spectrum software and an Olympus BX60 epifluorescence microscope.
Semi-quantitative kinetic GFP-NLS import assay
A GFP-NLS import assay was performed as described previously (43). Briefly, wildtype and mutant cells (gsp1-1, rna1-1, prp20-1, srp1-55 and rsl1 L63A) were transformed with the GFP2-IN NLS reporter plasmid. Cells were grown to early mid-log phase in SC ura−met− glu media at their permissive temperature of 25°C to induce expression of GFP2-IN NLS. Cells were then pelleted at 3000 r.p.m. (735 r.c.f.) for 1 min and resuspended in 1 ml of glucose-free SC media containing 10 mM sodium azide and 10 mM 2-deoxy-D-glucose. Cells were incubated at the non-permissive temperature of 37°C (18°C for srp1-55) for 45 min. The cells were then washed with 1 ml of ice-cold dH20 and resuspended in 100 μl of SC glu media at the non-permissive temperature to reinitiate nuclear import under nonpermissive conditions for each mutant. To measure the rate of nuclear import, a 2 μl aliquot was removed every 2.5 min and cells were analyzed and counted using a GFP-optimized filter (Chroma Technology) on an Olympus BX60 epifluorescence microscope. Approximately 100 cells were counted for each time point and were scored ‘nuclear’ if the nucleus was brighter than the surrounding cytoplasm.
Coimmunoprecipitation assay
A plasmid encoding Srp1-myc (S. cerevisiae importin-α) (pAC2326) was transformed into wildtype cells carrying plasmids encoding GFP2-IN NLS (pAC1804), GFP2-IN NLSmut (pAC2431), or GFP alone (N-fus GFP). Cultures (50 ml) were grown to log phase in SC ura− glu medium and then diluted into 500 ml of SC ura−met− glu to induce expression of GFP reporters. Cells were harvested by centrifugation at 3000 r.p.m. (1600 r.c.f.) for 10 min. Cell extracts were generated as described above for immunoblot analysis and 2 mg of total lysate was incubated for 1.5 h at 4°C with 30 μl of agarose-conjugated anti-Myc beads (9E10, Santa Cruz Biotechnology). The unbound fraction was collected by centrifugation for 1 min at 4000 r.p.m. (1310 r.c.f.). Beads were washed 4 times for 10 min in 1 ml of PBSMT followed by 2 washes in 1 ml of PBSM (phosphate-buffered saline, 5 mM MgCl2). Bound protein was eluted from the Myc beads with 45 μl of loading buffer (125 mM Tris-HCl, pH 6.8, 250 mM dithiothreitol, 5% SDS, 0.25% bromphenol blue, 25% glycerol). Unbound and bound fractions were resolved by SDS-PAGE and analyzed by immunoblot analysis. Proteins were detected using anti-GFP or anti-myc antibodies as described above for immunoblot analysis.
Recombinant protein expression and purification
Purified recombinant proteins used in these studies (GST, GST-IN NLS and a His6-tagged NLS-binding fragment of importin-α consisting of residues 89–530 (ΔIBB)) were expressed in E. coli BL21 (DE3) cells and purified either over glutathione-sepharose beads (GE Healthcare) or by nickel affinity chromatography as previously described (44).
In vitro binding assay
To assess the interaction between IN-NLS and importin-α, we employed a truncated form of importin-α, which lacks the N-terminal importin-β binding autoinhibitory domain (ΔIBB-importin-α) (45). This truncated form of importin-α mimics the import complex that forms when importin-β binds the IBB domain of importin-α to prevent the competition for the NLS binding pocket of importin-α (44–46). This ΔIBB-importin-α has a similar affinity for NLS cargo as importin-α in the context of the importin-α/β import complex (44). Purified GST-IN NLS protein (6 μg), GST-IN NLSmut, or GST control bound to GST-beads was incubated with 4 μg of ΔIBB-importin-α in 1× phosphate-buffered saline (PBS) containing 0.5 μg/ml BSA as a competitor. Proteins were mixed for 1.5 h at 4°C. Samples were then centrifuged at 4000 r.p.m. (1310 r.c.f.) for 1 min and the unbound fraction was collected in a fresh eppendorf tube. The beads were then washed 3 times in 1× PBS for 10 min at 4°C. Following the washes, sample buffer was added to elute the bound fraction and these fractions were analyzed on a 10% SDS-PAGE gel. After electrophoresis, the gel was stained in Coommassie Blue stain for 30 min and destained overnight. In order to assess how much importin-α was bound to GST-IN NLS, 4 μg of purified ΔIBB-importin-α was loaded on the gel (Input).
RESULTS
Ty1 retrotransposition is decreased in mutants of the classical nuclear protein import machinery
We exploited a previously described Ty1 retrotransposition assay (27) to assess the requirement for classical nuclear protein import factors in retrotransposition. Briefly, yeast cells were transformed with either the pAR100 (His+) or pSD600 (G418R) test plasmid (38), both of which carry the Ty1 element under control of the inducible GAL1 promoter. When cells are grown on medium containing galactose, retrotransposition is induced and, following the removal of the transposition plasmid, cells can be screened for either a His+ (pAR100) or G418R (pSD600) phenotype, indicative of successful retrotransposition (Figure 1A). We used this assay to screen a collection of previously characterized temperature-sensitive mutants that are defective in classical nuclear protein import. The genes screened include GSP1 (Ran), NTF2 (Ran-GDP import receptor), SRP1 (importin-α) and KAP95/RSL1 (importin-β). As a control, we used a strain deleted for KAP108/SXM1, a member of the importin-β family of transport receptors that does not contribute to classical nuclear protein import (47). Retrotransposition levels were severely decreased in gsp1-1 (48), ntf2-1 (49), srp1-55 (50) and rsl1-L63A (51) cells relative to wildtype cells (Figure 1B). Control Δsxm1 (52) cells show no defect in retrotransposition, indicating that not all nuclear import factors affect transposition. The apparent decrease in retrotransposition observed in this assay is not due to growth defects of these conditional mutant cell populations as each of these strains grew as well as control wildtype cells on the final selection plates (SC-his glu or SC + G418) when supplied with the appropriate test plasmid (Figure 1C). Furthermore, using a LacZ reporter assay, we established that gsp1-1, ntf2-1, srp1-55 and rsl1-L63A mutant cells induce expression of the LacZ gene from the GAL1 promoter at levels equal to wildtype cells indicating that these cells are competent to induce retrotransposition from the test plasmids (data not shown).
Ty1 proteins are made and processed properly in mutants of the classical nuclear protein import machinery
The Ty1 retrotransposon lifecycle is a complex process involving numerous cellular accessory proteins as shown in Figure 2A (38). The decrease in retrotransposition observed for nuclear protein import mutants could be explained by two possibilities: (i) the nuclear import of an essential Ty1 protein is affected in these mutants or (ii) the nuclear localization of a cellular protein involved in an early stage of the lifecycle, such as Ty1 transcription, is altered. As an initial test to distinguish between these two possibilities, Ty1 protein translation and processing was examined in each nuclear import mutant using immunoblot analysis. In order to assess Ty1 protein levels and processing, expression of the pAR100 or pSD600 transposition plasmid was induced in gsp1-1, ntf2-1, srp1-55, rsl1-L63A, and as controls in both wildtype and Δsxm1 cells and cell lysates were prepared. Lysates were then assayed for processed Ty1 proteins by immunoblotting. Figure 2B shows that the Ty1 proteins, Gag and RT, are made and processed in each of the mutants. Although some differences in expression and processing are observed, there is no correlation between levels of expression and defects in retrotransposition. The two forms of Gag observed have been previously described and are produced from two separate proteolytic cleavage sites during the protein processing steps of retrotransposition (53). Overall, results of this experiment indicate that both the RT and GAG proteins are properly translated and processed in these import mutants suggesting that a later stage of retrotransposition is affected with the most logical step being the nuclear import of Ty1 proteins.
Figure 2.
Analysis of Ty1 proteins in nuclear transport mutants defective for transposition. (A) A schematic of the Ty1 lifecycle: (1) Ty1 mRNA is transcribed; (2) the mRNA is processed and exported to the cytoplasm; (3) Ty1 proteins are translated and processed; (4) virus-like particles (VLPs) then form; (5) the mRNA is reverse transcribed into cDNA; (6) Ty1 IN and Ty1 cDNA are imported into the nucleus; and finally, (7) IN functions to integrate the Ty1 genome into the cellular genome. (B) Ty1 proteins are translated and properly processed in nuclear transport mutants that show transposition defects. gsp1-1, ntf2-1, srp1-55, rsl1 L63A and Δsxm1 cells were transformed with test plasmid and induced in media containing galactose to undergo retrotransposition. As a control, wildtype cells were also transformed with the test plasmid (+) and a URA (pRS316) vector (−) and induced for retrotransposition. As a further control, Δsxm1 cells, which show no retrotransposition defect, were also transformed with the test plasmid and induced for retrotransposition. Immunoblot analysis of Ty1 reverse transcriptase (RT) and Gag proteins was performed using antibodies against each Ty1 protein (see ‘Materials and Methods’ section). As a loading control, an antibody directed against Pgk1 (3-PhosphoGlycerate Kinase 1) was used (Molecular Probes).
Full-length IN and GFP2-IN NLS localize to the nucleus in wildtype cells
One key Ty1 protein that must enter the nucleus is the Ty1 IN protein, which mediates the integration of the Ty1 retrotransposon into the host genome (26,27). Previous studies have defined a C-terminal NLS in IN that is required for both IN nuclear localization and retrotransposition (31,32). To analyze nuclear transport properties of IN independent of intranuclear interactions, we generated a reporter consisting of the last 54 amino acids that contain the previously characterized nuclear targeting domain (Figure 3A). This minimal IN NLS was fused to the C-terminus of two tandem GFPs (GFP2-IN NLS) to create a reporter protein that would be too large for passive diffusion through NPCs to occur. It was critical to demonstrate that this IN reporter showed nuclear localization comparable to a full-length IN reporter. Therefore, we localized a GFP-LacZ-full-length IN (DSM1) (31) in parallel with our reporter plasmid, GFP2-IN NLS, as well as a modified GFP2-IN NLS construct, GFP2-IN NLSmut, which contains alanine substitutions at previously defined key lysine residues within the NLS (bolded residues in Figure 3A) (Figure 3B). As expected, both IN and IN NLS are localized exclusively to the nucleus whereas the GFP2-IN NLSmut is not (31). Hence, for all subsequent experiments the GFP2-IN NLS reporter was used rather than the full-length protein to ensure that any effects observed were the result of changes in nuclear targeting rather than intranuclear interactions.
Figure 3.
Localization and expression of GFP-IN, IN NLS and IN NLSmut in wildtype cells. (A) A schematic of the GFP-LacZ-Ty1 IN (32) and GFP2-IN NLS constructs is shown. Full-length IN is under control of the GAL1 promoter (DSM1); IN NLS and IN NLSmut are under control of the MET25 promoter (pAC1804 and pAC2431, respectively). Full-length IN contains three domains: an N-terminal zinc-finger domain, a central catalytic domain, and a C-terminal domain containing the IN NLS, consisting of basic regions 1 and 2 (BR1 and BR2). A GFP-fused truncated form of IN containing the last 54 amino acids (aa 582–636) (including BR1 and BR2 indicated in bold lettering—595SKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRI631) was used in most experiments and is referred to as GFP2-IN NLS. Previously defined key lysine residues are highlighted in bold. (B) GFP-fused IN and GFP2-IN NLS are targeted to the nucleus while GFP2-IN NLSmut is not. Cells expressing either full-length IN, GFP2-IN NLS, or GFP2-IN NLSmut (596KKR598 → AAA and 628KKR630 → AAA) were analyzed using direct fluorescence microscopy (GFP). Cells were stained with Hoechst to indicate the location of the nucleus and corresponding DIC images are shown.
GFP2-IN NLS is mislocalized to the cytoplasm in mutants of the classical nuclear import pathway
Conditional mutants of Ran (GSP1), as well as its GTPase activating factor (RNA1) and guanine exchange factor (PRP20), display defects in nuclear import of all cargoes that depend on karyopherin receptors (17,48). Therefore, we examined the localization of GFP2-IN NLS in temperature-sensitive gsp1-1, rna1-1 and prp20-1 cells that were shifted to the non-permissive temperature of 37°C for 3 h. Figure 4A shows that the localization of GFP2-IN NLS is nuclear in wildtype cells. In contrast, we find that GFP2-IN NLS is mislocalized to the cytoplasm in gsp1-1, rna1-1 and prp20-1 cells suggesting that IN nuclear localization is dependent on a functional Ran cycle.
Figure 4.
GFP2-IN NLS localization in classical nuclear protein import mutants. (A) GFP2-IN NLS is mislocalized to the cytoplasm in temperature-sensitive mutants of Ran and its regulatory proteins. Wildtype, gsp1-1, rna1-1 and prp20-1 cells expressing GFP2-IN NLS were grown to log phase at the permissive temperature and then shifted to 37°C for 3 h and analyzed by direct fluorescence microscopy. Hoechst dye was used to visualize the location of the nucleus. Corresponding DIC images are shown. (B) GFP2-IN NLS is mislocalized to the cytoplasm in a temperature-sensitive mutant of importin-α and β. Wildtype, srp1-55 (importin-α mutant), or rsll L63A (importin-β mutant), cells expressing either GFP2-IN NLS (top) or a control Nab2-GFP protein (bottom) were shifted to 37°C for 3 h and analyzed by direct fluorescence microscopy. Hoechst dye was used to visualize the location of the nucleus. Corresponding DIC images are shown.
To determine whether GFP2-IN NLS import also depends on the classical NLS receptors, importin-α (SRP1) and importin-β (RSL1), we localized GFP2-IN NLS in temperature-sensitive mutants of SRP1 and RSL1. GFP2-IN NLS was no longer strictly nuclear localized in srp1-55 and rsl1 L63A cells (Figure 4B), suggesting that importin-α/β contributes to proper nuclear localization of IN. As a control, Nab2, a cargo imported by an alternate karyopherin import receptor, Kap104 (54,55), is properly localized to the nucleus in the importin-α/β mutants (Figure 4B). For all studies, immunoblot analysis was performed to ensure expression of the full-length GFP2-IN NLS proteins in each of these mutants (data not shown).
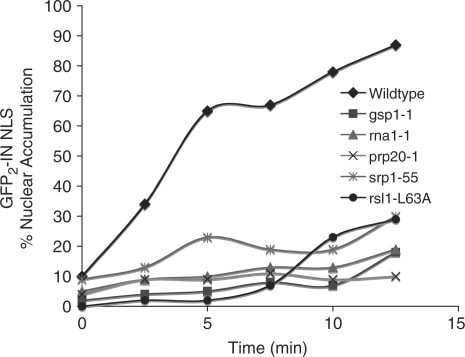
The initial rate of nuclear import of GFP2-IN NLS is dramatically decreased in mutants of the Ran cycle and the NLS receptors
Although our steady-state results reveal dramatic mislocalization of GFP2-IN NLS in gsp1-1, rna1-1 and srp1-55 cells, the effect is not as obvious in prp20-1 and rsl1 L63A cells (Figure 4A). As a more quantitative approach to investigate nuclear import of GFP2-IN NLS in these cells, we used an established kinetic import assay (43) to examine initial import rates. For this assay, expression of GFP2-IN NLS was induced at the permissive temperature (25°C) for each mutant. Cells were then incubated with azide and 2-deoxy-glucose to deplete the cell of energy, which causes a redistribution of nuclear proteins (43). Samples were then washed to remove azide and 2-deoxy-glucose and reinitiate protein import. The initial rate of import was then assessed by determining the number of cells with detectable accumulation of GFP2-IN NLS in the nucleus over a 12.5-min time course. For gsp1-1, rna1-1, prp20-1 and rsl1 L63A cells, initial import rates were measured at the non-permissive temperature of 37°C. For srp1-55 cells, initial import rates were measured at the non-permissive temperature of 18°C. As shown in Figure 5, we observed a dramatic decrease in the initial rate of nuclear import of GFP2-IN NLS in gsp1-1, rna1-1, prp20-1, srp1-55 and rsl1 L63A cells relative to wildtype cells. As a control, we also used Nab2-GFP as a cargo for this assay in both wildtype and srp1-55 cells and saw no difference in Nab2 import (data not shown). These data, taken together with our steady-state localization data (Figure 5), support our hypothesis that nuclear import of GFP2-IN NLS requires a functional Ran cycle as well as the import receptors importin α/β.
Figure 5.
The initial rate of GFP2-IN NLS import is reduced in classical nuclear protein import mutants. The initial rate of nuclear import of GFP2-IN NLS is dramatically reduced in gsp1-1, rna1-1, prp20-1, srp1-55 and rsl1 L63A cells relative to wildtype cells. Wildtype, gsp1-1, rna1-1, prp20-1, srp1-55 and rsll L63A cells containing GFP2-IN NLS were analyzed using a semi-quantitative kinetic import assay as described in the ‘Materials and Methods’ section (43). Import in wildtype cells was analyzed at both 37 and 18°C. Data obtained was identical at both temperatures and results are shown only for the 37°C sample. The percentage of cells with GFP2-IN NLS nuclear accumulation was plotted versus time to obtain an approximation of initial import rates.
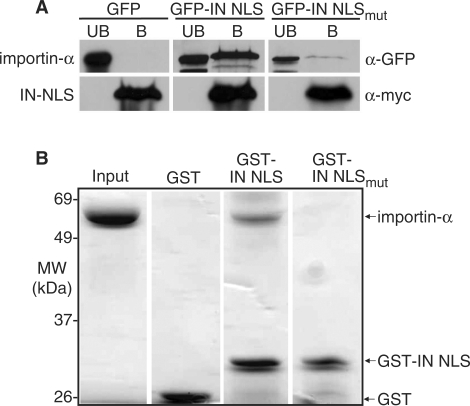
GFP2-IN NLS interacts with importin-α
To determine whether IN physically interacts with the classical NLS receptor importin-α, we tested whether the IN NLS reporter coimmunoprecipitates with importin-α. For this experiment, we expressed myc-tagged importin-α in wildtype cells also expressing either GFP2-IN NLS or, as a control, GFP2-IN NLSmut. We generated cell lysates and then purified importin-α-myc and assayed by immunoblotting for co-purification of GFP2-IN NLS. Results indicate that GFP2-IN NLS co-purifies with importin-α (Figure 6A). As a critical control, we show that GFP2-IN NLSmut does not co-purify with importin-α suggesting binding to importin-α is dependent upon previously characterized lysine residues (31,32) highlighted in Figure 3A. Although these binding data suggest that IN is bound by importin-α in cell lysate, this experiment does not provide evidence that IN NLS is bound directly to the import receptor.
Figure 6.
Ty1 IN interacts with importin-α. (A) IN NLS coimmunoprecipitates with importin-α from whole cell yeast lysate. Yeast cells expressing importin-α-myc were transformed with plasmids expressing GFP (control), GFP2-IN NLS, or GFP2-IN NLSmut. Cell lysates were prepared from induced cultures and epitope-tagged importin-α protein was purified using anti-myc beads as described in the ‘Materials and Methods’ section. GFP proteins present in the bound (B) and unbound (UB) fractions were detected with an anti-GFP antibody. An anti-myc antibody was used to ensure enrichment of importin-α-myc in the bound fraction. (B) Importin-α binds directly to IN NLS in vitro. Purified ΔIBB-importin-α was incubated with GST-IN NLS, or as controls, GST alone or GST-IN NLSmut. GST-fusion proteins and any associated proteins were purified on glutathione sepharose beads as described in the ‘Materials and Methods’ section. Bound fractions were collected and resolved on a 10% SDS-PAGE gel. As a control, 4 μg of purified ΔIBB-importin-α was loaded in the first lane (Input). ΔIBB-importin-α migrates at approximately 70 kDa; GST migrates at approximately 25 kDa; and GST-IN NLS migrates at approximately 35 kDa.
To determine whether IN NLS can bind directly to importin-α, we employed an in vitro binding assay using purified recombinant proteins. For these experiments, we used a truncated version of importin-α (ΔIBB importin-α) lacking the N-terminal 89 amino acid importin-β binding (IBB) domain (45). Previous studies have demonstrated that this truncated importin-α binds to NLS cargo with a similar affinity to importin-α in the context of the importin-α/β heterodimer (35,44,56). Briefly, purified GST-IN NLS, GST-IN NLSmut, or as a control, GST alone, was incubated with purified, recombinant His6-tagged ΔIBB importin-α. Bound fractions were separated on a denaturing polyacrylamide gel, which was then stained with Coommassie blue to visualize the purified proteins (Figure 6B). Results show that IN NLS binds to importin-α. As expected, importin-α does not bind to either the GST or GST-IN NLSmut control protein suggesting that the binding detected between importin-α and IN NLS is mediated by the atypical NLS.
DISCUSSION
This study was designed to elucidate the mechanism by which Ty1 IN gains access to the nuclear DNA in S. cerevisiae. Previous studies have shown that a nuclear targeting signal at the C-terminus of Ty1 IN is both necessary and sufficient for import of IN into the nucleus (31,32). However, this sequence does not match the consensus for a classical bipartite NLS. Typically, bipartite NLSs are characterized by two short stretches of basic amino acids separated by a linker region containing 10–12 residues (33,35). In contrast, IN NLS contains two stretches of basic residues separated by a longer linker consisting of 29 amino acids. Despite this unusually long spacer, IN NLS has been proposed to function as a bipartite NLS (31,32), suggesting IN uses the karyopherin, importin-α, as a means of entering the nucleus; however, it was also possible that this atypical bipartite NLS could mediate import via a different import pathway.
In this study we used in vivo and in vitro techniques to test the hypothesis that IN can exploit the classical cellular protein import machinery to access the nucleus. Our data indicate that IN NLS is able to interact with and utilize the classical import machinery. From our localization studies, our results suggest that proper nuclear localization of IN is dependent upon an intact Ran cycle. Mutations in importin-α also decrease the steady-state nuclear localization of IN but not to the same extent as Ran cycle mutants. Since the Ran cycle is critical not just for classical nuclear import mediated by the importin-α/β heterodimer but also for other karyopherin-mediated transport processes (57), our localization results may indicate that Ty1 could exploit additional Ran-dependent mechanisms for import. There is precedent for the use of multiple karyopherin pathways for nuclear import of critical cellular proteins such as histones (58) and key transcription factors such as TATA-binding protein (59). Therefore, although these localization studies indicate that nuclear import of IN can be mediated by the classical import receptors, IN import could also be achieved through another Ran-dependent mechanism, such as another member of the karyopherin family.
Although the C-terminus of Ty1 IN is not conserved in retroviral integrases like HIV, this analysis of Ty1 IN may have implications for how HIV IN can access the nucleus. Many studies have sought to characterize the import pathway for HIV IN and many of these conclusions conflict not only in the mechanism proposed, but also in the location of the presumed NLS (60–68). It is possible that HIV may have, in fact, evolved multiple mechanisms, to overcome host cell barriers and defenses. As an extension of this study, it is possible to imagine Ty1 IN, much like HIV IN, may have developed multiple routes of nuclear entry to ensure access to the host genome, which would allow for subsequent rounds of replication and the survival of the retroelement.
ACKNOWLEDGEMENTS
We thank D. Garfinkel for the GFP-LacZ plasmids, T. Menees for providing polyclonal Ty1 RT antibody, J. Boeke for providing polyclonal antibody 8B11, and members of the Corbett and Devine laboratories for stimulating discussions. This work was supported by grants from the NIH to AHC and SED. Funding to pay the Open Access publication charges for this article was provided by NSF grant #0749620.
Conflict of interest statement. None declared.
REFERENCES
- 1.Greene WC, Peterlin BM. Charting HIV's remarkable voyage through the cell: Basic science as a passport to the future therapy. Nat. Med. 2002;8:673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 2.Rijck JD, Vandekerckhove L, Christ F, Debyser Z. Lentiviral nuclear import: a complex interplay between virus and host. BioEssays. 2007;29:441–451. doi: 10.1002/bies.20561. [DOI] [PubMed] [Google Scholar]
- 3.Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. J. EMBO. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatziioannou T, Goff SP. Infection of non-dividing cells by Rous sarcoma virus. J. Virol. 2001;78:4902–4906. doi: 10.1128/JVI.75.19.9526-9531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 6.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Suntharalingam M, Wente SR. Peering through the pore. Nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 8.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 10.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 11.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 12.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 13.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 14.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 15.Moore MS. Ran and nuclear transport. J. Biol. Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 16.Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 17.Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J. Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 19.Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- 20.Jans DA, Ackermann MJ, Bischoff JR, Beach DH, Peters R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV40 T-antigen proteins. J. Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart M. Insights into the molecular mechanism of nuclear trafficking using nuclear transport factor 2 (NTF2) Cell Struct. Function. 2006;25:217–225. doi: 10.1247/csf.25.217. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rexach M, Blobel G. Protein import into the nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 24.Quimby BB, Lamtina T, L’Hernault S, Corbett AH. The mechanism of Ran import into the nucleus by NTF2. J. Biol. Chem. 2000;275:28575–28582. doi: 10.1074/jbc.M005055200. [DOI] [PubMed] [Google Scholar]
- 25.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 26.Eichinger DJ, Boeke JD. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 27.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 28.Garfinkel DJ, Boeke JD, Fink GR. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 29.Lauermann V, Boeke JD. Plus-strand strong-stop DNA transfer in yeast Ty retrotransposons. EMBO J. 1997;16:6603–6612. doi: 10.1093/emboj/16.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goetsch L, Byers B. Meiotic cytology of Saccharomycese cerevisiae in protoplast lysates. Mol. Gen. Genet. 1982;187:54–60. doi: 10.1007/BF00384383. [DOI] [PubMed] [Google Scholar]
- 31.Kenna MA, Brachmann CB, Devine SE, Boeke JD. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore SP, Rinckel LA, Garfinkel DJ. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontes MR, Teh T, Toth G, John A, Pavo I, Jans DA, Kobe B. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem. J. 2003;375:339–349. doi: 10.1042/BJ20030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 35.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J. Biol. Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetic. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 38.Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics. 2003;164:867–879. doi: 10.1093/genetics/164.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedures and some application. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karst SM, Rutz ML, Menees TM. The yeast retrotransposons Ty1 and Ty3 require the RNA Lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem. Biophys. Res. Commun. 2000;268:112–117. doi: 10.1006/bbrc.1999.2048. [DOI] [PubMed] [Google Scholar]
- 41.Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for Poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 43.Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fanara P, Hodel MR, Corbett AH, Hodel AE. Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 2000;275:21218–21223. doi: 10.1074/jbc.M002217200. [DOI] [PubMed] [Google Scholar]
- 45.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 46.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin a. Nat. Struct. Biol. 1999;6:301–304. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 47.Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol. Cell. Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 50.Harreman MT, Cohen PE, Hodel MR, Truscott GJ, Corbett AH, Hodel AE. Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J. Biol. Chem. 2003;278:21361–21369. doi: 10.1074/jbc.M301114200. [DOI] [PubMed] [Google Scholar]
- 51.Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J. Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seedorf M, Silver PA. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl Acad. Sci. USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merkulov GV, Lawler JF, Eby Y, Boeke JD. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J. Virol. 2001;75:638–644. doi: 10.1128/JVI.75.2.638-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee DC, Aitchison JD. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA- binding proteins and the role of Ran and RNA. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 55.Aitchison JD, Blobel G, Rout MP. Kap104p: A karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 56.Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- 57.Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2 doi: 10.1186/gb-2001-2-6-reviews3008. Epub Jun 5 2001) Reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 59.Morehouse H, Buratowski RM, Silver PA, Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl Acad. Sci. USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Armon-Omer A, Graessmann A, Loyter A. A synthetic peptide bearing the HIV-1 integrase 161-173 amino acid residues mediates active nuclear import and binding to importin alpha: characterization of a functional nuclear localization signal. J. Mol. Biol. 2004;336:1117–1128. doi: 10.1016/j.jmb.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 61.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl Acad. Sci. USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maertens G, Cherepanov P, Debyser Z, Engelborghs Y, Engelman A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 2004;279:33421–33429. doi: 10.1074/jbc.M404700200. [DOI] [PubMed] [Google Scholar]
- 64.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 65.Depienne C, Mousnier A, Leh H, Le Rouzic E, Dormont D, Benichou S, Dargemont C. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 2001;276:18102–18107. doi: 10.1074/jbc.M009029200. [DOI] [PubMed] [Google Scholar]
- 66.Devroe E, Engelman A, Silver PA. Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 2003;116:4401–4408. doi: 10.1242/jcs.00747. [DOI] [PubMed] [Google Scholar]
- 67.Ao Z, Fowke KR, Cohen EA, Yao X. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology. 2005;2:62. doi: 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hearps AC, Jans DA. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 2006;398:475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Traglia HM, Atkinson NS, Hopper AK. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol. Cell Biol. 1989;9:2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amberg DC, Fleischmann M, Stagljar I, Cole CN, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]