Abstract
Non-integrating gene vectors, which are stably and extrachromosomally maintained in transduced cells would be perfect tools to support long-term expression of therapeutic genes but preserve the genomic integrity of the cellular host. Small extrachromosomal plasmids share some of these ideal characteristics but are primarily based on virus blueprints. These plasmids are dependent on viral trans-acting factors but they can replicate their DNA molecules in synchrony with the chromosome of the cellular host and segregate to daughter cells in an autonomous fashion. On the basis of the concept of the latent origin of DNA replication of Epstein-Barr virus, oriP, we devised novel derivatives, which exclusively rely on an artificial replication factor for both nuclear retention and replication of plasmid DNA. In addition, an allosteric switch regulates the fate of the plasmid molecules, which are rapidly lost upon addition of doxycycline. Conditional maintenance of these novel plasmid vectors allows the reversible transfer of genetic information into target cells for the first time.
INTRODUCTION
Vectors employed for gene and immune therapy of various human diseases are based on recombinant nucleic acids. Most gene vectors include viral or metazoan genetic components such as promoters and enhancers, which provide the necessary cis-acting elements for expression of one or more genes of therapeutic interest. In addition, certain virus-based gene vectors also carry essential elements involved in packaging of genetic information into virus-like structures (1). In the recipient cell, gene vectors can be transiently present or maintained for a long period of time. In these cases, the genetic information is either rapidly lost through spontaneous degradation by cellular nucleases or maintained by integrating it into the chromosome of the recipient cell. Because a prolonged effect of the therapeutic gene product is often preferred, gene vectors are commonly employed, which promote their chromosomal integration. In particular, all retroviruses and adeno-associated viruses integrate randomly or at numerous potential predilections sites as proviruses and establish a persistent state (1). Similarly, non-viral gene vectors can also integrate chromosomally and in a random fashion but recent reports indicate targeted gene delivery via induced double-strand break in predetermined gene loci (2–5). Unfortunately, all integrating vectors can act as insertional mutagens, which might be destructive to the transduced cell. For example, oncogenic T-cell transformation was observed in a recent clinical trial with a retroviral vector (6). This problem can be avoided with gene vectors, which are stably maintained as extrachromosomal units in the recipient cell.
All extrachromosomal gene vectors are derived from recombinant plasmids, which carry autonomous replicons (7–9). Replicons are genetic units, which mediate initiation of DNA replication in the context of the cellular genome or on plasmid DNA. In metazoan cells, two extrachromosomal vector systems are well known, which replicate in synchrony with the host chromosome: oriP-based plasmids and a non-viral vectors system called pEPI (9,10). The plasmid replicon of Epstein-Barr virus (EBV), oriP, is the latent origin of DNA replication of this human herpes virus. Plasmids containing this element replicate akin to the genome of its host cell and are maintained extrachromosomally. Consequently, this plasmid replicon has been exploited as recombinant gene vector (7). oriP supports efficient DNA replication in cells selected to retain it at several copies when the viral gene product EBNA1 (Figure 1A) is provided. Recombinant plasmids containing oriP replicate once per cell cycle during S-phase and are efficiently segregated to daughter cells (11). Only two components, oriP in cis and EBNA1 in trans, are required, the cell contributes everything else.
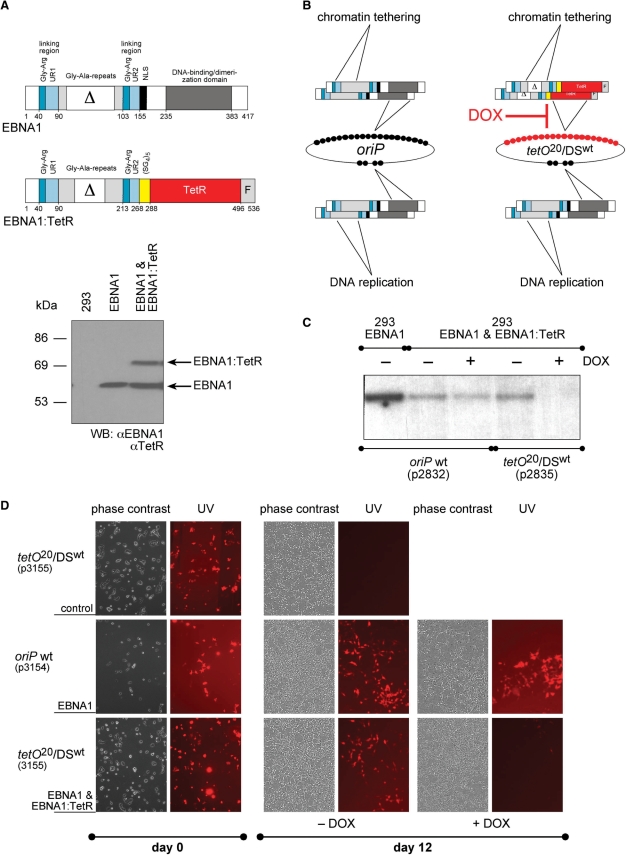
Figure 1.
Chimeric EBNA1:TetR confers conditional plasmid maintenance. (A) Design of the trans-acting factor EBNA1:TetR. Wild-type EBNA1 with designated functional domains and their amino acid residues is shown in comparison with the chimeric protein EBNA1:TetR composed of EBNA1's amino-terminal half joined via an artificial linking domain [(SG4)5] to the complete coding region of the tetR gene followed by three repeats of the motif F, a minimal transcriptional activation domain (24). Most of the central glycine–alanin repeats (Gly-Ala-repeats) and about half of them are deleted in EBNA1 and EBNA1:TetR, respectively, as indicated (Δ) without functional consequences (48). Western blot analysis of the parental HEK293 cell line and its derivatives expressing EBNA1 (center lane) and EBNA1 + EBNA1:TetR (right lane) was performed with antibodies directed against TetR and EBNA1. (B) Schematic overview of the oriP replicon and the hybrid replicon plasmid tetO20/DSwt. The two cis-acting elements of oriP, FR and DS, carry 20 and 4 EBNA1-binding sites as indicated by the number of black circles. The hybrid replicon plasmid tetO20/DSwt carries an array of 20 tetO-binding sites (red circles) to which EBNA1:TetR binds with high affinity. EBNA1:TetR binds as a dimer-like EBNA1, thus these trans-acting factors are shown as doublets. (C) The plasmids p2832 (oriP wt) and p2835 (tetO20/DSwt) were separately transfected into HEK293 cells expressing EBNA1 and EBNA1:TetR. The cells were selected with puromycin with or without DOX (2.0 µg/ml) for 8 days. Low-molecular weight DNA was isolated, linearized and digested with DpnI to cleave unreplicated plasmid DNAs. Full-length DpnI-resistant plasmid DNA was detected by Southern blot hybridization with a radioactive probe specific for the prokaryotic plasmid backbone. DOX had almost no effect on p2832. No plasmid signal was detected when the cells had been transfected with p2835 and kept under selection in the presence of DOX. The left lane shows p2832 (oriP wt) in the HEK293 cell line expressing EBNA1, only. (D) HEK293 cells, which express no viral protein, EBNA1 or both EBNA1 + EBNA1:TetR were transfected with two different plasmids p3154 (oriP wt) or p3155 (tetO20/DSwt) encoding mRFP and puromycin resistance and selected for 4 days (day 0). The selective pressure was removed and the cells were kept with or without DOX (2.0 µg/ml) for a period of 12 days. No or only very few cells appeared to express mRFP in parental HEK293 cells transfected with the two plasmids at that time point (top row and data not shown) indicating that they were rapidly lost in the absence of EBNA1 and EBNA1:TetR. In EBNA1 expressing HEK293 cells transfected with p3154 (oriP wt), a fraction of cells expressed mRFP in the presence or absence of DOX (middle row) after 12 days. HEK293 cells expressing both trans-factors and transfected with p3155 (tetO20/DSwt) were mRFP positive, only, when the cells had been cultivated in the absence of DOX (bottom row).
oriP consists of two essential cis-acting elements, the family of repeats (FR) and a structure called dyad symmetry element (DS) to both of which EBNA1 binds sequence specifically (Figure 1B). EBNA1 recruits components of the cellular replication machinery to DS where initiation of DNA replication takes place (11). The FR element is an array of 20 EBNA1-binding motifs dedicated to retain oriP in the nucleus because EBNA1 tethers oriP to cellular chromatin (12,13) (Figure 1B). Nuclear retention of oriP plasmids is considered mandatory for long-term plasmid maintenance and might contribute to partitioning in each cell cycle. Thus, DS and FR are functionally distinct elements dedicated to DNA replication and replicon nuclear retention, respectively. EBNA1 is characterized by its modular design. Its carboxy-terminal half mediates dimerization and DNA binding, its amino-terminal half, in particular, the two linking regions (Figure 1A) associate with cell chromosomes (13) and is essential for plasmid maintenance (11).
EBNA1's characteristics triggered an approach in which its carboxy-terminal dimerization and DNA-binding domain was fused to the cellular histone H1 or HMGA1a proteins conferring chromatin binding and association to mitotic chromosomes (14). These chimeric proteins were functional with respect to plasmid replication and nuclear retention presumably because EBNA1 contains AT-hook domains in its linking regions (15) similar to HMGA family members such as HMGA1a (16,17 and references therein) and histone H1 targets the same structural DNA motif as AT-hook domains. HMGA1a functionally interacts with the mammalian origin of DNA replication, ORC, (18) similar to EBNA1 (19). Targeting HMGA1a to specific sites on plasmid DNA generates artificial origins of DNA replication, which mediate DNA replication (18). We extended these initial observations and designed novel synthetic plasmids, which replicate and are retained in the nucleus of transduced cells upon selection similar to oriP but can be regulated at will at the level of its DNA.
MATERIAL AND METHODS
Cell lines
The HEK293 cell line is derived from primary human embryonal kidney cells transformed with human adenovirus type 5 DNA (20). 293-D is a clonal derivative established in our laboratory. Akata 27 is an EBV-negative derivative of Akata cells, a human Burkitt lymphoma cell line (21). Rat-1 cells are rat embryo fibroblasts (22). Cell line 143/tk− is a human cell line established from an osteosarcoma (23). A clonal HEK293 EBNA1 cell line was established by stable transfection of 293-D cells with two plasmids, p2816 and p2727, which express EBNA1 and neomycin phosphotransferase, respectively, and selection with 200 μg/ml G418. A HEK293 cell clone expressing both EBNA1 and EBNA1:TetR was established with 293-D cells. First, they were stably transfected with two plasmids, p2729.1 and p2727, which express EBNA1:TetR and neomycin phosphotransferase, respectively, and selected with 200 μg/ml G418. Second, the resulting EBNA1:TetR-positive cell clone was stably transfected with p2774 and selected with 100 μg/ml hygromycin. p2774 expresses both EBNA1 and hygromycin phosphotransferase and was a kind gift from B. Sugden. A HEK293 cell clone which expresses both wild-type EBNA1 and scTetR:HMGA1a was established on the basis of HEK293 EBNA1 cells, which were cotransfected with p3265.12 together with p3223.9 encoding scTetR:HMGA1a and puromycin resistance, respectively, and selected with 250 ng/ml puromycin. All clonal HEK293 derivatives were analyzed for the expression of EBNA1, EBNA1:TetR or scTetR:HMGA1a proteins by western blot immunostaining with the monoclonal antibody 1H4 directed against EBNA1 or a polyclonal rabbit antisera generated against TetR(B) protein. Akata cells and all HEK293-derived cell lines were maintained in RPMI1640 medium with 10% fetal calf serum, 100 units of penicillin per milliliter and 100 µg of streptomycin per milliliter at 37°C in a 5% CO2 atmosphere. Rat-1 and 143/tk− cells were maintained in DMEM with all supplements.
Plasmids
Plasmids carrying oriP, hybrid oriP replicons, or synthetic oriP-like replicons are summarized in Table 1 along with their key features. All plasmid DNAs were prepared with Jetstar 2.0 columns (Genomed, Löhne, Germany). Genetic modification of FR and DS elements were performed by cloning synthetic oligonucleotides at the appropriate locations by conventional techniques. DNA sequencing confirmed the sequence composition of the critical regions. The plasmids encoding EBNA1, EBNA1:TetR and scTetR:HMGA1a express their trans-genes from the immediate early human cytomegalovirus promoter. The expression plasmid encoding EBNA1:TetR, p2729.1, is based on pWHE120 (B + sB) encoding sctTA2i (24) in which the first tetR allele was replaced by the amino-terminal half of EBNA1. The expression plasmid encoding scTetR:HMGA1a, p3265.12, is based on pWHE120 (sB + B) encoding sctTA2 (24), in which the triple-F domain was replaced by the entire coding sequence of human HMGA1a (18). All plasmid DNA sequences are available upon request.
Table 1.
List of oriP- and oriP-like plasmids
| Plasmidsa | Retention element | Replication element | Genotypic marker | Phenotypic marker |
|---|---|---|---|---|
| p2832 oriP wt (12.2) | FR | DS | Puromycin | None |
| p2835 tetO20/FRwt (12.0) | 20xtetO | DS | Puromycin | None |
| p3154 oriP wt (12.9) | FR | DS | Puromycin | mRFP |
| p3155 tetO20/FRwt (12.7) | 20xtetO | DS | Puromycin | mRFP |
| p3230 oriP wt (8.8) | FR | DS | Hygromycin | GFPc |
| p3231 tetO20/FRwt (8.7) | 20xtetO | DS | Hygromycin | GFPc |
| p3293 tetO40/FRwt (8.9) | 40xtetO | DS | Hygromycin | GFPc |
| p3311 tetO20/tetO4 (8.7) | 20xtetO | 4xtetO | Hygromycin | GFPc |
| p3315 FRwt/tetO4 (8.8) | FR | 4xtetO | Hygromycin | GFPc |
| p3323 (pHEBNA)b (9.0) | FR | DS | Hygromycin | None |
| p3333 tetO20/tetO4 (11.1) | 20xtetO | 4xtetO | Hygromycin | GFPc |
| p3421 tetO20/tetO4 (8.7) | 20xtetO | 4xtetO opt.d | Hygromycin | GFPc |
aNumbers in parentheses denote the size of the plasmids in kilo base pairs.
bPublished in Yates et al. (49).
cDestabilized d2GFP.
dOptimized (opt.) compared to p3311. p3421 and p3311 differ in the distance of the two tetO pairs, which make up the DS-like element without apparent functional difference.
DNA transfections
DNA transfections into HEK293 cells and derivatives were performed with Polyfect (Qiagen, Hilden, Germany). Cells were seeded at ∼50% confluence into six-well cluster plates 1 day prior to transfection. For transfection, cells were placed in OptiMEM medium (Invitrogen, Karlsruhe, Germany) for 1 h and incubated with gene vector plasmid DNA embedded in lipid micelles (1 μg DNA, 2.5 μl Polyfect per well, 100 μl of RPMI1640 without supplements; preincubation for 15 min at room temparture) overnight. After 1–2 days, the cells were transferred into a 13-cm cell culture dish and cultivated in the presence of 80 μg/ml hygromycin, 250 ng/ml puromycin or 200 μg/ml G418.
Plasmid rescue in Escherichia coli
To determine the number of plasmid copies in transfected HEK293 cells low-molecular weight DNA was isolated (25) and digested with DpnI to cleave unreplicated plasmid DNAs, which retain the dam methylation pattern that the plasmids had acquired during propagation in E. coli. DNA (600 ng) was introduced into the E. coli DH10B strain by electroporation (1800 V, 25 µF, 100 Ω, 1 mm gap cuvettes, Genepulser (Biorad Laboratories Munich, Germany). Transformants were selected on agar plates containing 100 µg/ml ampicillin. Several colonies were picked and the DNA composition of the rescued plasmid was confirmed with restriction enzyme analysis.
Chromatin immunoprecipitation assay and real-time PCR analysis
For chromatin immunoprecipitation experiments, nuclei were prepared for each immunoprecipitation as described (26). For each sample 1 × 107 cells were harvested, washed with PBS and resuspended in 250 µl hypotonic buffer A [10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT and protease inhibitor mix Complete© (Roche Diagnostics, Mannheim, Germany)]. Cells were lysed by adding 0.04% Triton X-100 and incubated for 10 min on ice. Samples were centrifuged (4 min, 1300g, 4°C) to separate soluble cytosolic and nucleosolic proteins from chromatin. Nuclei were washed at a concentration of 1 × 108 nuclei/ml in ice-cold buffer A supplemented with 200 mM NaCl. After centrifugation (1300g, 5 min, 4°C) nuclei were carefully resuspended in 1 ml buffer A. Prewarmed buffer A (9 ml) supplemented with 1.1% formaldehyde was added and the nuclei crosslinked for 10 min at 37°C. Fixed nuclei were washed twice with PBS/0.5% NP40, resolved in 2.7 ml LSB (10 mM Hepes, pH7.9, 10 mM KCl and 1.5 mM MgCl2) and lysed by adding 300 µl 20% Sarkosyl. The chromatin was transferred onto a 40 ml sucrose cushion (LSB + 100 mM sucrose) and centrifuged (10 min, 4°C, 4000 g). The supernatant was removed and chromatin was resuspended in 2 ml TE and sonicated (Branson sonifier 250-D, 35% amplitude, 2 min in 1 s intervals). For partial DNA digestion, 2 mM CaCl2 and 8 units MNase (Roche) were added and incubated for 10 min at 37°C. The reaction was stopped by adding 5 mM EGTA. For immunoprecipitation, the extract was adjusted with 1/10 volume of 11xNET (50 mM Tris, 150 mM NaCl, 0.5 M EDTA and 0.5% NP40). Ten microliter of polyclonal rabbit antibody directed against TetR(B) protein or 50 µl supernatant of the monoclonal EBNA1-specific antibody 1H4 were added. The immunoprecipitation and purification of coprecipitated DNA was performed as described (19). Real-time PCR analysis was performed according to the manufacturer's instructions using the same parameters and the primer pair SC4 as described (19). Detailed protocols of the chromatin immunoprecipitation experiments are available at http://haema145.gsf.de/
Bromodeoxyuridine density transfer experiments
HEK293 cells stably transfected with either p3230 (oriP wt) or p3421 (tetO20/tetO4) were incubated in cell culture medium containing 5-bromodeoxyuridine (10 µM, BrdU) and deoxycytidine (100 μM) (27) for 16 h to label newly synthesized DNA. Total DNA was isolated from 1 × 107 cells, digested with BamHI and separated on CsCl equilibrium density gradients adjusted to the initial refractive index of 1.403. The gradients were spun in a Beckman SW41 rotor at 38 000 rpm for 48 h. To measure the incorporation of BrdU, 28 fractions were analyzed for the distribution of total cellular and plasmid DNAs. A quantity of 250 µl fractions was collected and their refractive index was determined. Samples were diluted, precipitated and further analyzed by quantitative PCR with the SC4 primer pair. Total cellular DNA was determined photometrically.
RESULTS
The prokaryotic Tet repressor fused to EBNA1 confers plasmid maintenance
We wanted to assess whether EBNA1's DNA-binding domain can be replaced with a heterologous but functionally similar domain and constructed a chimeric DNA-binding protein to mimic EBNA1's function to provide plasmid maintenance. As shown in Figure 1A and B, the entire open reading frame of the prokaryotic DNA-binding protein TetR(B) (28) replaced the EBNA1 DNA-binding domain. The tetR gene product forms homodimers similar to EBNA1 and binds to its cognate DNA motif tetO with very high affinity and specificity (29). We established a derivative of HEK293 cells, which stably expressed EBNA1 and the EBNA1:TetR chimera (Figure 1A). In order to allow binding of EBNA1:TetR to oriP, we replaced the 20 EBNA1-binding sites within FR with an identical number of tetO sites to which TetR binds (Figure 1B). This hybrid replicon plasmid tetO20/DSwt p2835 (Table 1, Figure 1C) also carries a selectable maker gene for puromycin resistance. The parental plasmid p2832 is identical to p2835 except that it contains wild-type (wt) oriP.
The plasmids p2832 (oriP wt) and p2835 (tetO20/DSwt) were separately transfected into HEK293 cells, which coexpressed both EBNA1 + EBNA1:TetR. The transfected cells were cultivated in the presence of selective concentrations of puromycin with (2 μg/ml) or without doxycycline (DOX; Sigma Aldrich, Deisenhofen, Germany) for a few days until the cells had reached confluence. As a reference, the wild-type oriP plasmid was also introduced into HEK293 cells, which expressed EBNA1, only, and selected with puromycin in the absence of DOX. Low-molecular weight DNA was isolated, linearized and cleaved with DpnI to fragment non-replicated, input plasmid DNA, which retained the dam methylation pattern that it had acquired during propagation in E. coli. As shown in Figure 1C, full-length DpnI-resistant plasmid DNA was detected by Southern blot analysis. Whereas DOX had almost no effect on oriP in HEK293 cells coexpressing EBNA1 and EBNA1:TetR, no signal was detected with p2835 (tetO20/DSwt) in the presence of DOX suggesting that DOX interfered with plasmid maintenance in this short-term experiment. In HEK293 cells, which express EBNA1, only, the copy number of the wild-type oriP plasmid was considerable higher in contrast cells, which coexpress EBNA1 and EBNA1:TetR (Figure 1C). This observation will be addressed below.
Next, we used two different plasmids which encode monomeric red fluorescence protein (mRFP) (30) to confirm our initial findings. p3154 carries wild-type oriP, whereas p3155 contains 20 tetO sites (tetO20/DSwt) (Figure 1C, Table 1). Three cell lines (parental HEK293 and derivatives expressing EBNA1 or both EBNA1 + EBNA1:TetR) were transfected and selected with puromycin for 2–4 days, only, until most of the remaining adherent cells expressed mRFP (day 0, Figure 1D). The selective pressure was removed and the cells were kept in media with or without DOX for 12 days. No or only very few cells appeared to express mRFP in parental HEK293 cells transfected with p3154 or p3155 at that period (Figure 1D top row and data not shown) indicating that both plasmids were rapidly lost in the absence of EBNA1 and EBNA1:TetR. In HEK293 EBNA1 cells transfected with p3154 (oriP wt), a fraction of cells still expressed mRFP in the presence or absence of doxycycline (+/− DOX in Figure 1D, middle row) after 12 days. HEK293 cells expressing both trans-factors (EBNA1 and EBNA1:TetR) and transfected with the hybrid replicon plasmid p3155 were mRFP positive, only, when the cells had been cultivated in the absence of DOX; addition of this drug led to loss of mRFP expression (Figure 1D, bottom row). This result confirmed that maintenance of p3155 is strictly dependent on EBNA1:TetR, which does not bind to tetO motifs in the presence of DOX (data not shown). As a consequence, hybrid oriP replicons cannot be maintained in the presence of the drug.
Long-term conditional maintenance of hybrid oriP plasmids
The amino-terminal part in the EBNA1:TetR chimera is expected to associate to chromatin and bind to ORC, which is essential for plasmid retention and DNA replication, respectively. Because oriP plasmids replicate and are stably maintained in the presence of HMGA1a:EBNA1 (14), we argued that the chimeric protein consisting of the coding sequence of HMGA1a fused to TetR (scTetR:HMGA1a, Figure 2A) (18) might be functional as well in retaining plasmid DNA with multiple tetO sites. It was also apparent that the plasmid copy number of both wild-type oriP and the 20xtetO plasmid p2835 was considerably lower in cells which coexpressed EBNA1 and EBNA1:TetR (Figure 1C) presumably because the linking region of EBNA1 and EBNA1:TetR (Figure 1A) caused heterotypic aggregations and functional interference (31). Thus, it seemed plausible that the fusion protein scTetR:HMGA1a would circumvent this problem. We made use of a dimeric single-chain (sc) tetR gene, which binds to tetO motifs as a single molecule in contrast EBNA1:TetR, which binds as homodimers (24).
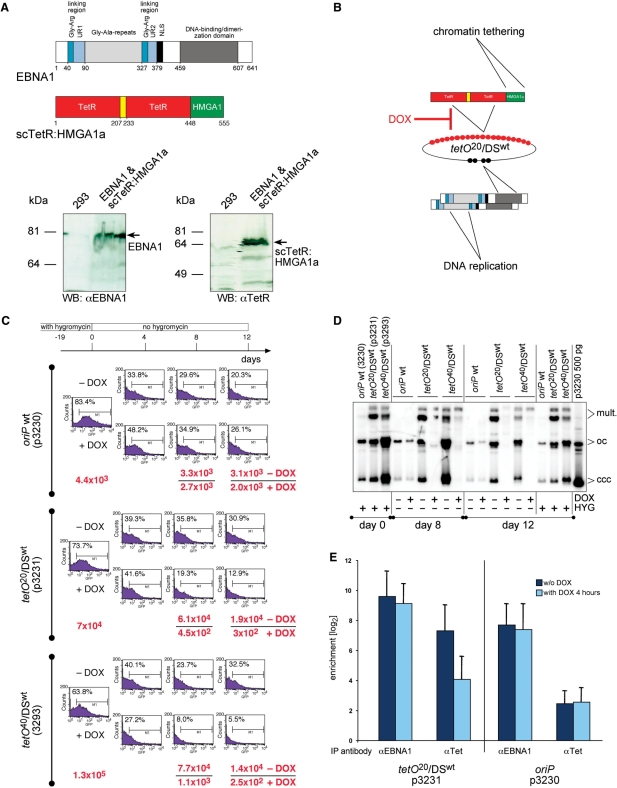
Figure 2.
scTetR:HMGA1a confers conditional plasmid maintenance. (A) The trans-acting factor scTetR:HMGA1a consists of a single-chain (sc) dimer of two tetR genes joined via an artificial linking domain (yellow) which was fused to the HMGA1a coding sequence. Western blot analysis of the parental HEK293 cells and its derivative expressing EBNA1 + scTetR:HMGA1a with TetR- and EBNA1-specific antibodies. (B) scTetR:HMGA1a and EBNA1 mediate chromatin tethering and DNA replication, respectively. (C) DOX downregulates vector-encoded GFP expression and causes induced loss of plasmid DNA. The oriP wt (p3230) plasmid and two hybrid replicon plasmids tetO20/DSwt (p3231) and tetO40/DSwt (p3293) were transfected into EBNA1 and scTetR:HMGA1a expressing HEK293 cells, which were then selected with hygromycin for 19 days. At that time point (day 0) selection was removed and the cells were further cultivated with or without DOX (2.5 g/ml) for 12 days. On days 0, 4, 8 and 12, cells were analyzed by FACS for the expression of plasmid-encoded GFP. Plasmid rescue experiments were performed on days 0, 8, and 12. Low-molecular weight DNAs were prepared and cleaved with DpnI and 600 ng of each DNA sample was electroporated into E. coli DH10B. The absolute numbers of ampicillin-resistant colonies were calculated and shown in red as indicated (−/+DOX). (D) Hybrid replicon plasmids are maintained as plasmids in the absence but precipitously lost in the presence of DOX. As in C, p3230 (oriP wt) and two plasmids p3231 (tetO20/DSwt) and p3293 (tetO40/DSwt) were transfected and selected. Low-molecular weight DNA was prepared on days 0, 8, and 12. DNA was also prepared from the same cells, which had been kept under continuous hygromycin selection (HYG). The DNAs were digested with DpnI to multiply cleave unreplicated DNAs and PmlI to cut cellular DNA, only. Southern blot hybridization were performed as in Figure 1C. The rightmost lane indicates the migration pattern of 500 pg E. coli-derived p3230 plasmid DNA indicating monomeric supercoiled, covalently closed and circular (ccc), monomeric open circles (oc) and multimeric (mult.) forms of plasmid DNA. The ccc and oc form DNAs indicate the presence of extrachromosomal plasmids for a period of 31 days throughout the entire experiment. Upon addition of DOX, the hybrid replicon plasmids were precipitously lost. In the absence of DOX, they were maintained at higher copy numbers than wild-type oriP. (E) ChIP experiments indicate DOX-induced loss of DNA binding of scTetR:HMGA1a but not EBNA1. From formaldehyde crosslinked chromatin of stably transfected cells as in C, EBNA1 and scTetR:HMGA1a were immunoprecipitated and the precipitates were analyzed for coprecipitated plasmid DNA molecules by quantitative real-time PCR. The histogram shows the results for p3231 (tetO20/DSwt) and p3230 (oriP wt) without or with DOX for 4 h. The columns indicate the difference between PCR data (mean values and standard deviations of three experiments) obtained with specific antibodies versus isotype control and preimmune serum on a logarithmic y-axis.
The scTetR:HMGA1a protein was stably expressed in EBNA1-positive HEK293 cells (Figure 2A), which were transfected with plasmids carrying oriP wt (p3230), or two different hybrid replicon plasmids with 20 (tetO20/DSwt; p3231) (Figure 2B) or 40 (tetO40/DSwt, p3293) copies of the tetO motif (Table 1). All three plasmids carry a destabilized GFP gene (32) as a short-lived phenotypic marker for simple detection by fluorescent-activate cell sorting (FACS). The transfected cells were selected with hygromycin for 19 days. At that time point (day 0) selection was removed and the cells were further cultivated with or without DOX for 12 days. GFP expression was determined by FACS on days 0, 4, 8 and 12. Low-molecular weight DNAs were isolated at three time points (day 0, 8 and 12) and analyzed by Southern blot hybridization. DNAs were also electroporated in E. coli in plasmid rescue experiments to quantify the number of plasmid DNA molecules by colony formation.
On day 0, the majority of the cells expressed GFP as expected (Figure 2C, FACS data), and bonafide plasmids could be detected and quantified in plasmid rescue experiments (Figure 2C, colony numbers) and by Southern blot hybridization (Figure 2D). It was immediately apparent that p3231 and p3293 plasmids (tetO20/DSwt and tetO40/DSwt) were present at higher copy numbers than p3230 (oriP wt) (Figure 2D), although the cells did not express higher amounts of GFP (Figure 2C). Upon removal of selective pressure, GFP expression gradually diminished in all samples and plasmid DNAs were gradually lost (Figure 2C) as reported previously for oriP plasmids (33). Very much in contrast, the hybrid replicon plasmids p3231 (tetO20/DSwt) and p3293 (tetO40/DSwt) were lost precipitously in the presence of DOX (Figure 2D). Chromatin immunoprecipitation experiments indicated that DOX induced the loss of DNA binding of scTetR:HMGA1a (but not EBNA1) within 4 h (data not shown, Figure 2E). When scTetR:HMGA1a rapidly dissociates from tetO-binding sites the plasmid DNA molecules presumably become instable and are lost during cell division or degraded by nucleases. Relative signal strengths in Southern blot analysis (Figure 2D and data not shown) and colony numbers (Figure 2C) indicated that DOX-induced plasmid loss was in the order of about factor 50–100 per week. Reduction of GFP expression appeared not as dramatic but a substantial loss of GFP expression could be documented (Figure 2C). DOX had a neglectable effect on p3230 (oriP wt). This and the two hybrid replicon plasmids were stably maintained for 31 days under hygromycin selection (Figure 2D and data not shown).
scTetR:HMGA1A confers DNA replication
We have previously shown that scTetR:HMGA1a recruits ORC to DNA and confers plasmid DNA replication similar to HMGA1a:EBNA1 (18). Toward this end we had constructed hybrid oriP-like plasmid in which the four EBNA1-binding sites that make up the DS element of oriP were precisely replaced by four tetO sites such as in p3315 (FRwt/tetO4, Figure 3A, left panel, Table 1). In this plasmid (and its derivatives, Table 1) all other flanking sequences that make the DS element remained untouched including the three TRF2-binding sites, which can influence efficient initiation of DNA synthesis in oriP (34). p3315 (FRwt/tetO4) and derivatives with tetO4 multimers (18) replicated faithfully in cells coexpressing both EBNA1 and scTetR:HMGA1a. p3315 plasmid DNA was only gradually lost in the presence of DOX over time in contrast to oriP-like plasmid such as tetO20/DSwt or tetO40/DSwt (compare Figure 2D and Figure 3B, left and middle panels) indicating that plasmid retention is more critical than DNA replication for the stable maintenance of extrachromosomal DNA in proliferating cells.
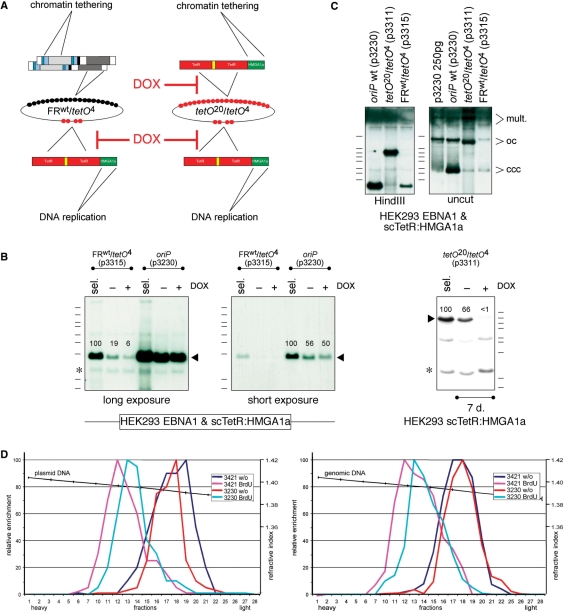
Figure 3.
scTetR:HMGA1a mediates both chromatin tethering and DNA replication. (A) Schematic overview of the hybrid FRwt/tetO4 and synthetic (tetO20/tetO4) replicon plasmids. (B) p3230 (oriP wt), p3315 (FRwt/tetO4) and p3311 (tetO20/tetO4) were stably transfected in HEK293 cells expressing EBNA1 + scTetR:HMGA1a; DNAs were prepared and digested with PmlI and HindIII. In the absence of hygromycin selection, addition of doxycycline (+DOX) caused a modest reduction in the copy number of p3315, but a dramatic loss of p3311; the drug did not affect p3230 copy numbers as indicated by relative signal strengths in percent. Asterisks (*) indicate background signals. (C) p3230 (oriP wt), p3315 (FRwt/tetO4) and p3311 (tetO20/tetO4) were stably introduced (sel.) as described in (B). Low-molecular weight DNAs were prepared, digested with PmlI and HindIII to cleave both plasmid and cellular DNA (left) or PmlI (right) to cleave cellular DNA, only. Southern blot hybridization was performed as in Figure 1C. All plasmids replicated and were maintained extrachromosomally as indicated by signals characteristic of covalently closed circles (ccc) and open circles (oc) DNA molecules together with multimeric forms (mult.). (D) Synthetic, oriP-like plasmids replicate once per cell cycle in Meselson–Stahl density transfer experiments. scTetR:HMGA1a-expressing HEK293 cells were stably transfected with p3230 (oriP wt) or p3421 (tetO20/tetO4) and newly synthesized DNA was labeled with 5-bromodeoxyuridine and separated on CsCl density equilibrium gradients. All DNAs shifted from light, unlabeled DNA (1.70 g/cm3) to heavylight, hemisubstituted DNA (1.73 g/cm3). Plasmid DNA in the gradient fractions (left panel) was quantified by real-time PCR in comparison with chromosomal DNA (right panel). The data indicate that wild-type oriP and synthetic oriP-like plasmids replicated in synchrony with cellular DNA.
Our experiments shown in Figure 2 indicated that scTetR:HMGA1a can replace EBNA1 in order to efficiently retain extrachromosomal plasmid molecules. Our previous data (18) and experiments shown in Figure 3B proved that scTetR:HMGA1a can functionally confer DNA replication of extrachromosomal plasmid DNA. In a combinatorial approach, we generated synthetic plasmids, which lack all EBNA1-binding sites but harbor tetO motifs arranged in two clusters. tetO20/tetO4 (p3311) plasmid DNA (Figure 3A, right panel) was introduced into EBNA1 negative, scTetR:HMGA1a-expressing HEK293 cells, which were selected with hygromycin for 2–3 weeks. p3311 plasmid DNA was maintained extrachromosomally as an artificial replicon comparable to oriP wt and p3315 (FRwt/tetO4) (Figure 3C). Plasmid DNA of p3311 became almost undetectable when, in the absence of hygromycin selection, DOX was added for 7 days (Figure 3B, right panel) whereas p3230 copy numbers were not affected (Figure 3B, middle panel). As expected, synthetic, tetO20/tetO4 oriP-like plasmids replicated once per cell cycle in synchrony with cellular DNA identical to oriP wt plasmid DNA in Meselson–Stahl density transfer experiments (Figure 3D).
Conditional plasmid maintenance in cells of different species
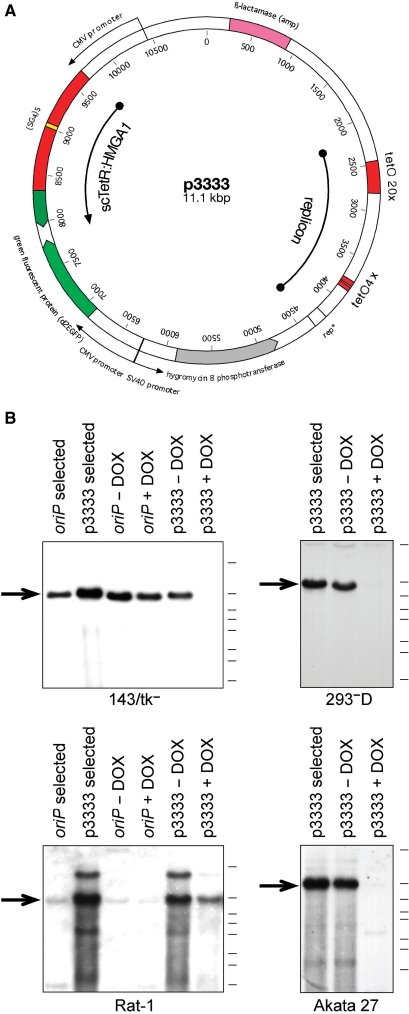
We constructed a prototypic all-in-one synthetic plasmid p3333, which encompasses the two clusters of tetO sites (tetO20/tetO4) and encodes scTetR:HMGA1a (Figure 4A). p3333 also carries a selectable marker gene, which confers resistance against hygromycin in transduced cells. p3333 was stably introduced into three different human cell lines including HEK293 cells, fibroblasts (143/tk−) and B cells (Akata 27, an EBV-negative derivative of Akata cells). Rat-1 cells, a murine fibroblast cell line, were also included, in which small and middle-sized oriP plasmids cannot be stably maintained as indicated in Figure 4B. The transduced cells were selected with hygromycin for 2–3 weeks, when the selective pressure was alleviated. In all four cell lines and in the absence of selection (−DOX), p3333 was maintained at stable copy numbers; they were dramatically reduced or became undetectable upon addition of doxycycline (+DOX) within 1 week (Figure 4B). In case of Rat-1 cells we cannot exclude the possibility that part of the detected signals might stem from tandemly integrated p3333 DNA. Previously, similar experiments have been overinterpreted but it is apparent that the signal strength of p3333 DNA is considerably diminished in the presence of DOX, which can only result from the loss of non-integrated, extrachromomal plasmid molecules.
Figure 4.
Stable maintenance and induced loss of an all-in-one synthetic oriP-like plasmid. (A) Genetic map of p3333, the cis-acting replicon and the trans-acting factor scTetR:HMGA1a are indicated together with the marker genes encoding destabilized GFP and hygromycin B phosphotransferase. (B) The vector plasmid p3333 replicates and is maintained extrachromosomally in four different cell lines in a DOX-dependent manner. As expected the oriP plasmid pHEBNA (49) (p3323, Table 1) does not stably replicate in Rat-1 murine cells.
DISCUSSION
For the first time we have established extrachromosomal gene vector plasmids, whose maintenance can be controlled in cis by a small-molecular weight compound. The independent and separable functions of the oriP elements DS and FR individually dedicated to DNA replication and nuclear replicon retention (reviewed in 11,19,35,36) indicate that EBNA1 plays a dual but distinct role when it binds to either DS or FR. Our combinatorial approach support that view. Synthetic, oriP-like tetO20/tetO4 plasmids such as p3311 and p3333 replicated and were maintained extrachromosomally long term. DOX, which abrogated binding of EBNA1:TetR or scTetR:HMGA1a to tetO motifs (Figure 2E and data not shown), induced rapid loss of plasmid DNAs presumably by nuclease degradation. In contrast, plasmids in which only the DS element was replaced by tetO4 such as p3315 (FRwt/tetO4) were passively lost after addition of DOX because they do not replicate in the presence of this drug (18). Our observations suggest that the attachment of plasmid DNA molecules to the host cell chromatin is more critical for plasmid stability than DNA replication.
Because these synthetic oriP-like plasmids rely solely on TetR fusions proteins and do not require EBNA1 they are virtually non-viral in terms of their plasmid features (Figure 4). Our results also support HMGA1a as a cellular protein that can functionally replace EBNA1 with respect to its central contribution(s) to DNA replication (18,19,37,38). This finding indicates that HMGA1a could be a cellular candidate of the factor that fosters ORC's recognition of replication origins in mammalian genomes (18).
Tetracycline-regulated gene expression has been demonstrated in vivo (39) indicating that gene vectors, which can be regulated in cis are likely to be functional in vivo as well. Mutants of the tetR gene with a reverse phenotype bind to tetO motifs exclusively in the presence of the drug (40,41) and are expected to be functional in our system, too. We assume that TetR fusions with the reverse phenotype would allow establishment of oriP-like plasmids only in the presence of tetracycline or its derivatives, which is an even more stringent condition for plasmid gene vectors regulated in cis.
We have developed a first generation of gene vector plasmids, which can be regulated in cis. Such gene vector plasmids can also be packaged into a viral particle if the necessary packaging signals are provided (42–44). Additional genes of interest (Figure 4) or even gene loci of therapeutic interest can be added because the packaging capacity of DNA-based plasmid vectors or viral vectors can be large, exceeding 100 kb (45,46 and references therein). Somatic gene and immune therapy are in need of gene vector systems, which are maintained long term, can be controlled by simple means and do not alter the recipient cell(s) genetically (8,47). Our novel gene vector system offers such advantages and should contribute to the feasibility of innovative therapeutic approaches in the future.
ACKNOWLEDGEMENTS
We are grateful to Bill Sugden for helpful discussions, suggestions and comments on the manuscript. We thank Natalie Grober for technical assistance and Christian Berens for TetR-specific antibodies. Funding of our work was supported by Deutsche Forschungsgemeinschaft (Transregio TR 36 and SFB 455 to W.H., SSP 1230 to A.S. and W.H.) and by Public Health Service (Grant CA70723 to W.H.). Funding to pay the Open Access publication charges for this article was provided by the Helmholtz Center Munich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 2.Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 3.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 4.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 6.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 7.Sugden B, Leight ER. EBV's plasmid replicon: an enigma in cis and trans. Curr. Top. Microbiol. Immunol. 2001;258:3–11. doi: 10.1007/978-3-642-56515-1_1. [DOI] [PubMed] [Google Scholar]
- 8.Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol. Ther. 2006;14:613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Baiker A, Maercker C, Piechaczek C, Schmidt SB, Bode J, Benham C, Lipps HJ. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 10.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Sugden B. Origins of bidirectional replication of Epstein-Barr virus: models for understanding mammalian origins of DNA synthesis. J. Cell. Biochem. 2005;94:247–256. doi: 10.1002/jcb.20324. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor P, Frappier L. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 2003;77:6946–6956. doi: 10.1128/JVI.77.12.6946-6956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung SC, Kang MS, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl Acad. Sci. USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 17.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 18.Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl Acad. Sci. USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 21.Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 23.Graham FL, Bacchetti S, McKinnon R, Cordell B, Goodman H. Transformation of mammalian cells with DNA using the calcium technique. Wistar Inst. Symp. Monogr. 1979;1:3–25. [Google Scholar]
- 24.Krueger C, Berens C, Schmidt A, Schnappinger D, Hillen W. Single-chain Tet transregulators. Nucleic Acids Res. 2003;31:3050–3056. doi: 10.1093/nar/gkg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell. Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- 27.Meuth M, Green H. Induction of a deoxycytidineless state in cultured mammalian cells by bromodeoxyuridine. Cell. 1974;2:109–112. doi: 10.1016/0092-8674(74)90099-3. [DOI] [PubMed] [Google Scholar]
- 28.Postle K, Nguyen TT, Bertrand KP. Nucleotide sequence of the repressor gene of the TN10 tetracycline resistance determinant. Nucleic Acids Res. 1984;12:4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinschmidt C, Tovar K, Hillen W, Porschke D. Dynamics of repressor-operator recognition: the Tn10-encoded tetracycline resistance control. Biochemistry. 1988;27:1094–1104. doi: 10.1021/bi00404a003. [DOI] [PubMed] [Google Scholar]
- 30.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J. Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 33.Kirchmaier AL, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindner SE, Zeller K, Schepers A, Sugden B. EBNA1's Affinity for it's origin of DNA synthesis is a determinant of the origin's replicative efficiency. J. Virol. 2008 doi: 10.1128/JVI.00332-08. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerhardt J, Jafar S, Spindler MP, Ott E, Schepers A. Identification of new human origins of DNA replication by an origin-trapping assay. Mol. Cell. Biol. 2006;26:7731–7746. doi: 10.1128/MCB.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 38.Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl Acad. Sci. USA. 2001;98:10085–10089. doi: 10.1073/pnas.181347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schonig K, Schwenk F, Rajewsky K, Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002;30:e134. doi: 10.1093/nar/gnf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gossen M, Freundlieb S, Bender G, Mueller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 42.Delecluse HJ, Pich D, Hilsendegen T, Baum C, Hammerschmidt W. A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc. Natl Acad. Sci. USA. 1999;96:5188–5193. doi: 10.1073/pnas.96.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreppel F, Kochanek S. Long-term transgene expression in proliferating cells mediated by episomally maintained high-capacity adenovirus vectors. J. Virol. 2004;78:9–22. doi: 10.1128/JVI.78.1.9-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hettich E, Janz A, Zeidler R, Pich D, Hellebrand E, Weissflog B, Moosmann A, Hammerschmidt W. Genetic design of an optimized packaging cell line for gene vectors transducing human B cells. Gene Ther. 2006;13:844–856. doi: 10.1038/sj.gt.3302714. [DOI] [PubMed] [Google Scholar]
- 45.White RE, Wade-Martins R, James MR. Infectious delivery of 120-kilobase genomic DNA by an Epstein-Barr virus amplicon vector. Mol. Ther. 2002;5:427–435. doi: 10.1006/mthe.2002.0557. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Sebastian S, Gimenez-Cassina A, Diaz-Nido J, Lim F, Wade-Martins R. Infectious delivery and expression of a 135 kb human FRDA genomic DNA locus complements Friedreich's ataxia deficiency in human cells. Mol. Ther. 2007;15:248–254. doi: 10.1038/sj.mt.6300021. [DOI] [PubMed] [Google Scholar]
- 47.Manzini S, Vargiolu A, Stehle IM, Bacci ML, Cerrito MG, Giovannoni R, Zannoni A, Bianco MR, Forni M, Donini P, et al. Genetically modified pigs produced with a nonviral episomal vector. Proc. Natl Acad. Sci. USA. 2006;103:17672–17677. doi: 10.1073/pnas.0604938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates JL, Camiolo SM. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- 49.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]