Abstract
The consequences of PARP-1 disruption or inhibition on DNA single-strand break repair (SSBR) and radio-induced lethality were determined in synchronized, isogenic HeLa cells stably silenced or not for poly(ADP-ribose) polymerase-1 (PARP-1) (PARP-1KD) or XRCC1 (XRCC1KD). PARP-1 inhibition prevented XRCC1-YFP recruitment at sites of 405 nm laser micro irradiation, slowed SSBR 10-fold and triggered the accumulation of large persistent foci of GFP-PARP-1 and GFP-PCNA at photo damaged sites. These aggregates are presumed to hinder the recruitment of other effectors of the base excision repair (BER) pathway. PARP-1 silencing also prevented XRCC1-YFP recruitment but did not lengthen the lifetime of GFP-PCNA foci. Moreover, PARP-1KD and XRCC1KD cells in S phase completed SSBR as rapidly as controls, while SSBR was delayed in G1. Taken together, the data demonstrate that a PARP-1- and XRCC1-independent SSBR pathway operates when the short patch repair branch of the BER is deficient. Long patch repair is the likely mechanism, as GFP-PCNA recruitment at photo-damaged sites was normal in PARP-1KD cells. PARP-1 silencing elicited hyper-radiosensitivity, while radiosensitization by a PARP inhibitor reportedly occurs only in those cells treated in S phase. PARP-1 inhibition and deletion thus have different outcomes in terms of SSBR and radiosensitivity.
INTRODUCTION
The poly(ADP-ribose) polymerase (PARP) superfamily in higher eukaryotes is composed of 17 members (1). PARP-1 (Mr = 113 000), the founding member, is the most abundant and most studied protein of the family. PARP-1 binds to DNA strand interruptions (2) through two zinc fingers located at its NH2-terminal end (3). Binding at sites of single-strand breaks (SSBs) results in PARP-1 dimerization and triggers elongation and transfer of long linear or branched chains of poly(ADP-ribose) (PAR) onto various nuclear acceptors (heteromodification), including PARP-1 itself (automodification) at the expense of NAD+ (4). In addition to early sensing of SSBs (5–7), PARP-1 is involved in many cellular pathways including DNA replication (8,9), transcription (10), chromatin remodelling (11) and cell death (12,13).
SSBs are repaired through the base excision repair (BER) pathway (14) operating via either the short patch (SPR) or long patch repair (LPR) sub-pathways (15) differing by the size of the repair patch (one nucleotide for the SPR, up to 15 nucleotides for the LPR) and the enzymes involved. The proliferating cell nuclear antigen (PCNA) reportedly controls the LPR pathway. PCNA is loaded by the replication factor C (RFC) and allows the replicative DNA polymerases δ/ɛ to be clamped in place (16,17). PCNA also stimulates the activity of endonuclease I (FEN-I) to remove flaps (18), and recruits DNA ligase I (Lig I) (19,20). The major player in the SPR sub-pathway is XRCC1, a scaffold protein with no known enzymatic activity, but however essential for the recruitment of polymerase β and DNA ligase IIIα (Lig III) (21). XRCC1 is loaded at sites of SSBs by PARP-1 through the interaction of one of its BRCT domains with the PAR chains formed during PARP-1 automodification (5,21). For this reason, PARP inhibitors impair XRCC1 recruitment at sites of DNA damage (22).
PARP inhibitors were shown to induce a large increase in radiosensitivity specifically in the S phase of the cell cycle, due to the collision of unrepaired DNA lesions with replication forks (23) in which altered regulation of a complex involving PARP-1 and DNA topoisomerase I might play a role (24). In contrast PARP-1 null, 3T3 mouse embryonic fibroblasts (MEF) showed hypersensitivity to ionizing radiation (IR) independently of the cell-cycle phase (6). PARP-1 inhibition and deletion thus have different outcomes. To shed light on this issue, we analyzed the SSBR kinetics by alkaline filter elution in PARP-1 (PARP-1KD) or XRCC1 (XRCC1KD) knockdown (KD) and control HeLa cells synchronized in the S or G1 phases of the cell cycle. The same cells were transfected with plasmids encoding fluorescent conjugates of PARP-1, XRCC1 or PCNA, in order to visualize protein movement after the induction of SSBs induced by laser microirradiation at 405 nm. In PARP-1 proficient cells, PARP inhibition by 4-amino-1,8-naphthalimide (ANI) slowed down SSBR 10-fold and inhibited XRCC1 recruitment at DNA damage sites. Under these experimental conditions, the complete religation of SSBs was however seen in G1 cells but not in the S phase. In contrast, PARP-1KD cells synchronized in S phase were able to rejoin SSBs as rapidly and as completely as controls, while SSBR was delayed in G1. These data suggest the existence of a PARP-1-independent repair pathway that acts more rapidly in S phase than in G1. The LPR sub-pathway is the likely mechanism as PCNA recruitment at DNA damage sites induced by laser microirradiation was not affected by the absence of PARP-1. However, in the same way as in 3T3 PARP-1−/− MEFs, PARP-1KD cells were considerably more sensitive than PARP-1 proficient cells to the killing effect of radiation.
MATERIALS AND METHODS
Reagents
Products and their suppliers were as follows: [2-14C]thymidine and BioMax films, GE Healthcare–Amersham Biosciences (Orsay, France); detergents, tetrapropylammonium hydroxide, methyl methanesulfonate (MMS), proteinase K, protease inhibitors, phosphatase inhibitors and mouse monoclonal anti-α-tubulin antibody, Sigma-Aldrich Chemicals (Saint Quentin Fallavier, France); ANI, Acros Organics (Geel, Belgium); other chemicals and solvents, Merck (Darmstadt, Germany); polycarbonate filters (Nuclepore, 2.0 µm pore size), Whatman (Banbury, Oxon, UK); nitrocellulose membrane (0.2 µm pore size), Schleicher & Schuell (Dassel, Germany); hygromycin B, lipofectamine 2000, and products and antibiotics for cell culture, Invitrogen (Cergy-Pontoise, France); ECL Western blotting substrate and M-PER reagent for protein extraction, Pierce (Perbio Science, Brebières, France); mouse monoclonal primary antibodies directed against PARP-1 (clone C2-10) and Lig III (clone 7), Becton-Dickinson (Le-Pont-de-Claix, France), and against XRCC1, Trevigen (Gaithersburg, Maryland); goat anti-mouse, HRP-conjugated secondary antibodies, Jackson ImmunoResearch Laboratories (Soham, Cambridgeshire, UK).
Cell culture
HeLa cells were grown in plastic flasks or on round coverslips (videomicroscopy experiments) in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin under 5% CO2 in air. KD and control clones were grown in the presence of 125 µg/ml hygromycin B. Synchronization of cells at the G1–S junction was achieved using a double thymidine block. Cell cycle progression was monitored by dual parameter flow cytometry using a FACStarPLUS cytofluorometer (Becton-Dickinson) with BrdUrd pulse labeling (10 µM, 15 min) of S phase cells as described previously (25).
Gene silencing in HeLa cells by pEBV-based vectors
siRNA design and cloning in pEBVsiRNA vectors carrying a hygromycin B resistance cassette and establishment of stable knockdown and control HeLa clones were carried out as previously described (26,27). Control HeLa cells carried the pBD650 plasmid that expressed an inefficient shRNA sequence (26). BD650 cells (hereafter referred to as as ‘control cells’) expressed normal levels of PARP-1 and XRCC1. KD HeLa cells were cultivated for more than 200 days in culture with levels of the targeted proteins remaining below the limits of detection by western blot. The RNAi sequences for PARP1 (NM_001618) and for XRCC1 (NM_006297) were nucleotides 2068–2086 and nucleotides 1832–1850, respectively.
Expression plasmids and transfection
pEGFP-NLS-PCNA, pEGFP-PARP-1, pRFP-XRCC1 and XRCC1-pEYFP plasmids were kindly provided by Drs P. Kannouche (FRE2939 CNRS, Institut Gustave-Roussy, Villejuif, France), V. Schreiber (UMR7175 CNRS, ESBS, Illkirch, France), K. W. Caldecott (Genome Damage and Stability Centre, University of Sussex, Brighton, UK) and P. Radicella (IRCM, CEA-DSV, Fontenay-aux-Roses, France), respectively. After amplification, purification of the plasmids was carried out using a Macherey-Nagel kit (Düren, Germany). Twenty-four hours before transfection cells were plated at 50% confluence onto 18 mm round coverslips and transfected 48 h before the live-cell experiments with lipofectamine 2000 according to the manufacturer's recommendations.
Radiation and MMS survival
γ-Ray irradiation of HeLa cells for the determination of clonogenic radiation survival was performed at room temperature in culture medium using an IBL-637 (137Cs) irradiator (CIS-Biointernational) at a dose-rate of 0.70 Gy/min. Each measurement was performed in triplicate. Following treatment cells were allowed to grow as colonies for 10–15 days, fixed, stained with Coomassie blue and counted. Colonies of less than 50 cells were disregarded. The colony count relative to mock-irradiated cells (S) was adjusted for best fit to an exponential equation (eq. 1) or to the classical linear-quadratic equation (eq. 2),
| 1 |
| 2 |
where D is the radiation dose and α and β adjustable parameters characterizing radiation response. Calculations were made through nonlinear least-squares regression taking all data points into account, using Kaleidagraph software (Synergy Software, Reading, Pennsylvania, USA). In experiments using synchronized HeLa cells, the cellular multiplicity, i.e. the number of cells (n) per potential colony-forming unit was measured by digital microscope examination of the culture flasks at the time of irradiation. The single-cell surviving fraction (SCSF) was calculated using the discrete distribution equation (28):
| 3 |
where Sexp is the experimental cell survival determined from bulk colony scoring and an the fraction of colony-forming units containing n cells.
The response of cells to MMS was determined by a clonogenic assay in the same way as above. Contact with MMS was for 1 h (37°C, 5% CO2).
Analysis of SSB repair by alkaline filter elution
Radiation-induced SSBs were measured by alkaline filter elution over polycarbonate filters according to Filipski and Kohn (29). This method allows titration of direct SSBs, alkali-labile and abasic sites altogether. Cells were grown in the presence of [2-14C]thymidine (0.02–0.06 µCi/ml) for two doubling times. Radioactive thymidine was removed and cells submitted or not to a double thymidine block. Two hours (S) or 15 h (G1) after release from the second thymidine block the culture flasks were exposed to 10 Gy γ-rays at a dose-rate of 2.95 Gy/min (room temperature) in the presence or not of ANI and returned to the incubator for varying times (up to 6 h). For the determination of the initial yield of SSBs, irradiation was performed on ice to prevent SSB rejoining. When present, ANI (30 µM from a 3 mM stock solution, 1% DMSO final concentration) was introduced 30 min before irradiation and was present for the whole length of postirradiation incubation. The medium was removed at the end of the incubation period, the flasks rapidly rinsed once with ice-cold PBS, and maintained on ice for not >10 min before harvesting with the aid of a cell scraper. 6 × 105 cells were deposited onto polycarbonate filters, lyzed and DNA fragments eluted and counted as described (30). Each measurement was performed in duplicate.
Western blotting
Total cells extracts were prepared from ≥107 cells using M-PER reagent with a cocktail of protease and phosphatase inhibitors. Protein concentrations were measured by the Bradford method using the Bio-Rad protein assay and the extracts heated to 95°C (5 min) in Laemmli loading buffer and separated on 7.5% (PARP-1, Lig III) or 10% (XRCC1) SDS–PAGE gels. After migration proteins were electrotransferred to nitrocellulose membranes. The membranes were blocked with 5% bovine serum albumin for 1 h at room temperature, then incubated with the primary antibodies overnight at 4°C and with secondary antibodies coupled to horseradish peroxidase for 1 h at room temperature. Membranes were probed with an enhanced chemiluminescence reagent and re-hybridized with anti-α-tubulin antibody.
Live-cell videomicroscopy
Photodamage experiments were performed on a Leica SP5 confocal system, attached to a DMI6000 stand using a 63×/1.4 objective, under a controlled environment (37°C, 5% CO2). All recordings were made using the appropriate sampling frequency (512 × 512 images, line average of 4 and zooming set to 8) and an argon laser line (488 nm for eGFP, 514 nm for YFP) or a 561 nm diode (RFP) adapted to the fluorescent protein of interest. In the first step, two images were acquired within a time lag of 2–3 s at low enough laser energy not to induce any photodynamic damage. The 405 nm laser line (diode) was then set to maximum output for 500 ms and focused onto a single spot of constant size (176 nm) within the nucleus to perform a point of photodamage with a constant amount of energy. Recruitment of the protein of interest was then monitored by fluorescence using the same setting as for the predamage sequence. Nomarski contrast images were taken in parallel. Images were captured at 2–5 s interval, except in a few instances where GFP-PARP-1 and GFP-PCNA foci images were taken each at 5 min interval to avoid photobleaching.
All images were processed using the freely available software ImageJ (http://rsb.info.nih.gov/ij/) (31) complemented with the LOCI bioformat plugin (http://www.loci.wisc.edu/ome/formats.html) to open images generated by the Leica SP5 confocal system. A macro was written to automate data extraction from images. Briefly, it consisted of retrieving two regions of interest (ROI), namely the photodamage spot and the nucleus area excluding the spot, and quantifying the total intensity within these ROIs. The latter was used to correct fluorescence intensity for the observational photobleaching. Intensity within the former ROI was normalized to 1, based on quantifications before photodamage, then plotted against time to get the recruitment kinetics. The size of the spots in each cell was evaluated using ImageJ software; firstly, using the threshold function to segment the image based on intensities and define the position of the spot; secondly, using the magic wand tool to define their contour and thirdly, using the measure function to retrieve their widest diameter (Ferret's diameter).
RESULTS
PARP inhibition impairs SSBR in PARP-1 proficient cells with different outcomes in S and G1 phases
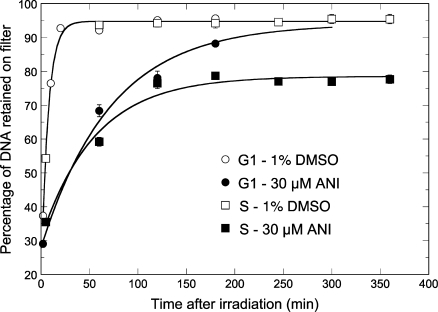
The SSBR kinetics following exposure to IR was analyzed by alkaline elution. This method was preferred over the comet assay because, by a combination of accurate temperature control and instant arrest of DNA repair upon addition of lysis buffer directly onto the cell layer, it allows a more precise determination of the SSBR kinetics and a powerful statistical analysis over a large cell population. In a typical experiment, [2-14C]thymidine labeled control cells were synchronized at the G1–S junction by a double thymidine block, irradiated in S or G1 phase without or with the PARP inhibitor ANI, returned to the incubator and finally harvested and lyzed at defined times for the determination of residual SSBs. The results are shown in Figure 1. SSBR in the absence of ANI proceeded to completion in <20 min (t1/2 = 4.9 ± 0.3 min), irrespective of whether cells were in G1 or S phase. ANI slowed down SSBR by ca. 10-fold in both phases. In G1 phase, SSBR reached completion after 5 h repair. However, in cells treated in S phase as much as 26% of the initial load of SSBs remained unrejoined from 2 to 6 h post-irradiation, demonstrating a clear block in SSBR. We have previously shown that this block is correlated with the induction of a large number of de novo DNA double-strand breaks (DSBs) that are suppressed by aphidicolin, indicating that they originate from the collision of unrepaired SSBs with replication forks (23).
Figure 1.
| 4 |
PARP inhibition hampers the recruitment of XRCC1 at sites of laser-induced DNA photodamage
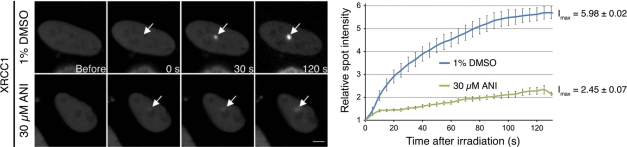
The 405 nm laser microirradiation (see Materials and methods section) was used to generate localized base damage, SSBs and DSBs (32,33) in the nucleus of HeLa cells transiently expressing YFP-tagged XRCC1. Using confocal microscopy, we found that XRCC1-YFP accumulated at sites of microirradiation (Figure 2). ANI (30 µM) lowered by over 12-fold the amount of XRCC1-YFP recruited at these spots. These data are in close agreement with El-Khamisy et al. (21) and Mortusewicz et al. (22) and confirm that poly(ADP-ribosyl)ation is mandatory for the recruitment of the XRCC1 protein by PARP-1 at sites of DNA damage.
Figure 2.
Live cell imaging of microirradiated HeLa cells transiently expressing XRCC1-YFP. A confocal microscope with a laser microbeam was used to induce and follow the evolution of localized DNA damage in the nucleus of target cells (37°C, 5% CO2 in air). Accumulation of XRCC1-YFP was observed immediately after microirradiation (white arrows). ANI reduced the amount of XRCC1-YFP recruited by 12-fold. The scale bar (lower right) represents 5 µm.
PARP inhibition results in the accumulation of PARP-1 and PCNA at sites of laser microirradiation
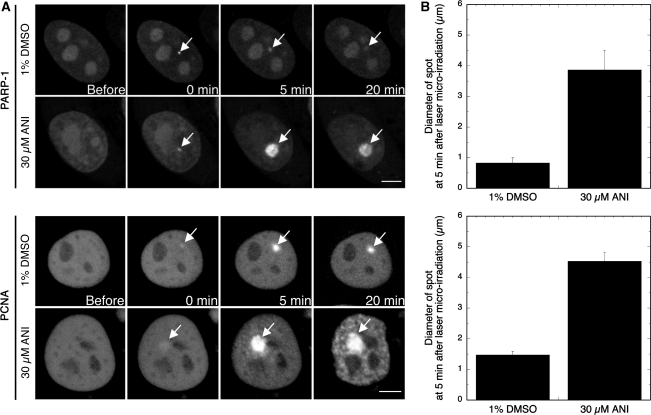
To further decipher the effect of PARP inhibition on the fate of microlaser-induced DNA damage, the time-dependent recruitment of GFP-tagged PARP-1 and PCNA in response to laser microirradiation was determined without or with ANI. In the absence of ANI, we observed an immediate recruitment of GFP-PARP-1 (<10 s) at damage sites. These foci dissipated in ca. 20 min (Figure 3). In contrast, in the presence of ANI GFP-PARP-1 formed intense spots of fluorescence that were ca. 4-fold larger than the size of the foci observed at 5 min post-irradiation without ANI and were still at their maximum 15 min later (Figure 3). A similar accumulation pattern was seen for GFP-PCNA in the presence of ANI (Figure 3). These observations suggest that, while the catalytic activity of PARP-1 is not required for the recruitment of PARP-1 and PCNA at sites of DNA damage, the inhibition of PARP-1 auto-poly(ADP-ribosyl)ation leads to the accumulation of considerable amounts of PARP-1 and PCNA at damage sites and delays their dissociation.
Figure 3.
Effect of ANI on the recruitment of GFP-PARP-1 and GFP-PCNA after 405 nm laser microirradiation. (A) Control cells were transfected with the GFP-PARP-1 or the GFP-PCNA plasmids and grown on coverslips for 48 h. Thirty minutes prior to irradiation, cells were exposed to 1% DMSO or 30 µM ANI, 1% DMSO until the end of the experiment. Cells were microirradiated with a 405 nm laser and the time-course of the reaction was followed for 20 min (37°C, 5% CO2 in air). Images were captured at a relatively large interval (5 min) to avoid photobleaching. Similar figures were obtained over 10 successive experiments. Scale bar (lower right), 5 µm. The (B) shows the size of the spots at their maximum intensity (average over 10 cells).
SSBR is efficient in cells lacking PARP-1
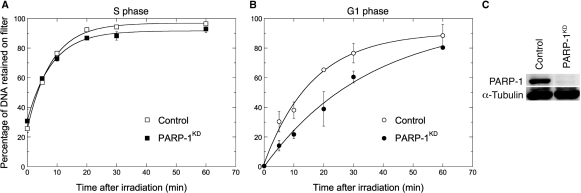
The level of PARP-1 expression in the PARP-1KD clone used was below the limit of detection in western blot experiments, while PARP-1 expression was at normal levels in control cells (Figure 4). Using immunofluorescence assays PAR synthesis was not detectable in the PARP-1KD cells following exposure to H2O2 (1 mM, 10 min) or 50 Gy γ-rays (data not shown), but was clearly detectable in the control cells.
Figure 4.
PARP-1 expression and rejoining of radiation-induced SSBs in synchronized HeLa cells expressing (control) or lacking PARP-1 (PARP-1KD). (A and B) Alkaline elution analysis of the kinetics of SSB rejoining after 10 Gy irradiation. Both cell lines were synchronized in S or G1 phase by a double thymidine block (separate experiments). The processing of cells was as in Figure 1. The solid lines were fitted to an exponential time-dependent equation [Equation (4), see legend to Figure 1] and gave α = 0.123 ± 0.007/min (t1/2 = 5.6 ± 0.3 min), R∞ = 96.8% (open square); α = 0.121 ± 0.008/min (t1/2 = 5.7 ± 0.4 min), R∞ = 91.6% (closed square); α = 0.0612 ± 0.0095/min (t1/2 = 11.6 ± 1.8 min), R∞ = 90.6% (open circle); α = 0.0282 ± 0.0048/min (t1/2 = 25.3 ± 4.3 min), R∞ = 99.6% (closed circle). (C) Western blot analysis of PARP-1 expression in these two cells lines. PARP-1 and α -tubulin were probed on the same membrane. Bars, SD.
Both control and PARP-1KD cells were synchronized by a double thymidine block, allowed to progress into S (2 h) or G1 phase (14 h), and exposed to γ-rays for the determination of the time-dependence of SSBR using alkaline filter elution (Figure 4). SSB rejoining in S phase went to completion and was as fast in PARP-1KD cells as in controls. In contrast, in G1 phase, PARP-1 silencing delayed SSBR by 2.2-fold based on t1/2 values (see legend to Figure 4). This result suggests the existence of an alternative, PARP-1 independent SSBR pathway operating more rapidly in the S phase than in G1.
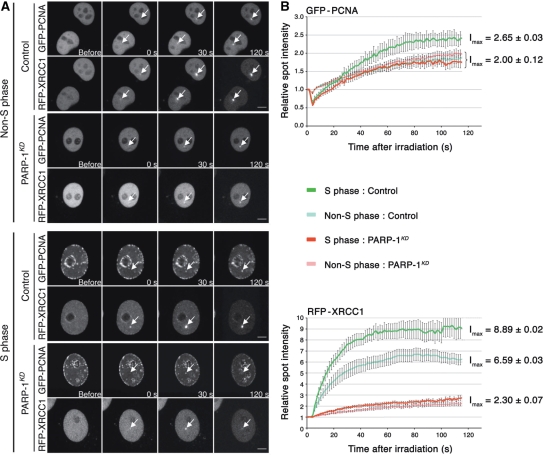
To determine whether this alternative pathway was XR-CC1/Lig III independent, both control and PARP-1KD cells were co-transfected with two plasmids coding for RFP-XRCC1 and GFP-PCNA proteins and exposed to laser microirradiation as described earlier. Spontaneous formation of GFP-PCNA foci at replication forks was used to differentiate S- from non-S-phase cells under the microscope as described by Mortusewicz et al. (34). XRCC1 recruitment at damaged sites was observed in control cells; the formation of XRCC1 foci was comparatively faster in cells in S phase compared to those in other phases of the cell cycle (Figure 5). XRCC1 foci were barely detectable in PARP-1KD cells in both S and non-S phases, thus confirming that PARP-1 is mandatory for XRCC1 recruitment.
Figure 5.
Analysis of XRCC1 and PCNA recruitment at DNA damage sites in cells expressing (control) or lacking PARP-1 (PARP-1KD). (A) Live cell imaging of microirradiated control and PARP-1KD cells expressing RFP-XRCC1 and GFP-PCNA proteins. Cells were transfected with both plasmids 48 h before the experiments. To evaluate the recruitment of these proteins to DNA damage sites, a time-course analysis from to 0 to 120 s was performed. The distribution of PCNA foci was used to determine the cell cycle phase (S or non-S phase) of each cell analyzed, according to Mortusewicz et al. (20). Scale bar (lower right), 5 µm. (B) Quantitative time-lapse analysis of RFP-XRCC1 and GFP-PCNA recruitment at sites of microirradiation in control and PARP-1KD in S or non-S phase. The relative spot intensity was calculated as described under Materials and methods section. Each value represents the mean fluorescence intensity from an average over 10 cells. Bars, SEM. Data points were fitted to an exponential equation to calculate the maximum fluorescence intensities (Imax) at the plateau of the reaction.
To check whether the PCNA-dependent sub-pathway was functional in cells lacking PARP-1, the kinetics of PCNA recruitment at sites of laser microirradiation was determined in control and PARP-1KD cells. In control cells, the amount of PCNA recruited at damaged sites was ca. 25% higher in S relative to non-S phase. In PARP-1KD cells, PCNA recruitment appeared to be independent of the phase of the cell cycle and was similar to that for non-S phase controls (Figure 5). Thus, the recruitment of PCNA does not dependent to a significant extent on the PARP-1 status.
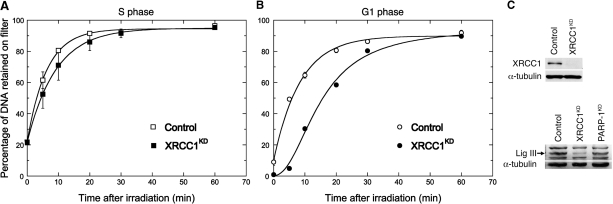
SSBR is functional in HeLa cells lacking XRCC1
Western blot analysis showed that residual XRCC1 expression in XRCC1KD cells was below the limit of detection (Figure 6). To determine their SSB rejoining capacity, XRCC1KD cells were analyzed by alkaline elution following γ-ray exposure in S phase or in G1. SSBR in S phase was nearly as fast in XRCC1KD as in control cells (Figure 6). However, in XRCC1KD cells irradiated in G1, SSBR was delayed by ca. 2.5-fold based on the t1/2 values (see legend to Figure 6), in agreement with data obtained using XRCC1-deficient rodent cells (35).
Figure 6.
Comparison of radio-induced SSB rejoining in control and XRCC1KD cells synchronized in S or G1 phase. (A and B) Alkaline elution analysis of the SSB rejoining kinetics. The [2-14C]thymidine-labeled cells were synchronized by a double thymidine block, irradiated in S or G1 phase (10 Gy) and processed for alkaline elution (separate experiments). The results were analyzed as described in Figure 1 and gave: α = 0.161 ± 0.011/min (t1/2 = 4.32 ± 0.30 min), R∞ = 94.4% (open square); α = 0.110 ± 0.003/min (t1/2 = 6.30 ± 0.18 min), R∞ = 94.8% (closed square); α = 0.117 ± 0.008/min (t1/2 = 5.9 ± 0.4 min), R∞ = 89.9% (open circle). XRCC1KD cells in G1 (closed circle) fitted a sigmoidal function (t1/2 = 15.1 ± 0.8 min, R∞ = 95.1%) suggesting that repair was delayed by a few minutes following radiation. (C) Western blot analysis (30 µg protein per lane) of the expression of XRCC1 and Lig III in Control and XRCC1KD cells, and of Lig III in PARP1KD cells. Bars, SD.
To examine whether the pathway downstream of XRCC1 was inactivated, the expression of Lig III was determined by western blotting in XRCC1KD cells. A major reduction of the expression of Lig III was observed in these cells (Figure 6), in agreement with Caldecott et al. (36) who showed that XRCC1 tightly associates with and stabilizes Lig III. This confirms that an alternative SSBR mechanism can operate when the PARP-1/XRCC1/Lig III pathway is inactivated by deletion of PARP-1 or XRCC1.
XRCC1 and PARP-1 silencing elicits differential radiosensitivity
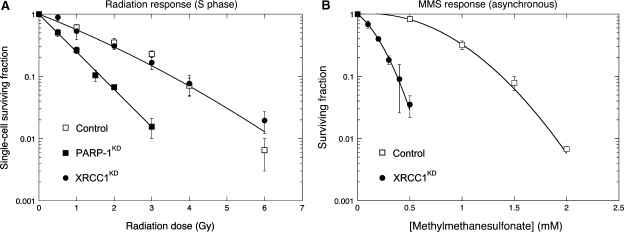
Radiation survival was determined in control, XRCC1KD and PARP-1KD cells using clonogenic assays. In a first experiment, HeLa cells were synchronized at the G1–S junction, allowed to progress into S phase for exactly 2 h, exposed to graded doses of γ-rays and grown as colonies for 10 (controls) or 15 days (PARP-1KD, XRCC1KD). PARP-1 silencing resulted in a 2.5-fold increase in radiosensitivity (Figure 7) as calculated from the initial slope (α) of the survival curves [Equation (2), see Materials and methods section]. In contrast, the radiation response of XRCC1KD cells was indistinguishable from that of control cells (Figure 7). Radiation survival was also determined in asynchronous cultures (cells mostly in G1 phase) of XRCC1KD and PARP-1KD and similar results were obtained (data not shown).
Figure 7.
Radiation survival in control, PARP-1KD and XRCC1KD HeLa cells synchronized in S-phase, and MMS survival in asynchronous control and XRCC1KD cells. (A) Cells were synchronized by a double thymidine block, allowed to progress in S phase for 2 h and exposed to graded doses of γ-rays. Survival curves were drawn for best fit to experimental data using an exponential equation [PARP-1KD, Equation (1)] or the linear-quadratic model (Control and XRCC1KD, Equation (2), see Materials and methods section). The experimental data for control and XRCC1KD lay within the same envelope of statistical deviation. Results: α = 0.547 ± 0.068/Gy, β = 0.0298 ± 0.0314/Gy2 (open square, closed circle); α = 1.381 ± 0.060/Gy (closed square). Bars, SD. (B) The sensitivity of cells to MMS was determined by a clonogenic assay using asynchronous cells. Contact with MMS was for 1 h. The survival curves were drawn for best fit to experimental data using a linear-quadratic equation. Results: α = 0.03/mM, β = 1.46/mM2 (open circle); α = 3.00/mM, β = 8.14/mM2 (closed circle). Bars, SD.
As the response to MMS is regarded as the gold standard for characterizing XRCC1-deficient cells, the dose-dependence of MMS survival was determined in asynchronous XRCC1KD HeLa cells. In agreement with other authors (37), XRCC1KD cells were 5-fold more sensitive to the cytotoxic effect of MMS as calculated from the IC50 values (Figure 7).
DISCUSSION
The aim of this study was to compare in isogenic human cell lines the effect of PARP inhibition versus RNAi-induced KD of PARP-1 or XRCC1 on: firstly, the kinetics of γ-ray induced SSBR determined by alkaline elution; second, the recruitment of PARP-1, XRCC1 and PCNA at sites of DNA damage induced by 405 nm laser microirradiation and third, radiation susceptibility. Interestingly, PARP inhibition and PARP-1 KD had different impacts on these three endpoints.
In the present study, inhibition of the PARP-1 catalytic activity by ANI slowed by about 10-fold, the rate of SSBR in cells in both the G1 and S phases of the cell cycle. However, it should be noted that in cells treated in G1, ANI did not prevent complete rejoining of SSBs. This result provides an explanation as to why no modification of radiation sensitivity by ANI was previously shown in this phase of the cell cycle (23). In contrast, SSBR did not reach completion in cells treated in S phase and about 26% of SSBs remained unrepaired for over 6 h. The time course of SSBR under these conditions paralleled that of de novo formation of DSBs resulting from the collision of unrepaired lesions with stalled replication forks, which has previously been correlated with induced radiosensitization specifically in S phase (23). In agreement with Mortusewicz et al. (22), ANI also hindered the recruitment of YFP-tagged XRCC1 at sites of laser-induced damage. In contrast, ANI did not impair the recruitment of GFP-tagged PARP-1 but appeared to block the reversal of this process and resulted in an accumulation of large amounts of PARP-1 at the damage sites. Such an accumulation is consistent with the fact that PARP-1 auto-poly(ADP-ribosyl)ation is necessary for its dissociation from SSBs after the recruitment of XRCC1 (5,14,22,38). PARP inhibition also resulted in the recruitment of a large amount of PCNA at damage sites, showing that PCNA recruitment does not depend on poly(ADP-ribosyl)ation, but that the reversal of PCNA foci does. Our results, therefore, suggest that the inhibition of PARP-1 activity by ANI blocks the SPR and considerably slows down the LPR through the occupancy of SSBs by excess PCNA and inactive PARP-1.
In contrast, during S phase SSBR in cells lacking PARP-1 or XRCC1 was as fast as in control cells. In cells synchronized in G1, however, PARP-1 or XRCC1 silencing delayed SSBR by 2.2- to 2.5-fold, respectively, although repair reached completion in <2 h in both instances. This observation clearly demonstrates that cells defective in the SPR sub-pathway as a result of PARP-1 or XRCC1 silencing, are able to use an alternative, efficient mechanism for SSBR, and that this sub-pathway operates more rapidly in S phase. In other words, it appears that there is a greater requirement for XRCC1 and PARP-1 for SSBR during the G1 phase, compared to S phase, and that this may, at least in part, reflect a novel XRCC1- and PARP-1-independent S phase process. A few years ago, Vodenicharov et al. (39) using a DNA plasmid assay to test DNA repair in cells exposed to the alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine or X-rays, also showed that SSBR is efficient in mouse fibroblasts lacking PARP-1. Moreover, defects in SPR due to mutations in the BRCT II domain of XRCC1, which affect its interaction with Lig III, are not sufficient to abolish SSBR in S phase (40). While PARP-1, together with FEN-1, is able to stimulate strand displacement and DNA synthesis by Pol β during LPR in in vitro assays (41–43), the results presented here suggest that, in living cells, LPR is able to substitute for the SPR sub-pathway when the latter is compromised by a lack of PARP-1 and/or XRCC1. This hypothesis is supported by a plethora of evidence. First, under physiological conditions DNA synthesis associated with LPR in S phase is carried out by replicative polymerases instead of Pol β. Second, normal SSBR occurs in S phase in Pol β null mouse fibroblasts (44). And third, it has been proposed that in case of XRCC1 deficiency the PCNA-dependent LPR sub-pathway can substitute for SPR in S and G2 phases (45,46). In support of this scheme, we demonstrate that the recruitment of PCNA at sites of laser microirradiation does not depend on the presence of PARP-1. Taken together, these data and ours suggest that rapid SSBR occurs under control of PCNA through the LPR in human cells during S phase when SPR is not functional, either because of a lack of XRCC1 and Lig III recruitment due to suppression of PARP-1, or to a downregulation of Lig III expression following loss of XRCC1. The substitution of the LPR for the SPR sub-pathway could be more favorable in S phase because most of the LPR effectors including RFC, PCNA, DNA polymerases δ/ε, FEN-1 and Ligase I participate in the replication process, so are located in close proximity to the damaged DNA. This might be the reason for the observed differences in the SSBR kinetics in S versus G1 phase in cells defective in the SPR for lack of PARP-1 or XRCC1.
Even though HeLa cells in which the expression of PARP-1 is suppressed are able to rejoin SSBs as rapidly as controls in S phase, the lack of PARP-1 results in a large increase of radiosensitivity in this phase of the cell cycle (Figure 7) and in asynchronous cultures as well (data not shown). The same effect was observed in PARP-1−/− mouse 3T3s (6,47). In contrast, in our hands the radiosensitivity of XRCC1KD cells was indistinguishable from that of controls in S phase, nor was a difference found in asynchronous cells (data not shown). This correlates with the fact that SSBR reaches completion in XRCC1KD cells but is inconsistent with earlier reports showing that XRCC1-mutant Chinese hamster ovary cells are more radiosensitive than parent cells and that this defect is corrected by re-expression of wild-type XRCC1 (35). Residual expression of XRCC1 undetectable by western blot in XRCC1KD cells could provide an explanation to the lack of enhanced radiosensitivity following XRCC1 silencing. However, the large difference in the sensitivity to MMS between XRCC1KD and control cells (Figure 7) clearly indicates a XRCC1 deficient phenotype. This suggests that, at least in our model system, invalidation of the SPR sub-pathway does not alter the radiation susceptibility and raises the question of the mechanism that underlies the large increase of radiosensitivity observed in HeLa cells lacking PARP-1. There would not appear to be a straightforward answer to this question. A defect in DSB repair might be linked to this radiosensitivity. PARP-1 indeed plays a role in the switch between homologous recombination and non-homologous end-joining (NHEJ) (48). A defect in the activation of the protein kinase ATM could also be evoked since PARP-1 has been shown to form a complex with ATM (49). This complex is important for the ATM-dependent phosphorylation of p53, SMC1 and H2AX (50). PARP-1 has also been proposed to be required for the rapid recruitment of polynucleotide kinase, MRE11 and NBS1 proteins at sites of DSBs (51,52). However, whether the modification of these mechanisms results in the increase in radiosensitivity seen when PARP-1 expression is suppressed, remains to be established. Indeed, the physical interaction of PARP-1 with polynucleotide kinase, ATM or MRE11 relies on poly(ADP-ribosyl)ation while, as known from earlier studies, PARP inhibitors (23) or PARP suppression (24,53) do not impair the rejoining of drug- or radiation-induced DSBs yet they compromise the reactivation of stalled replication forks. Studies of local DNA damage by proton microbeam irradiation also showed that radio-induced poly(ADP-ribosyl)ation synthesis and phosphorylation of histone H2AX, were independent events (7). A direct role of PARP-1 in the back-up NHEJ (B-NHEJ) pathway that occurs in the context of inactivated DNA-PKcs (54) cannot be an explanation either as DNA-PKcs was functional in the cell lines used here. Furthermore, while B-NHEJ requires XRCC1/Lig III we demonstrate that suppression of XRCC1 does not confer any significant increase of radiosensitivity. Thus, while the absence of PARP-1 or XRCC1 suggests a switch of the mechanism of SSBR from SPR to LPR, only the silencing of PARP-1 modulates radiosensitivity. PARP-1 does have many other roles in addition to the control of the first steps of the SPR sub-pathway. In particular, PARP-1 binds nucleosomes and plays a major role in the condensation of chromatin and transcriptional repression (11,55,56). This activity involves the C- and N-terminal domains of PARP-1 but occurs independently of the enzyme's catalytic activity (11,56) and it was proposed (57,58), based on the susceptibility of chromatin to digestion by deoxyribonuclease I or micrococcal nuclease, that the structure of chromatin is altered in cells lacking PARP-1. We would thus suggest as a working hypothesis that altered regulation of chromatin conformation following suppression of PARP-1 expression might result in enhanced susceptibility to radiation-induced damage.
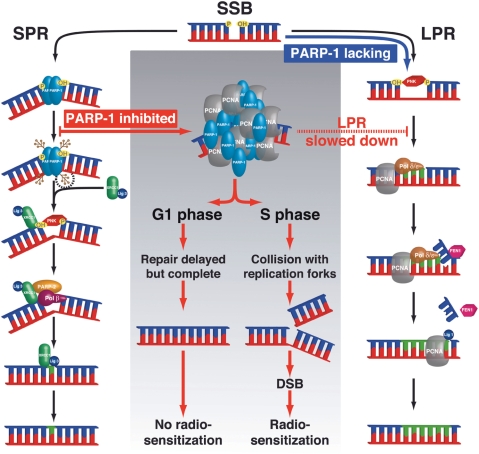
In the light of the results reported here, we propose a model (Figure 8) detailing the repair options that can function in cells when PARP-1 is either depleted or inhibited during S-phase and their consequences. Although the exact role of PARP-1 in the LPR is still unclear, it seems not to be as essential in LPR as it is in SPR. Accordingly, we propose that LPR can substitute for SPR when PARP-1 or XRCC1 is lacking and thus allow SSBs to be repaired. However, in control cells PARP inhibition leads to the accumulation of both PARP-1 and PCNA at damaged sites and slows down SSBR by as much as 10-fold, possibly due to steric hindrance modulating the recruitment of the LPR effectors. The mechanisms that control the recruitment of PCNA under these conditions remain to be established.
Figure 8.
Scheme summarizing the differential effect of PARP-1 inhibition versus silencing on SSB repair via the BER pathway. The experimental evidence would suggest that SSB rejoining proceeds via the LPR sub-pathway when the SPR is deficient because of lack of PARP-1. In contrast, inhibition of poly(ADP-ribosyl)ation in PARP-1 proficient cells was found to result in the accumulation of PARP-1 and PCNA in the vicinity of DNA damaged sites, with a 10-fold reduction of the bulk rate of SSBR. This does not impact on radiosensitivity in the G1 phase of the cell cycle, probably because cells have enough time to perform SSB repair. However, under such conditions in S phase collision of unrepaired SSBs with replication forks results in a large increase of radiosensitivity due to the formation of a large number of DSBs.
ACKNOWLEDGEMENTS
We wish to thank Drs Valérie Schreiber (UMR 7175 CNRS, ESBS, Illkirch, France), Patricia Kannouche (FRE 2939 CNRS, Institut Gustave-Roussy, Villejuif, France), Pablo Radicella (IRCM, CEA-DSV, Fontenay-aux-Roses, France) and Keith W. Caldecott (Genome Damage and Stability Centre, University of Sussex, Brighton, UK) for kindly providing us the plasmids coding the fluorescent-tagged proteins GFP-PARP-1, GFP-PCNA, XRCC1-YFP and RFP-XRCC1, respectively. C.G. is recipient of a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche. This work was supported by financial aid from the Institut National de la Santé et de la Recherche Médicale and Institut Curie. Denis Biard acknowledges a grant from Electricité de France. Funding to pay the Open Access publication charges for this article was provided by Institut Curie.
Conflict of interest statement. None declared.
REFERENCES
- 1.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin RC, Gill DM. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J. Biol. Chem. 1980;255:10502–10508. [PubMed] [Google Scholar]
- 3.de Murcia G, Ménissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 4.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 5.Masson M, Niedergang C, Schreiber V, Muller S, Ménissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernet M, Ponette V, Deniaud-Alexandre E, Ménissier-de Murcia J, de Murcia G, Giocanti N, Mégnin-Chanet F, Favaudon V. Poly(ADP-ribose) polymerase, a major determinant of early cell response to ionizing radiation. Int. J. Radiat. Biol. 2000;76:1621–1629. doi: 10.1080/09553000050201118. [DOI] [PubMed] [Google Scholar]
- 7.Tartier L, Spenlehauer C, Newman HC, Folkard M, Prise KM, Michael BD, Ménissier-de Murcia J, de Murcia G. Local DNA damage by proton microbeam irradiation induces poly(ADP-ribose) synthesis in mammalian cells. Mutagenesis. 2003;18:411–416. doi: 10.1093/mutage/geg015. [DOI] [PubMed] [Google Scholar]
- 8.Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Ménissier-de Murcia J. Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 1998;26:1891–1898. doi: 10.1093/nar/26.8.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simbulan-Rosenthal CM, Rosenthal DS, Boulares AH, Hickey RJ, Malkas LH, Coll JM, Smulson ME. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry. 1998;37:9363–9370. doi: 10.1021/bi9731089. [DOI] [PubMed] [Google Scholar]
- 10.Oei SL, Herzog H, Hirsch-Kauffmann M, Schneider R, Auer B, Schweiger M. Transcriptional regulation and autoregulation of the human gene for ADP-ribosyltransferase. Mol. Cell Biochem. 1994;138:99–104. doi: 10.1007/BF00928449. [DOI] [PubMed] [Google Scholar]
- 11.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase-1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol. Res. 2005;52:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 14.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 15.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 16.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson SH, Hubscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 17.Pascucci B, Stucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J. Biol. Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- 18.Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 19.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 20.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noël G, Godon C, Fernet M, Giocanti N, Mégnin-Chanet F, Favaudon V. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1,8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Mol. Cancer Ther. 2006;5:564–574. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 24.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 25.Hennequin C, Giocanti N, Balosso J, Favaudon V. Interaction of ionizing radiation with the topoisomerase I poison camptothecin in growing V-79 and HeLa cells. Cancer Res. 1994;54:1720–1728. [PubMed] [Google Scholar]
- 26.Biard DS, Despras E, Sarasin A, Angulo JF. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: application to NER gene silencing. Mol. Cancer Res. 2005;3:519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 27.Biard DS. Untangling the relationships between DNA repair pathways by silencing more than 20 DNA repair genes in human stable clones. Nucleic Acids Res. 2007;35:3535–3550. doi: 10.1093/nar/gkm195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rockwell S. Effects of clumps and clusters on survival measurements with clonogenic assays. Cancer Res. 1985;45:1601–1607. [PubMed] [Google Scholar]
- 29.Filipski J, Kohn KW. Ellipticine-induced protein-associated DNA breaks in isolated L1210 nuclei. Biochim. Biophys. Acta. 1982;698:280–286. doi: 10.1016/0167-4781(82)90158-0. [DOI] [PubMed] [Google Scholar]
- 30.Balosso J, Giocanti N, Favaudon V. Additive and supraadditive interaction between ionizing radiation and pazelliptine, a DNA topoisomerase inhibitor, in Chinese hamster V-79 fibroblasts. Cancer Res. 1991;51:3204–3211. [PubMed] [Google Scholar]
- 31.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophot. Int. 2004;11:36–42. [Google Scholar]
- 32.Walter J, Cremer T, Miyagawa K, Tashiro S. A new system for laser-UVA-microirradiation of living cells. J. Microsc. 2003;209:71–75. doi: 10.1046/j.1365-2818.2003.01117.x. [DOI] [PubMed] [Google Scholar]
- 33.Hashiguchi K, Matsumoto Y, Yasui A. Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells. Nucleic Acids Res. 2007;35:2913–2923. doi: 10.1093/nar/gkm115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smulson M, Istock N, Ding R, Cherney B. Deletion mutants of poly(ADP-ribose) polymerase support a model of cyclic association and dissociation of enzyme from DNA ends during DNA repair. Biochemistry. 1994;33:6186–6191. doi: 10.1021/bi00186a018. [DOI] [PubMed] [Google Scholar]
- 39.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 42.Dantzer F, de La Rubia G, Ménissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 43.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 44.Pascucci B, Russo MT, Crescenzi M, Bignami M, Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005;33:280–288. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 46.Mourgues S, Lomax ME, O’Neill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ménissier de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez-Bendala J, Masutani M, McWhir J. Down-regulation of PARP-1, but not of Ku80 or DNA-PK(cs), results in higher gene targeting efficiency. Cell Biol. Int. 2006;30:389–393. doi: 10.1016/j.cellbi.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Aguilar Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta A, Valenzuela T, Martinez-Romero R, Quiles-Perez R, Ménissier de Murcia J, de Murcia G, Ruiz de Almodovar M, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia (ATM) signaling network is modulated by a novel PAR-dependent pathway in the early response to DNA damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 51.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 52.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2007;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 53.Noël G, Giocanti N, Fernet M, Mégnin-Chanet F, Favaudon V. Poly(ADP-ribose) polymerase (PARP-1) is not involved in DNA double-strand break recovery. BMC Cell Biol. 2003;4:7. doi: 10.1186/1471-2121-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 55.Huletsky A, de Murcia G, Muller S, Hengartner M, Ménard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J. Biol. Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 56.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 57.Ding R, Pommier Y, Kang VH, Smulson M. Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J. Biol. Chem. 1992;267:12804–12812. [PubMed] [Google Scholar]
- 58.Ding R, Smulson M. Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influences on genomic stability, chromatin organization, and carcinogen cytotoxicity. Cancer Res. 1994;54:4627–4634. [PubMed] [Google Scholar]