Abstract
Analysis of the human protein-tyrosine phosphatase (PTP) PTPRJ mRNA detected three in-frame AUGs at the 5′-end (starting at nt +14, +191 and +356) with no intervening stop codons. This tandem AUG arrangement is conserved between humans and the mouse and is unique among the genes of the classical PTPs. Until now it was assumed that the principal open reading frame (ORF) starts at AUG356. Our experiments showed that: (i) translation of the mRNA synthesized under the PTPRJ promoter starts predominantly at AUG191, leading to the generation of a 55 amino acid sequence preceding the signal peptide; (ii) the longer form is being likewise correctly processed into mature PTPRJ; (iii) the translation of the region between AUG191 and AUG356 inhibits the overall expression, a feature which depends on the sequence of the encoded peptide. Specifically, a sequence of 13 amino acids containing multiple arginine residues (RRTGWRRRRRRRR) confers the inhibition. In the absence of uORF these previously unrecognized characteristics of the 5′-end of the mRNA present a novel mechanism to suppress, and potentially to regulate translation.
INTRODUCTION
It has been noted that a wide variety of proteins, including some protein kinases, growth factors, oncogenes, receptors and transcription factors are expressed from messengers, which are poorly translated (1). The mRNAs of these proteins are characterized by a long 5′ leader (5′UTR) with high GC content, potentially strong secondary structure and the presence of short upstream open reading frames (uORFs). Recent genome wide analyses have revealed that uAUGs and uORFs are quite common (2,3). Generally the translation of these mRNAs follows the standard route for eukaryotes. The 43S scanning complex, composed of the 40S ribosomal subunit, Met-tRNAi and translation initiation factors, is attached to the m7G cap at the 5′-end of the mRNA. Unwinding the regions with secondary structure, the scanning proceeds towards the 3′-end, and when an AUG triplet in a favorable context is encountered, the 60S ribosomal subunit is recruited and translation initiates. The presence of an uORF impairs the translation of the principal reading frame as the ribosomes need to reinitiate at the downstream AUG. Alternatively, the sequence environment of the upstream AUG (uAUG) may diverge from the one which is optimal for recognition by the scanning complex [A/G]CCaugG (4). In this case, some of the 40S subunits will start translation at the uAUG, while others will continue scanning (‘leaky scanning’).
It is generally assumed that the role of the uORF is to secure low levels of expression of proteins which are harmful to the cell when abundant (5–7). In addition, regulatory functions of uORFs have also been identified, for example for the CAAT enhancer binding proteins alpha and beta and for the SCL transcription factor (8,9).
The receptor-like protein-tyrosine phosphatase J (PTPRJ, also designated DEP-1, CD148), a candidate tumor suppressor protein with potent anti-proliferative and anti-migratory activity, is differently expressed in different cell types and at different cell densities (10,11). By dephosphorylating yet only partially characterized cellular substrates it can interfere with signal transduction downstream of several growth factor receptors, and exerts anti-transforming activity in cancer cell lines of different origin (12–19). Therefore regulation of PTPRJ expression may represent an important level of controlling cellular tyrosine phosphorylation, and deregulation of expression may contribute to carcinogenesis. Although important, the basic mechanisms of PTPRJ expression regulation have not been explored until now.
Some of the structural features of the mRNAs discussed above are shared by the mRNA of PTPRJ, in particular a long, GC rich 5′ leader (GC 82%). However, this region contains three AUGs, all of which are in the reading frame of the main protein, with no intervening stop codons.
Experiments addressing the mechanisms of PTPRJ expression regulation showed that translation of the mRNA starts predominantly at AUG191, 55 codons upstream of the AUG356, the start of the signal peptide. We discovered properties of the 5′ leader, which have hitherto not been described in other genes. In the tandem arrangement of the in-frame AUGs, the codons between them are poorly translated, resulting in lower expression. These results uncover a previously unrecognized mechanism of suppressing and potentially regulating translation, which may be relevant not only to PTPRJ.
MATERIALS AND METHODS
Firefly luciferase reporter constructs
Reporter constructs with ATGLuc present. ‘In-frame’ (InF) and ‘Out-of-frame’ (OutF) constructs
The genomic region of human PTPRJ containing the putative promoter and the 5′ leader was amplified from BAC DNA (details in Supplementary Data and Supplementary Figure 1). The fragment (1762 bp, GenBank EF219146) was cloned into the NheI and BglII sites of pGL3-Basic Vector (Promega, Mannheim, Germany), which lacks eukaryotic promoter or enhancer. The construct p1.7_InF(pGL3) contained nucleotides from −1419 to +343 of PTPRJ (+1 is the transcription start site, NM_002843). The arrangement of the ATGs in this clone was the same as in PTPRJ—all three ATGs (ATG14, ATG191 and ATGLuc) were in one reading frame. Clone pNar_InF(pGL3) contained sequences −323 to +343. The pNar(17)_OutF clone was the same as pNar_InF(pGL3) but with additional 17 nucleotides in the region between +343 and ATGLuc. The additional nucleotides changed the reading frame. The clone with deleted 3′ region – pNar-Nar_InF(pGL3), contained sequences from −323 to +82. For details on all cloning steps, see Supplementary Data.
Constructs with PTPRJ fused to firefly luciferase
The construct pNar_Luc_Fused(pGL3) expressed the firefly luciferase fused to the first five N-end amino acids of PTPRJ (starting with AUG356). The construct was prepared by PCR and contained PTPRJ sequences from −323 to +370, followed by the firefly luciferase sequences coding amino acids 2–550. All mutations of the ATGs were performed by PCR and the respective clones were sequenced. ATG14 was mutated to TTG (Leu); ATG191 to AGG (Arg), and ATG356 to ATT (Ile).
Constructs with PTPRJ 5′ leader and luciferase transcribed from the CMV promoter
The constructs in pcDNA3.1(+) (Invitrogen, Karlsruhe, Germany) contained PTPRJ sequences from +171 to +370 followed by firefly luciferase codons from 2 to 550 and the stop codon. In these constructs, designated as (191–356)(Luc)(pcDNA3), the firefly luciferase was transcribed under the strong CMV promoter and translated from the AUG191 and AUG356 of PTPRJ.
Constructs with frame-shift mutations and with optimized codons
Frame-shift mutations in the region between +191 and +356 (without introducing a stop codon) were performed by PCR. We inserted one A (after +200) in the codon 4 (AUG191 is codon 1) or deleted one C (+340) in the 5th codon preceding AUG356. The distance between the frame shifts was 139 nt, and, when both frame-shift mutations are present in one template, 47 codons were altered. The optimization of codons for human translation and synthesis of the fragment was done by GENEART AG (Regensburg, Germany) by assembling synthetic oligonucleotides and PCR products, and was verified by DNA sequencing. For further details and sequences of the wild type region ATG191–ATG236, the double frame-shift, and the optimized codons, see the Supplementary Data and Supplementary Figure 4.
Constructs with deleted codons in the 3′-end of the region AUG191–AUG356
Constructs with deleted codons (vector pcDNA3.1) were created by site-directed mutagenesis. They contained sequences from +171 to +270 (codons 1–26), followed by four codons of the signal peptide and codons 2–550 of the firefly luciferase. In these clones, designated as (191-Δ)(Luc)(pcDNA3), the transcription is driven by the CMV promoter, and the translation by AUG191 (AUG356 is modified to CAU, encoding His). Two double frame-shift mutants were also constructed—one altering the sequence of codons 4 through 13, and the other altering the sequence of codons 4 through 26 (for the sequence of the wild type and the frame-shifted codons, see Supplementary Figure 5).
PTPPRJ cDNA clones
The human PTPRJ cDNA has been described previously (10); accession U10886. The cDNA was inserted in the EcoRI site of the expression vector pcDNA3 (Invitrogen, Kalsruhe, Germany) and contained at the C-end a HA tag (3×). Mutations of the ATGs and the double frame-shift mutation were prepared by PCR and verified by sequencing.
Cell culture, transfection and antibodies
HEK293 and HeLa cells cells were maintained in DMEM/F12 1:1 medium, and HCT116 cells in McCoy's medium, all supplemented with 10% FBS. All transfections were done with PEI (polyethylenimine, Aldrich, Cat. No. 40872-7, transfection protocol in Supplementary Data). Cells were lysed in buffer with 1% Triton X100, 20 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 5% glycerol, supplemented with protease inhibitors. Immunoprecipitation was performed with goat anti-firefly luciferase polyclonal antibody from Chemicon (Cat. No. AB3256, Millipore, Schwalbach, Germany) cross-linked to Protein G Sepharose beads with dimethylpimelimidate (DMP) according to standard procedures. To determine the synthesis of HA-tagged PTPRJ, HEK293 cells growing in 6-well plates were transfected with constructs in pcDNA3, together with a plasmid encoding EYFP-tagged SHP1 (20) for control of transfection efficiency. The anti-HA monoclonal antibody was from Cell Signaling Technology (cat. No 2362, Frankfurt, Germany) and the anti-SHP1 antibody was from Santa Cruz (sc-287, Heidelberg, Germany).
Determination of dual firefly—Renilla luciferase activity
To determine the expression of the firefly luciferase, HEK293 cells were grown in 96-well plates (flat bottom, clear, Greiner Bio-One, Frickenhausen, Germany, cat. No 655098) coated with 10 µg/ml poly-L-lysine (Sigma-Aldrich, Deisenhofen, Germany). Transfection (0.2 µg DNA per well) was with firefly luciferase constructs (or vector pGL3-Basic alone). To determine the efficiency of transfection and to normalize values plasmid DNA coding for Renilla luciferase was added (pRL-TK in ratio 1:16 with constructs in pGL3 or pRL-CMV in ratio 1:40 with constructs in pcDNA3.1. Both Renilla expressing plasmids are from Promega, Mannheim, Germany). After 24 h the cells were washed with PBS, lysed with Passive Lysis Buffer (Promega, Mannheim, Germany) and the dual-luciferase activity was measured (21). The expression of constructs was measured in at least three independent experiments (8–12 wells in each experiment). The values were normalized as a ratio of firefly to Renilla luciferase activity. Where indicated the fold increase of construct activity over the activity of the promoter-less pGL3 is presented.
Determination of RNA levels by Northern hybridization and real-time RT–PCR
HEK293 cells were transfected with constructs with PTPRJ 5′ leader and firefly luciferase transcribed from the CMV promoter (vector pcDNA3.1) together with pRL-CMV DNA (Promega, Mannheim, Germany) for normalization. Total RNA was isolated with the RNeasy Mini kit (Qiagen, Hilden, Germany), fractionated on 1% agarose-formaldehyde gels, blotted on Hybond N membranes (GE Healthcare, Freiburg, Germany) and hybridized to DIG-labelled PCR fragment in 20% SDS, 0.25 M sodium phosphate, 50% formamide buffer at 42° (22). For labelling details, see Supplementary Data. Detection of the hybrids on the blots was with anti-DIG POD Fab fragments (Roche, Penzberg, Germany) according to the manufacturers instructions.
For real-time RT–PCR, the RNA preparations were additionally treated with Amplification Grade DNase I (Invitrogen, Karlsruhe, Germany) according to the protocol of the manufacturer. The inactivation of the DNase I was checked on supercoiled plasmid DNA. cDNA was prepared by the SuperScript First-Strand Synthesis System for RT–PCR with oligo(dT)12–18 (Invitrogen, Karlsruhe, Germany). Amplification of the target cDNA was performed with the QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) using primers specific for firefly luciferase cDNA and Renilla luciferase cDNA. The level of residual contamination with plasmid template DNA was checked with primers specific for the beta-lactamase resistance gene (for primer sequences, see Supplementary Data).
RESULTS
The structural features of the first exon of PTPRJ are highly conserved
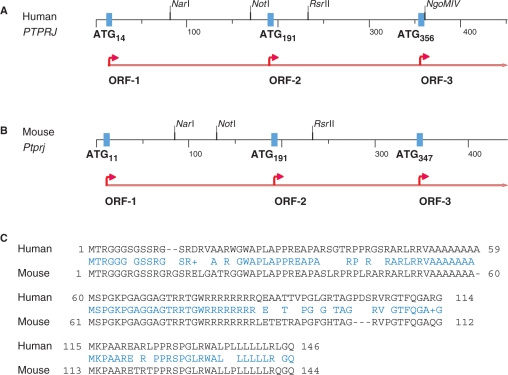
Examination of the 5′-end of PTPRJ mRNA (NM_002843) showed the presence of three AUGs which are in the reading frame of the main protein. Comparison of the first exon of the human PTPRJ and its mouse homolog (NM_008982) revealed that they share a common pattern of organization and several remarkable similarities (Figure 1A and B). First, the length of both exons is almost the same (451 nt human, mouse 442 nt); they code for the 5′ leader of the mRNA, and for 32 amino acids of the predicted signal peptide. Second, both exons contain three tandem AUGs in one reading frame, with no intervening stop codons. The relative position and the context of the AUGs are also conserved (Table 1). Third, in case translation starts at AUG14 (+14 to +16), or at AUG191 (+191 to +193), the conservation of the amino acid sequence is also high (77% identity, Figure 1C). The high degree of conservation in the PTPRJ 5′ leader, notably the conservation of the position of AUGs, their context and the encoded amino acids suggested that this arrangement might be of functional importance (for conservation at nucleotide sequence level, see Supplementary Figure 2).
Figure 1.
Pattern of organization of the first exon of the PTPRJ gene (A) in humans (NM_002843) and (B) in the mouse (NM_008982). The open reading frames are indicated. Note that ATG14 (ATG11), ATG191 (ATG191) and ATG356 (ATG347) are in-frame with no intervening stop codons, resulting in the same amino-acid sequence downstream of ATG356 (ATG347) when translation starts at any of them. (C) Conservation of the amino acid sequences; from top to bottom—the amino acid sequences starting from AUG14 (AUG11), from AUG191 (ATG191), and the 32 amino acids of the predicted signal peptide, starting from AUG356 (AUG347). Identical residues are in blue, ‘+’ represents conservative change.
Table 1.
Context of the AUG codons
Translation of the mRNA synthesized from the PTPRJ promoter may start at AUG356 as well as at AUG191
We cloned the genomic region of the human PTPRJ (1762 bp), spanning the transcription start site into the firefly luciferase reporter vector pGL3 (see Materials and Methods section). The reporter activity analysis of several 5′ deletion constructs showed that the PTPRJ core promoter is located within about 300 nucleotides upstream of the transcription start (Supplementary Figure 3).
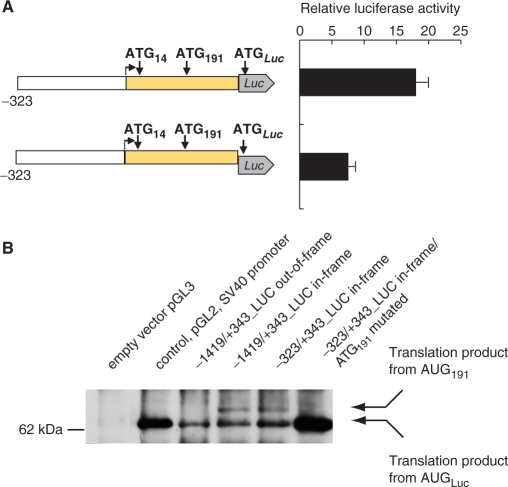
We went on to test the functional significance of the above described conserved AUG arrangement. First we tested the possibility of translation starting upstream of AUG356 (+356 to +358), the codon, which in the public database is marked as the translation start. For this we compared the activity of various constructs with different arrangement of ATG14, ATG191 and ATGLuc (the ATG of the firefly luciferase). The results showed invariably that the reporter activity was strongly reduced when the upstream AUGs (AUG14 and AUG191) were not in-frame with AUGLuc (Figure 2A). This demonstrated that initiation of translation takes place in the 5′ leader, at AUG191 (or at AUG14). This translation then proceeds not in the reading frame of the luciferase, thus preventing its expression. The residual reporter activity is presumably due to leaky scanning directed towards the ATGLuc, as both AUG14 and AUG191 are not in a perfect context (Table 1).
Figure 2.
In PTPRJ promoter/luciferase reporter constructs translation can start at AUG191 as well as at AUG356. (A) Inhibition of reporter activity when AUG191 and AUGLuc are not in-frame. Firefly luciferase reporter constructs were transiently transfected into HEK293 cells, and activity was measured (details in Materials and Methods section). Relative luciferase activity represents the ratio of firefly luciferase and Renilla luciferase activity (pRL-TK) normalized to empty vector controls (pGL3-Basic). The frame shift is indicated by a shifted firefly luciferase gene. (B) Synthesis of extended firefly luciferase from various constructs. HEK293 cells were transfected with the indicated constructs. Cell extracts were subjected to immunoprecipitation with anti-firefly luciferase antibodies (covalently coupled to beads), and subsequently immunoblotted with anti-firefly luciferase antibodies. Size marker, and the relative positions of the expected translation products are indicated.
Based on these observations, we tested directly the ability of AUG14 and AUG191 to initiate translation. First, we analyzed the length of the firefly luciferase synthesized by the reporter constructs. The use of AUG14, or AUG191 as start codons would give rise to longer translation products than translation starting at the AUGLuc. HEK293 cells were transfected with different constructs, lysed after 24 h, luciferase was immunoprecipitated and analyzed by western blotting. As shown in Figure 2B, two luciferase proteins were synthesized when the AUGs in the reporter were in-frame. The sizes of the luciferase bands corresponded to one product synthesized from AUGLuc (63 kDa), and another synthesized from AUG191 (69 kDa). This directly indicates that AUG191 initiates translation and that scanning is leaky. If AUG191 was not in tandem with AUGLuc only the synthesis of firefly luciferase with a size of 63 kDa was supported. AUG14 appeared to be largely inactive in these constructs, since a protein product of 76 kDa, corresponding to translation starting at AUG14 could not be detected.
Comparison of the amounts of protein with low and higher molecular mass, shows prevalence of the 63 kDa products (starting presumably at ATGLuc). This is in discrepancy with results presented on Figure 2A, which indicated robust translation starting upstream of ATGLuc. We considered that low abundance of the form with higher molecular mass might be caused by differential immunoprecipitation and/or by high susceptibility to protease degradation of the protruding N-tail of the extended protein. This seemed to be indeed the case, as further analysis of luciferase translation products directly in cell lysates revealed (Figure 7C).
Figure 7.
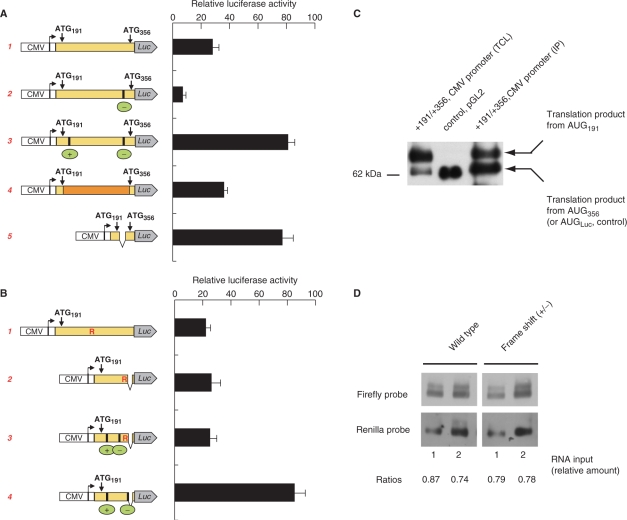
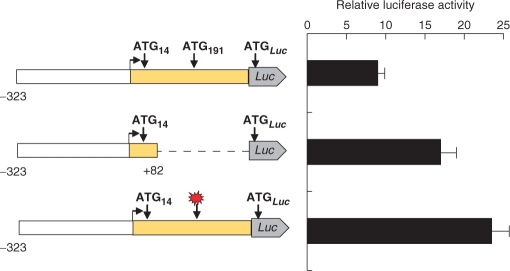
Activity of reporter firefly luciferase constructs driven by the CMV promoter. (A) 5′ truncated sequence of the PTPRJ 5′ leader containing ATG191 and ATG356 was fused to the firefly luciferase (CMV promoter from vector pcDNA3.1). In the region between ATG191 (codon 1) and ATG356 (codon 56) (from top to bottom): 1, the nucleotide and the encoded amino-acid sequence was left unchanged (wild type); 2, single nucleotide at codon 51 was deleted (−); 3, the amino acid sequence was altered by the indicated frame-shift mutations caused by inserting a single nucleotide at codon 4, and deleting a single nucleotide at codon 51. Note that the combined frame-shift mutations alter the amino-acid sequence of the translation product between the mutations, but the reading frame of the luciferase is maintained and luciferase can be produced. 4, A synthetic nucleotide sequence encoding the wild type amino acids, but with optimized codons was inserted between ATG191 and ATG356; 5, the region between codons 4 and 51 was deleted without altering the reading frame (for construct sequences, see Supplementary Figure 4). The firefly luciferase reporter activity was normalized to Renilla luciferase expression also driven by a CMV promoter. (B) 5′ truncated sequence of the PTPRJ leader containing ATG191 was fused to the firefly luciferase (ATG356 was modied to CAT). In the region between ATG191 and firefly luciferase (from top to bottom): 1, the nucleotide and the encoded amino acid sequence was left unchanged (wild type); the position of the arginine stretch is indicated (R). 2, the leader sequence was 3′ truncated after codon 26 (Arg), the reading frame was maintained; 3, the amino acid sequence of the region was altered by frame-shifts at codons 4 and 13. 4, the amino acid sequence of the region was altered by frame-shifts at codons 4 and 27 (for construct sequences, see Supplementary Figure 5). (C) Size of firefly luciferase products synthesized after transfection of HE293 cells with (191–356)Luc(pcDNA3) construct [construct 1, in (A)]. The sizes of the products in total cell lysates (TCL) before and after immunoprecipitation (IP) were compared to the size of control luciferase coded by pGL2 (Promega). (D) Reporter mRNA levels in HEK 293 cells transfected with firefly luciferase fusion constructs (vector pcDNA3.1) with PTPRJ wild type or frame-shifted leader sequence as shown in (A). Total RNA isolated after transfection was loaded in two lanes (amount of RNA differed by factor of two), separated, blotted and hybridized with a DIG-labelled firefly luciferase DNA probe. To normalize, a Renilla luciferase expression construct (CMV promoter) was cotransfected, the blot was stripped and reprobed with a DIG-labelled Renilla DNA fragment (see Materials and Methods section).
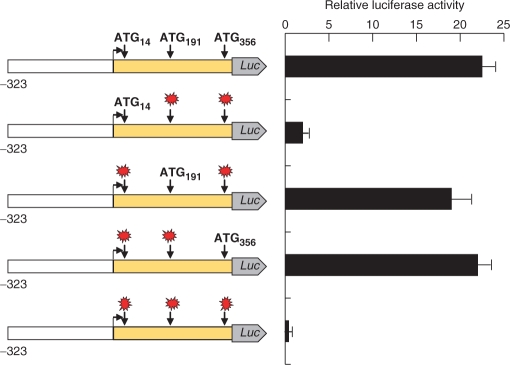
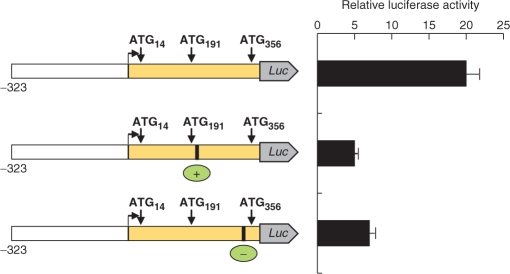
To estimate the relative ability of each AUG codon in the PTPRJ mRNA to sustain translation, we designed a fusion construct, containing all three 5′-end ATGs. This construct contained (from 5′ to 3′) the PTPRJ core promoter (−323 to −1), the 5′ leader (+1 to +355), and the sequence coding for the first five amino acids of the signal peptide (+356 to +370). This sequence was fused to the sequence of the firefly luciferase (codons 2–550) (see Materials and Methods section). Mutations were introduced to disrupt two AUGs at a time, leaving one AUG intact, and the constructs were subjected to expression analysis. As shown in Figure 3, translation can start at any of the AUG codons, albeit with different efficiency. Translation from AUG14 was the weakest, apparently due to its poor sequence context (Table 1), and the short distance from the m7G cap. The potential of AUG191 or AUG356 to drive expression of reporter luciferase seems approximately equal. Our further results showed that translation starting at AUG191 is attenuated. The lack of marked difference in expression starting from AUG191 or AUG356, implies that AUG356 is in a weaker sequence context; more ribosomes start at AUG191, but the overall translation is not higher.
Figure 3.
Contribution of the different AUGs to the activity of reporter constructs containing the PTPRJ promoter and the 5′ leader, fused to the firefly luciferase. Activity of the fusion constructs (vector pGL3, core PTPRJ promoter) with point mutations in the ATGs (indicated by asterisks). Relative luciferase activity represents the ratio of firefly luciferase and Renilla luciferase activity (pRL-TK) normalized to empty vector controls (pGL3-Basic).
Translation starting at AUG191 results in a correctly processed protein
PTPRJ is a receptor-like protein-tyrosine phosphatase (mature protein 1302 amino acids, calculated molecular mass 150 kDa). It contains a single intracellular catalytic domain, a single transmembrane domain, and a number of fibronectin type III repeats in its extracellular domain. An N-end signal peptide of 35 amino acids has been predicted (10). The mature PTPRJ is heavily glycosylated, and migrates as a 180–200 kDa protein.
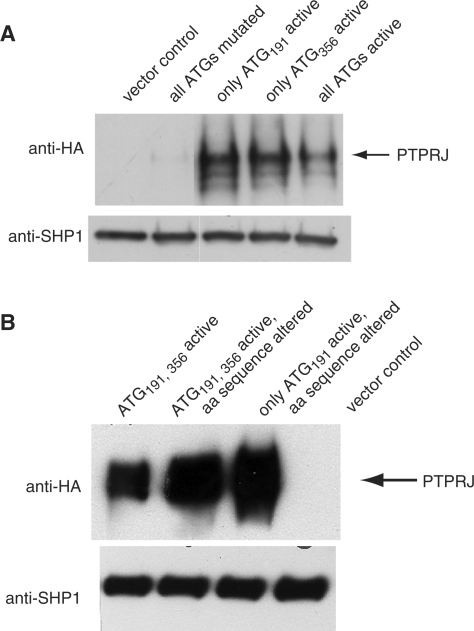
We investigated the possibility of PTPRJ synthesis from the different AUGs in the 5′ leader, employing HA-tagged PTPRJ cDNA constructs in pcDNA3. The ATGs in the cDNA were point mutated, leaving either AUG191 or AUG356 intact. The results again showed (Figure 4A) that both AUG191 and AUG356 have the capacity to support translation. The mobility of PTPRJ synthesized from AUG191 or from AUG356 is the same, showing a similar degree of glycosylation. The protein bands are fairly broad, and bands of lesser size are also detectable, reflecting the different extent of glycosylation of protein molecules present in the cell. Apparently the presence at synthesis of additional 55 amino acids at the N-end does not prevent correct processing of PTPRJ, which involves binding of the signal peptide to the recognition particle, the subsequent correct translocation into the ER lumen, cleavage of the signal peptide, and glycosylation. It was of interest to know whether the specific sequence of the additional amino acids at the N-end of PTPRJ may play a role in the process of signal recognition. To test this we designed an expression construct with two frame-shift mutations (plus and minus) in the region between AUG191 and AUG356. These combined mutations change the amino-acid sequence upstream of AUG356, but leave the reading frame for the signal peptide and the mature PTPRJ protein intact. Judging from mobility (Figure 4B), processing and protein glycosylation of PTPRJ were not severely affected by this manipulation. We conclude that the additional amino acids preceding the signal peptide are compatible with correct processing,
Figure 4.
Expression of HA tagged PTPRJ with different 5′ leader sequences driven by the CMV promoter in pcDNA3. HEK293 cells were transiently transfected with the indicated constructs expressing PTPRJ with HA-epitope at the C-terminus. To normalize for transfection efficiency, co-transfection with hSHP-1 expression plasmid was performed (SHP-1 is not expressed endogenously in HEK293 cells). Cell extracts were subjected to SDS–PAGE and immunoblotting using anti-HA antibodies and anti-hSHP-1 antibodies. (A) cDNA constructs with differentially inactivated ATGs were compared, as indicated. All lanes were on the same blot with identical exposure and image processing, but rearranged for better clarity. (B) The role of the amino-acid sequence downstream of ATG191 was tested by altering the sequence between ATG191 and ATG356.
Translation starts predominantly at AUG191
The results on the expression of PTPRJ from the cDNA constructs as well as on the expression of the luciferase reporters showed that translation could start at AUG191, as well as at AUG356. Further experiments were performed to estimate what fraction of scanning complexes would start translation from AUG191. For this we again introduced single nucleotide frame-shift mutations (one insertion or one deletion) positioned in the region between AUG191 and AUG356. These frame-shift mutation deviate the translation starting at AUG191 into a reading frame, different from that of the luciferase. In this case, it is reasonable to assume that luciferase can be produced only by initiation of translation at AUG356 (resulting from leaky scanning at AUG191). Both frame-shift mutations caused a severe reduction of reporter activity, indicating that more than two-third of the scanning complexes start translation at AUG191 (Figure 5). The prevalence of translation from AUG191 was confirmed by analyzing the translation products by immunoblotting of the lysates of transfected cells (see section ‘Translation of the codons between AUG191 and AUG356 slows down expression’).
Figure 5.
Preferred initiation of translation at ATG191. Wild-type construct, construct with plus frame shift mutation (insertion of a single nucleotide at codone 4; AUG191is codone 1), and a construct with minus frame-shift mutation (deletion of a single nucleotide in codon 51) were transiently transfected and luciferase activity was determined (vector pGL3, core PTPRJ promoter). Relative luciferase activity represents the ratio of firefly luciferase and Renilla luciferase activity (pRL-TK) normalized to empty vector controls (pGL3-Basic).
Attenuation of PTPRJ expression by a sequence downstream of AUG191
The highly conserved features of the 5′-end of the PTPRJ mRNA, and the start of translation from the AUG191 suggest a functional significance of this region of the messenger. The 5′-end of the mRNA is very GC-rich (82%) and highly structured. The predicted secondary structure of nucleotides +1 to +358 with overall free energy dG = −186.70 (23) is presented in Supplementary Figure 6. To investigate the role of this region, we prepared a deletion mutant missing nucleotides from +83 to +343, eliminating part of the structured 5′ leader as well as the conserved ATG191. The results showed that the reporter activity of this construct is increased (Figure 6). We considered the possibility that the effect of the +83 to +343 deletion is solely due to loss of secondary structure, making scanning easier. To test this, ATG191 was eliminated by point mutation (to AGG, coding Arg). As supposed the single nucleotide change did not affect significantly the RNA secondary structure (dG = −183.10).
Figure 6.
Inhibition of expression by translation initiating at AUG191. Firefly luciferase reporters (vector pGL3, core PTPRJ promoter) with non-mutated sequence, with deletion downstream of +84 and with point mutated AUG191 (indicated by an asterisk) were transiently transfected and activity was compared. Relative luciferase activity represents the ratio of firefly luciferase and Renilla luciferase activity (pRL-TK) normalized to empty vector controls (pGL3-Basic).
Unexpectedly, however, this mutation increased reporter activity as it was the case with the deletion (Figure 6). To explain these results we made the following assumption, which we tested further. Translation of the codons that follow AUG191 may not be efficient, presumably leading to pausing of translating ribosomes and lowering the total expression. Thereby scanning through this region may be more favorable than translation itself. In case the translation from AUG191 is abolished, the scanning complexes continue unrestrained towards the downstream AUG.
Translation of the codons between AUG191 and AUG356 slows down expression
To investigate further the causes for the inefficient translation downstream of AUG191, and to exclude that the effects are promoter-specific, we generated several reporters in the pcDNA3 vector. The constructs downstream of the CMV promoter contained PTPRJ sequences from +171 to +370, which encode ATG191, the downstream region (wild type or modified), ATG356, plus the codons of the next 4 amino acids of PTPRJ, fused to the codons of the firefly luciferase for amino acids 2–550 (see Materials and Methods section). In these reporters the luciferase mRNA is transcribed from the strong cytomegalovirus promoter, however, translation is dependent on the tandem AUG191 and AUG356 (and the region between them). We compared the activity of several constructs: the wild-type construct, a construct with one frame-shift mutation, a construct in which the sequence of 47 amino acids was altered by introducing two point mutations, of which the first one altered, and the second one restored the reading frame, and a construct in which these 47 amino acids were deleted (Supplementary Figure 4). As expected, the activity of the construct with one frame-shift mutation was low, again demonstrating that translation initiation starts predominantly at AUG191. As shown in Figure 7A, the reporter activity increased 2–3-fold when the amino acid sequence of the region between AUG191 and AUG356 was altered or deleted. This indicated that translation of the codons following AUG191 reduces the overall expression of the reporter. The similar level of expression of these two constructs (3 and 5) shows additionally that the elimination of the region with potentially strong secondary structure has no great effect on translation.
The luciferase fusion constructs under the CMV promoter were also employed to test the length of the produced luciferase proteins by immunoblotting. Transient transfection with CMV promoter-driven constructs produced enough reporter luciferase to be detected by immunoblotting directly in the cell lysates (as opposed to products driven by the core PTPRJ promoter, see section ‘The region between AUG191 and AUG356 is difficult to translate’). The results presented in Figure 7C show that the protein with higher molecular mass, corresponding to translation from AUG191 is preferentially produced. However, after immunoprecipitation the ratio of products is changed, disfavoring the form with higher molecular mass. Presumably either immunoprecipitation of this form is inefficient, or during immunoprecipitation degradation of the additional amino acids takes place (see also Figure 2B).
We also checked the levels of the construct mRNAs in the cell. As detected by Northern blotting, mRNA levels were not affected by introduction of the two point frame-shift mutations (Figure 7D). Consistent with these data, real-time RT-PCR revealed no significant changes of the mRNA levels (Mean Ct firefly luciferase—25.1, and 25.2, control Renilla luciferase—26.3, and 26.4 for transfection with the wild type and the double frame-shift mutant, respectively). Thus, the increase in activity by altering the amino-acid sequence can be attributed solely to changes in translation efficiency, and not to accompanying changes in mRNA synthesis or stability.
One possibility for the increase in reporter activity would be the presence of rare codons in the reading frame, which were eliminated by the frame-shift. Examples of translation inhibition by the occurrence of rare codons have been reported for viral genes, expressed in mammalian cells (24,25). Inspection of the PTPRJ sequence from +191 to +356 showed a marked codon bias (see Supplementary Figure 4). Out of 55 codons, 14 codons are rare (http://www.kazusa.or.jp/codon/). To test the possible relevance of rare codons for inefficient translation, we used a reporter with unchanged amino acids, but the nucleotides between ATG191 and ATG356 were replaced with synthetic DNA encoding the same amino acids but with codons optimized for human expression (see Materials and Methods section, and Supplementary Figure 4). At the same time the overall secondary structure of the region was left largely unchanged (for the 1–284 nt of the mRNA transcribed from the pcDNA3 constructs dG = −112.83 for the wild type and dG = −104.01 for the mRNA with optimized codons). The results showed, however, that ‘optimizing’ the codons by replacing the rare codons leads only to modest increase in reporter activity (note the similarity in reporter expression of constructs 1 and 4, Figure 7A).
Another possible reason for inefficient translation could be the properties of the nascent peptide itself. The activity of the reporter construct encoding an altered sequence of a stretch of 47 amino acids was clearly elevated (compare activity of constructs 1 and 3, Figure 7A). Deletion of these amino acids also resulted in enhanced luciferase expression (construct 5, Figure 7A). It was of importance to know which region of the nascent 55 amino-acid peptide would contribute specifically to the translation attenuation. To narrow this region, we prepared a construct encoding only the 26 N-terminal amino acids of the presumably inhibitory peptide (Supplementary Figure 5). This construct showed low luciferase reporter activity similar to that of the construct containing the entire 55 amino-acid sequence (Figure 7B). This indicated that the down-modulation is a property of the N-terminal part of the sequence encoded downstream of AUG191.
To define more precisely the amino-acid sequence responsible for translation attenuation we changed the sequence of 10 amino acids, starting with codon 4 and preceding the arginine stretch. This modification did not lead to significant increase in luciferase expression (Figure 7B and Supplementary Figure 5). However, changing the codons from 4 through 26, again by introducing two frame-shift mutations (plus and minus) resulted in an increase of reporter activity as it was observed with the altered full-length 55 amino-acid sequence (Figure 7B, construct 4 and Figure 7B, construct 3).
These results indicate that translation attenuation is caused primarily by only 13 amino acids, which include a homopolymer stretch of eight arginines, encoded by codons 19–26.
Taken together, the use of AUG191 results in inefficient translation of the PTPRJ mRNA. The impairment is caused presumably by poorly translatable codons between AUG191 and AUG356, and more specifically by codons 14–26.
Cell specific and cell-density dependent differences in expression driven by the tandem AUGs are not due to differences in translation attenuation
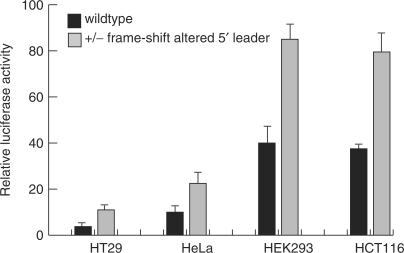
The existence of a poorly translated region at the 5′-end of mRNA, which can diminish expression, provides a possibility for translation control. To test for possible regulation we checked the expression of some of our reporter constructs in several cell lines, and at different cell densities. As shown in Figure 8, luciferase expression in the different cell types was rather different, with low expression in HT29 cells, intermediate expression in HeLa cells, and high expression in HEK293 and HCT116 cells. It should be emphasized that the increased expression in some cell lines cannot be explained by differential promoter activity as the reporter firefly luciferase and the control Renilla luciferase in these experiments are expressed under the same CMV promoter. However, the ratios of expression from wild type and mutated reporter constructs were not altered significantly. Therefore, while our data suggest differences in efficiency of initiation of translation at AUG191 in the different cell lines, the translation attenuation by the sequence encoded between AUG191 and AUG356 can be observed in all of them, indicating the generality of this mechanism for down-regulation. Since PTPRJ expression is elevated in some cell lines at high culture densities, we also explored the possibility that changes in translation efficiency would contribute to this phenomenon. These experiments revealed that, while reporter activity was clearly increased in cell cultures at high density, the attenuation of translation from AUG191 is still apparent in dense cells (Supplementary Figure 7). This finding indicates that upregulation of PTPRJ expression in dense cells is not caused by a release from attenuated translation.
Figure 8.
Translational repression by the PTPRJ 5′ leader sequence in different cell lines. The CMV-promoter driven constructs 1 (wildtype) and 3 (+/− frame-shifted 5′ leader), depicted in Figure 7A, were transfected in the indicated cell lines and reporter activity was measured relative to a cotransfected pRL-CMV Renilla luciferase expression construct.
DISCUSSION
PTPRJ mRNA translation starts mainly at AUG191
The GC-rich 5′ region of the PTPRJ mRNA is highly conserved among mammals. We were particularly attracted by the observation that a tandem arrangement of in-frame AUGs present in the leader sequence in the first exon is highly conserved between the human and the mouse (Figure 1). Therefore, we explored the putative regulatory importance of this arrangement. It should be noted that many of the 5′UTRs of the human PTPs contain uORFs with yet unexplored relevance for expression regulation. However none of them contains a similar tandem AUG arrangement. Only in case of hCD45/PTPRC an in-frame upstream AUG was detected, which was, however, only two codons apart from the proposed translation start site (for details, see Supplementary Tables 1 and 2). Nevertheless, the tandem AUG arrangement is unlikely to be unique to PTPRJ. A recent search of available cDNA human and mouse sequences has revealed that some of the conserved AUG codons in the 5′ leader are not followed by stop codons and are in the same reading frame as the main protein (3). The relevance of such tandem arrangements has, however, to the best of our knowledge not yet been studied.
Our experiments showed that AUG191 is the preferred starting codon in the context of the endogenous promoter. Still, apparently not all ribosomes seem to recognize it (leaky scanning). The least active starting codon is AUG14 presumably due to its proximity to the m7G cap and its unfavorable context (Table 1).
AUG191 will direct the synthesis of the same PTPRJ mature protein of 1302 amino acids, but the N-end of (Pro-) PTPRJ is supposed to be 55 amino acids longer than the previously predicted precursor. This has apparently no effect on the functioning of the PTPRJ as our results showed that the N-end extended protein is glycosylated in the same way as the PTPRJ, which starts directly with a signal peptide of 35 amino acids (Figure 2B). Apparently, in each form the N-end region of the protein is recognized by the signal recognition particle, cleaved by the signal peptide peptidases, the mature protein being correctly glycosylated. Moreover, our results indicate that the exact sequence of the additional 55 amino acids is not a precondition for the protein maturation. These results are in agreement with previous data showing that the signal peptide retains its function when situated downstream of the N-end of a protein (26).
The region between AUG191 and AUG356 is difficult to translate
The analysis of the activity of the various reporter constructs indicated that the region downstream of AUG191 is not efficiently translated, and translation of the 55 codons at the N-end seems to be a rate-limiting step in the overall protein expression. The alteration of 47 codons of the same region by shifting and restoring the reading frame resulted in marked increase in reporter activity. Importantly, this effect on translation does not depend on the promoter. Essentially the same results were obtained when transcription was driven not by the endogenous PTPRJ promoter, but by the strong cytomegalovirus promoter. Only a modest increase in the expression was observed when the wild-type sequence was replaced by a synthetic fragment encoding the same amino acids, but with ‘optimized’ codons. This suggests that the presence of rare codons is not the primary reason for the observed translation inhibition. Moreover, our results showed that the attenuation of translation depends more specifically on 13 amino acids at the N-end of the protein. Based on these findings we propose that the translation inhibition is due to the sequence of the nascent peptide. Examples of translation regulation through interaction between the nascent peptides and the ribosome are well known for prokaryotes (27). The translation inhibition exerted in cis- by some uORF in eukaryotes depends also on their amino-acid sequence. In these cases, according to the accepted model, the nascent uORF encoded peptide interacts with the ribosome and prevents its release at the uORF termination codon (5,7). In the mRNA of the β2 adrenergic receptor, the uORF encodes a peptide which supposedly interacts with the messenger, and thereby inhibits translation (28). For mammalian genes, to the best of our knowledge, observations of regulatory peptides encoded by domains within the main coding sequence have not yet been reported. In Arabidopsis, however, a stretch of 11–13 amino-acid residues, located 80 residues from the N-terminus of the cystathionine gamma-synthetase CGS1 gene, causes a nascent peptide-mediated translation elongation arrest most likely at the step of ribosome translocation (29).
We do not know yet the exact mechanism by which the nascent stretch of 13 amino acids encoded by the PTPRJ 5′ leader causes translational attenuation. Several mechanisms are feasible. One is the interaction of the nascent residues with the ribosome, interfering either with some of the elongation steps (the peptidyl transfer and the translocation are candidates) or with the exit of the polypeptide from the ribosome through the exit tunnel (30–32). Another possibility is an elongation arrest by interaction of the nascent chain with mRNA as proposed for the peptide synthesized by the uORF of the β2 adrenergic receptor mRNA. It is relevant to note that the only general feature of the nascent peptides interacting with the ribosome is that they are not acidic, some being highly basic (27). The amino acids which are involved in translation attenuation of PTPRJ comprise a conserved stretch of 13 residues (RRTGWRRRRRRRR) containing 10 Arg residues. It is tempting to speculate that the properties of this stretch of amino acids are important for inefficient translation by binding to the ribosome or the mRNA. The inhibiting peptide of the β2 adrenergic receptor mRNA has revealed the critical role of three consecutive arginine residues for inhibition (28). In Escherichia coli the translation of regions particularly rich in arginine is impaired (33). Still another reason for impeded translation of the primary ORF could be the existence of significant ribosome frameshifting, which might possibly take place along sequences between AUG191 and AUG356 (for examples of +1 and −1 frameshifting in eukaryotes see http://recode.genetics.utah.edu/).
Attenuating the translation of PTPRJ may be important to maintain the cellular levels of a critical regulatory and possibly harmful protein low. Potentially, it could also present the basis for regulation, assuming that the translation constraint can be overcome in certain differentiation states or under certain conditions of cell stimulation. While our experiments showed a marked cell specific degree of expression of the reporter constructs (Figure 8), they indicated that attenuation of translation via the described mechanism operates in a similar manner in all analyzed cell lines. Also, we did not yet observe differences in the extent of translational repression at different cell densities, or in cells stimulated with a variety of growth factors or cytokines (Supplementary Figure 7, and R. Godfrey, unpublished data). Further work is required to address the possibility that expression can be modulated by a mechanism involving the tandem AUGs and the translational repression by the specified stretch of amino acids. Alternatively, the described inhibition of translation may be constitutive and merely serve to achieve low levels of PTPRJ protein, thereby permitting a more effective expression control by other mechanisms, such as transcriptional activation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants to F.D.B. from the DFG SFB604 (A1), BMBF FKZ 01EA0103, DFG Bo 1043/7-1, and the EC training network MRTN-CT-2006-035830. We thank Dr Cornelis Calkhoven for discussion, and Susann Müller and Petro Zhupanyn for generation of some constructs. Funding to pay the Open Access publication charges for this article was provided by a LOM grant of the Medical Faculty.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kozak M. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1991;1:117–125. [PMC free article] [PubMed] [Google Scholar]
- 2.Crowe ML, Wang XQ, Rothnagel JA. Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides. BMC Genomics. 2006;7:16. doi: 10.1186/1471-2164-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol. Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachs MS, Geballe AP. Downstream control of upstream open reading frames. Genes Dev. 2006;20:915–921. doi: 10.1101/gad.1427006. [DOI] [PubMed] [Google Scholar]
- 8.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 9.Calkhoven CF, Müller C, Martin R, Krosl G, Pietsch H, Hoang T, Leutz A. Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev. 2003;17:959–964. doi: 10.1101/gad.251903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Östman A, Yang Q, Tonks NK. Expression of DEP-1, a receptor-like protein-tyrosine-phosphatase, is enhanced with increasing cell density. Proc. Natl Acad. Sci. USA. 1994;91:9680–9684. doi: 10.1073/pnas.91.21.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Autschbach F, Palou E, Mechtersheimer G, Rohr C, Pirotto F, Gassler N, Otto HF, Schraven B, Gaya A. Expression of the membrane protein tyrosine phosphatase CD148 in human tissues. Tissue Antigens. 1999;54:485–498. doi: 10.1034/j.1399-0039.1999.540506.x. [DOI] [PubMed] [Google Scholar]
- 12.Kovalenko M, Denner K, Sandström J, Persson C, Gross S, Jandt E, Vilella R, Böhmer F, Östman A. Site-selective dephosphorylation of the platelet-derived growth factor beta-receptor by the receptor-like protein-tyrosine phosphatase DEP-1. J. Biol. Chem. 2000;275:16219–16226. doi: 10.1074/jbc.275.21.16219. [DOI] [PubMed] [Google Scholar]
- 13.Palka HL, Park M, Tonks NK. Hepatocyte growth factor receptor tyrosine kinase met is a substrate of the receptor protein-tyrosine phosphatase DEP-1. J. Biol. Chem. 2003;278:5728–5735. doi: 10.1074/jbc.M210656200. [DOI] [PubMed] [Google Scholar]
- 14.Jandt E, Denner K, Kovalenko M, Östman A, Böhmer FD. The protein-tyrosine phosphatase DEP-1 modulates growth factor-stimulated cell migration and cell-matrix adhesion. Oncogene. 2003;22:4175–4185. doi: 10.1038/sj.onc.1206652. [DOI] [PubMed] [Google Scholar]
- 15.Kappert K, Paulsson J, Sparwel J, Leppanen O, Hellberg C, Östman A, Micke P. Dynamic changes in the expression of DEP-1 and other PDGF receptor-antagonizing PTPs during onset and termination of neointima formation. FASEB J. 2007;21:523–534. doi: 10.1096/fj.06-6219com. [DOI] [PubMed] [Google Scholar]
- 16.Keane MM, Lowrey GA, Ettenberg SA, Dayton MA, Lipkowitz S. The protein tyrosine phosphatase DEP-1 is induced during differentiation and inhibits growth of breast cancer cells. Cancer Res. 1996;56:4236–4243. [PubMed] [Google Scholar]
- 17.Trapasso F, Iuliano R, Boccia A, Stella A, Visconti R, Bruni P, Baldassarre G, Santoro M, Viglietto G, Fusco A. Rat protein tyrosine phosphatase eta suppresses the neoplastic phenotype of retrovirally transformed thyroid cells through the stabilization of p27(Kip1) Mol. Cell. Biol. 2000;20:9236–9246. doi: 10.1128/mcb.20.24.9236-9246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Trapasso F, Yendamuri S, Dumon KR, Iuliano R, Cesari R, Feig B, Seto R, Infante L, Ishii H, Vecchione A, et al. Restoration of receptor-type protein tyrosine phosphatase eta function inhibits human pancreatic carcinoma cell growth in vitro and in vivo. Carcinogenesis. 2004;25:2107–2114. doi: 10.1093/carcin/bgh224. [DOI] [PubMed] [Google Scholar]
- 19.Balavenkatraman KK, Jandt E, Friedrich K, Kautenburger T, Pool-Zobel BL, Östman A, Böhmer FD. DEP-1 protein tyrosine phosphatase inhibits proliferation and migration of colon carcinoma cells and is upregulated by protective nutrients. Oncogene. 2006;25:6319–6324. doi: 10.1038/sj.onc.1209647. [DOI] [PubMed] [Google Scholar]
- 20.Biskup C, Böhmer A, Pusch R, Kelbauskas L, Gorshokov A, Majoul I, Lindenau J, Benndorf K, Böhmer FD. Visualization of SHP-1-target interaction. J. Cell Sci. 2004;117:5165–5178. doi: 10.1242/jcs.01397. [DOI] [PubMed] [Google Scholar]
- 21.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 22.Engler-Blum G, Meier M, Frank J, Muller GA. Reduction of background problems in nonradioactive northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhao KN, Gu W, Fang NX, Saunders NA, Frazer IH. Gene codon composition determines differentiation-dependent expression of a viral capsid gene in keratinocytes in vitro and in vivo. Mol. Cell. Biol. 2005;25:8643–8655. doi: 10.1128/MCB.25.19.8643-8655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedmann M, Huth A, Rapoport TA. Internally transposed signal sequence of carp preproinsulin retains its functions with the signal recognition particle. FEBS Lett. 1986;194:139–145. doi: 10.1016/0014-5793(86)80065-5. [DOI] [PubMed] [Google Scholar]
- 27.Lovett PS, Rogers EJ. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J. Biol. Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 29.Onouchi H, Nagami Y, Haraguchi Y, Nakamoto M, Nishimura Y, Sakurai R, Nagao N, Kawasaki D, Kadokura Y, Naito S. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005;19:1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 31.Nakatogawa H, Ito K. Intraribosomal regulation of expression and fate of proteins. Chembiochem. 2004;5:48–51. doi: 10.1002/cbic.200300751. [DOI] [PubMed] [Google Scholar]
- 32.Mankin AS. Nascent peptide in the ‘birth canal’ of the ribosome. Trends Biochem. Sci. 2006;31:11–13. doi: 10.1016/j.tibs.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Gerchman SE, Graziano V, Ramakrishnan V. Expression of chicken linker histones in E. coli: sources of problems and methods for overcoming some of the difficulties. Protein Expr. Purif. 1994;5:242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.