Abstract
APE1/Ref-1 is thought to be a multifunctional protein involved in reduction–oxidation (redox) regulation and base excision DNA repair, and is required for early embryonic development in mice. APE1/Ref-1 has redox activity and AP endonuclease activity, and is able to enhance DNA-binding activity of several transcription factors, including NF-κB, AP-1 and p53, through reduction of their critical cysteine residues. However, it remains elusive exactly how APE1/Ref-1 carries out its essential functions in vivo. Here, we show that APE1/Ref-1 not only reduces target transcription factors directly but also facilitates their reduction by other reducing molecules such as glutathione or thioredoxin. The new activity of APE1/Ref-1, termed redox chaperone activity, is exerted at concentration significantly lower than that required for its redox activity and is neither dependent on its redox activity nor on its AP endonuclease activity. We also show evidence that redox chaperone activity of APE1/Ref-1 is critical to NF-κB-mediated gene expression in human cells and is mediated through its physical association with target transcription factors. Thus, APE1/Ref-1 may play multiple roles in an antioxidative stress response pathway through its different biochemical activities. These findings also provide new insight into the mechanism of intracellular redox regulation.
INTRODUCTION
The reduction–oxidation (redox) states of cysteine residues, which can change reversibly within cells, often greatly influence the various properties of proteins, such as protein stability, chaperone activity, enzymatic activity and protein structure (1–5). It has been shown that the redox states of cysteine residues of several transcription factors, most of which are located in the DNA-binding domains, affect their DNA-binding activity through redox mechanisms (6–18). For example, a cysteine residue in the highly conserved Rel homology DNA-binding domain (RHD) of NF-κB, such as Cys-62 of p50, needs to be in a reduced state for efficient DNA binding (19–22). We have shown in a previous report that Cys-62 of p50 is highly oxidized in the cytoplasm, and that after stimulation, it is converted to the reduced form in the nucleus to gain DNA-binding activity (23). Thus, redox regulation is an important mode of regulation for several transcription factors.
Glutathione (GSH) and thioredoxin (Trx) are two major intracellular redox systems. GSH is the most abundant tripeptide thiol in mammalian cells that is present at millimolar concentrations and plays a pivotal role in the maintenance of cellular reducing environment and defense against oxidative stress (24–29). On the other hand, Trx is a highly conserved disulfide reductase that catalyzes reduction through the NADPH-dependent thioredoxin reductase (TrxR) system. The Trx system plays an important role in the redox regulation of multiple intracellular processes, including DNA synthesis, cell growth and resistance to oxidative stress and apoptosis (30).
Furthermore, APE1/Ref-1 (for AP endonuclease 1/redox factor 1) is known to contribute to redox regulation. APE1/Ref-1 was originally identified as a DNA repair enzyme with apurinic/apyrimidinic (AP) endonuclease activity and was shown to be important for the base excision repair pathway (31–33). Subsequently, it was reported that APE1/Ref-1 reduces a redox-sensitive cysteine residue of the transcription factor AP-1 and thereby facilitates its DNA-binding and transcriptional activities (6,7). APE1/Ref-1 is now known to enhance DNA-binding activity of several transcription factors, including NF-κB (6,20,23,34), Egr-1 (8), HIF-1α (9–11), HLF (9), Pax-5, Pax-8 (12–15) and p53 (16,17), by regulating their redox states. Thus, APE1/Ref-1 is thought to be a multifunctional protein involved in both base excision DNA repair and redox regulation of transcription factors.
Despite accumulating evidence for the role of APE1/Ref-1 in redox regulation, the underlying mechanism is poorly understood. Although initial studies suggested that Cys-65 of human APE1/Ref-1 (Cys-64 of mouse APE1/Ref-1) is critical for redox regulation of AP-1 (35), subsequent analyses showed that this cysteine residue is not involved in redox regulation (36,37). Thus, we initiated biochemical analyses to elucidate the mechanism of APE1/Ref-1 redox regulation, with a focus on possible interplay among APE1/Ref-1 and other redox systems such as GSH and Trx. Here, we report an unexpected finding that APE1/Ref-1 not only reduces transcription factors such as NF-κB or AP-1 directly, but also assists their reduction by other reducing molecules such as GSH or Trx independently of the cysteine residues of APE1/Ref-1. Using reporter gene assays, we also show that APE1/Ref-1 augments NF-κB-mediated transcription independently of its cysteine residues. These results suggest that APE1/Ref-1 acts as a ‘redox chaperone’ that facilitates the reduction of redox-sensitive transcription factors by other reducing molecules.
MATERIALS AND METHODS
Preparation of bacterial recombinant proteins
The pET-14b (Novagen, Madison, WI, USA)-based expression plasmids of full-length human APE1/Ref-1, p50, p50 C62S, p65 and Trx, and pGEX-APE1/Ref-1 have been described previously (23,34). For the APE1/Ref-1 C/S mutant, site-directed mutagenesis was carried out by the PCR-based overlap extension technique, and the resulting cDNA fragment was cloned into pET-14b or pGEX-6T-1 (GE Healthcare, Buckinghamshire, UK). The cDNAs encoding p52 and c-Rel were obtained by RT–PCR using human total RNA isolated from Jurkat cells and were introduced, respectively, into pET-14b. Expression vectors of p65 RHD, p52 RHD and c-Rel RHD were prepared by inserting the corresponding DNA fragments into the NdeI–BamHI sites of pET-14b. To construct expression plasmids for the bZIP domains of c-Jun and c-Fos, the DNA fragments encoding amino acids 222–331 of human c-Jun and 118–221 of human c-Fos were amplified by PCR and cloned into pET-14b. Bacterial expression and purification of histidine-tagged proteins were performed as described previously (23).
Glutathione S-transferase pull-down assays
To prepare 35S-labeled proteins, pET-14b p50, p65 RHD, p52 RHD, c-Rel RHD, c-Jun bZIP and c-Fos bZIP were transcribed and translated using TNT T7 quick coupled transcription/translation system (Promega, Madison, WI, USA). Escherichia coli strain BL21 (DE3) lysates containing 1 μg of glutathione S-transferase (GST), GST-APE1/Ref-1 WT or GST APE1/Ref-1 C/S were incubated with 30 μl of glutathione Sepharose 4B resin (GE Healthcare) for 1 h at 4°C. After washing three times with 300 μl of 0.1HgKEN (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.1% NP-40, and 10% glycerol), the resin was incubated with 2 μl of 35S-labeled proteins in 300 μl of 0.1HgKEN for 1 h at 4°C. After the resin was washed with 300 μl of 0.1HgKEN six times, bound proteins were eluted with 30 μl of Laemmli buffer, resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and visualized by Coomassie blue staining and autoradiography.
Electrophoretic mobility shift assays
Immediately prior to use, purified recombinant transcription factors were oxidized with diamide, and unreacted diamide was removed from protein samples by affinity purification, as described (23). Recombinant proteins were then incubated in 10 μl of 0.1HgKEN with 1 μg of poly (dI-dC) (dI-dC) at 37°C for 30 min. Where indicated, reduced l-glutathione (Sigma-Aldrich, St. Louis, MO, USA) or TrxR from rat liver (Sigma) and NADPH (Sigma) were included in the reactions. Where indicated, anti-APE1/Ref-1 or anti-Trx was also included, and the reactions were incubated on ice for an additional 30 min. Then, 1 pmol of 32P-labeled probe was added, and the reactions were further incubated on ice for 20 min. The reaction mixture was then subjected to native 4% polyacrylamide gels in 0.5 × Tris borate–EDTA buffer at 4°C. The oligonucleotide probe containing an NF-κB- or AP-1 site was prepared by annealing the following oligonucleotides: sense, 5′-AGTTGAGGGGACTTTCCC-3′ and antisense, 5′-GCCTGGGAAAGTCCCTC-3′ for NF-κB; sense, 5′-GAGCCGCAAGTGACTCAGCGCGGGGCGTGTG-3′ and antisense, 5′-GGCGTTCACTGAGTCGCGCCCCGCACACGTCC-3′ for AP-1. In Figure 6D, nuclear extracts were prepared from transfected 293T cells as described before (38), and subjected to electrophoretic mobility shift assay (EMSA).
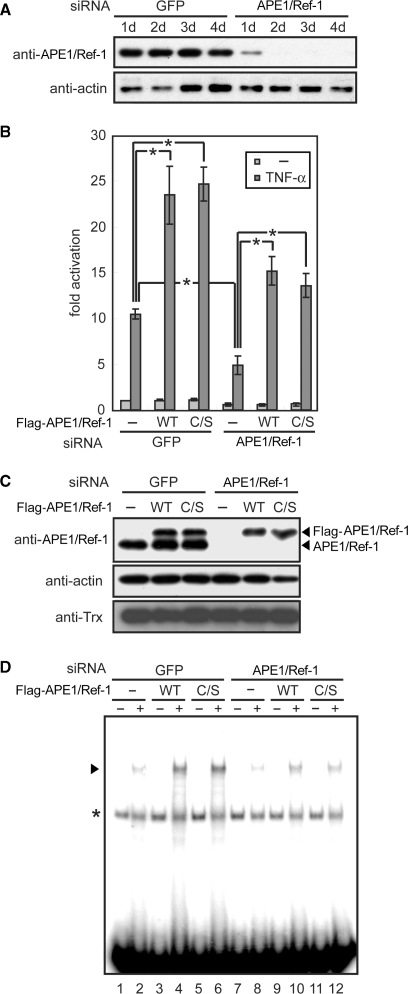
Figure 6.
APE1/Ref-1 activates NF-κB-dependent transcription independently of its cysteine residues in living cells. (A) siRNA-mediated knockdown of APE1/Ref-1. The 293T cells were transfected with siRNA for APE1/Ref-1 or control GFP and various days later, harvested for immunoblot analysis. (B–D) The 293T cells were transfected with various combinations of siRNA, NF-κB-driven and control reporter plasmids, and siRNA-resistant APE1/Ref-1 expression plasmids. Where indicated, TNF-α was added 24 h before harvest. Cell lysates were subjected to luciferase assays (B), immunoblotting (C) and EMSA (D). Asterisk in (D) indicates a nonspecific band. Results in (B) are means ±SD from three independent experiments. *P < 0.01, unpaired t-test.
Fluorescence assays
Oxidized transcription factors were incubated in 0.1HgKEN for various times at 37°C in the presence or absence of reduced l-glutathione or TrxR from rat liver and NADPH. Then, to modify reduced cysteine residues, reaction mixtures were incubated with 2 mM fluorescence-5-maleimide (F5M, Molecular Probes, Eugene, OR, USA) on ice for 5 min. Reactions were terminated by addition of l-Cys to a final concentration of 20 mM, and unreacted F5M was removed by acetone precipitation. Precipitated proteins were resuspended in Laemmli buffer, separated by SDS–PAGE, and analyzed with a fluorescence scanner as described previously (23).
AP endonuclease assays
AP endonuclease activity of APE1/Ref-1 was examined by the conversion of supercoiled, depurinated pBluescript II SK (+) to a nicked form. Depurination of the DNA was performed as described previously (7). Depurinated DNA (200 ng) in a 20 μl reaction volume was incubated with APE1/Ref-1 at the indicated concentrations for 15 min at 37°C in 10 mM Tris–HCl pH 8.0, 5 mM MgCl2, 1 mM EDTA and 0.01% NP-40. Reaction products were then electrophoresed on a 0.8% agarose gel and visualized by ethidium bromide staining.
Knockdown using siRNA
The siRNA duplex for APE1/Ref-1 was prepared by annealing two oligonucleotides; sense, 5′-GUCUGGUACGACUGGAGUACC-3′ and antisense, 5′-UACUCCAGUCGUACCAGACCU-3′. The siRNA duplex for green fluorescent protein (GFP) was purchased from Dharmacon (Lafayette, CO, USA). 293T cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 μg/ml penicillin and 100 μg/ml streptomycin. For knockdown of endogenous APE1/Ref-1, 1 day before transfection, 293T cells were seeded on 6-well plates, and 125 pmol of siRNA was transfected using Lipofectamine 2000 reagent (Invitrogen). After 24 h, transfected cells were seeded at an appropriate density and further grown for 1–4 days. The cells were then lyzed with RIPA buffer (10 mM Tris–HCl pH 8.0, 140 mM NaCl, 0.1% SDS, 1% Triton-X-100, 0.1% sodium deoxycholate, 1 mM EDTA and 0.5 mM EGTA), incubated on ice for 30 min and centrifuged. The supernatants were mixed with Laemmli buffer, subjected to SDS–PAGE and immunoblotted using anti-APE1/Ref-1 and anti-actin (MAB1501, Chemicon, Temecula, CA, USA) antibodies. Anti-APE1/Ref-1 antibody was prepared from rabbit using purified bacterial recombinant full-length APE1/Ref-1 as an antigen.
Reporter gene assays
The pHygEF2-flag-Ref-1 WT anti-siRNA and pHygEF2-flag-Ref-1 C/S anti-siRNA are expression plasmids for Flag-tagged, siRNA-resistant APE1/Ref-1. To confer resistance to siRNA, two synonymous mutations were introduced by the overlap extension technique into the siRNA-targeted sequence of APE1/Ref-1 as follows (substituted nucleotides underlined): 5′-GTCTGGTACGACTGGAGTACC-3′. For reporter gene assays, 293T cells were seeded on 6-well plates 1 day before transfection and transfected with 125 pmol of siRNA, 0.125 μg of pNHκBHL containing four NF-κB-binding sites and the firefly luciferase gene (39), and 0.05 μg of pEF-Rluc containing the control Renilla luciferase gene (40) using Lipofectamine 2000 reagent. Twenty-four hours later, one of the siRNA-resistant APE1/Ref-1 expression plasmids was transfected again into the cells. Six hours after the second transfection, the cells were reseeded on new 24-well plates at a 1:8 dilution. Twenty-four hours after the second transfection, the cells were treated with 10 ng/ml TNF-α (Pepro Tech, Rocky Hill, NJ, USA) and cultured for another 24 h. The cell extracts were prepared, and firefly and Renilla luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega) and Lumat LB9501 (Berthold Technologies, Bad Wildbad, Germany).
RESULTS
APE1/Ref-1 has two distinct effects on DNA-binding activity of NF-κB p50
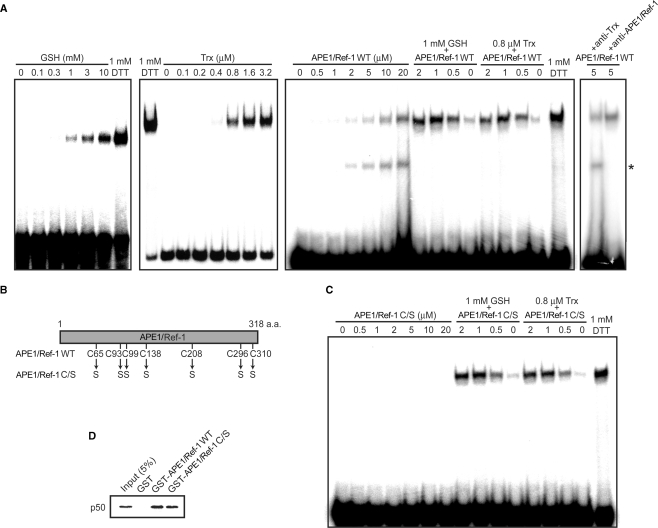
Previously, we and others have shown that Cys-62 of NF-κB p50, the key cysteine residue involved in its DNA binding is redox-regulated by APE1/Ref-1 and Trx (20,22,23,34). In this study, we investigated potential interplay among different redox systems present in cells using p50 reduction as a model. As a measure of the redox states of Cys-62, DNA-binding activity of p50 was measured by EMSA using recombinant p50 that was oxidized with diamide prior to use. Whereas oxidized p50 showed little DNA binding without additional factors, it generated a shifted band in the presence of 1 mM DTT (Figure 1A). GSH and Trx increased p50 DNA binding in a concentration-dependent manner. As reported previously (20,23,34), APE1/Ref-1 also increased p50 DNA binding at relatively high concentrations, that is, at concentrations >50-fold higher than that of p50. Remarkably, APE1/Ref-1, when used together with a limiting concentration of GSH or Trx, strongly increased p50 DNA binding at concentrations as low as 0.5 μM. Similar observation was previously obtained with APE1/Ref-1 and Trx (20).
Figure 1.
APE1/Ref-1 has two distinct effects on DNA-binding activity of NF-κB p50. (A) NF-κB p50 (0.05 μM) was incubated with the indicated concentrations of GSH, Trx and APE1/Ref-1 WT either individually or in combination for 30 min, and then EMSA was performed with NF-κB probe. When Trx was used, 13–16 nM TrxR and 0.2 mM NADPH were also included in binding reactions. Where indicated, anti-APE1/Ref-1 or anti-Trx was included in the reactions. Asterisk indicates the APE1/Ref-1-DNA complex. (B) Schematic structure of APE1/Ref-1 WT and C/S. (C) EMSA were performed using APE1/Ref-1 C/S as in (A). (D) Protein–protein interactions of APE1/Ref-1 with p50. GST pull-down assays were performed as described in Materials and methods section.
We speculated that in the presence of APE1/Ref-1 and GSH or Trx, APE1/Ref-1 is mainly involved in p50 reduction, and that GSH or Trx may facilitate p50 DNA binding by converting oxidized APE1/Ref-1 to a reduced state. We therefore sought to determine cysteine residues of APE1/Ref-1 critical to p50 reduction. To our surprise, APE1/Ref-1 C/S, in which all the seven cysteine residues were substituted to serine, increased p50 DNA binding as efficiently as APE1/Ref-1 WT in the presence of GSH or Trx (Figure 1B and C, and data not shown). Therefore, contrary to the above proposition, redox activity of APE1/Ref-1 is not involved in the activation of p50 DNA binding under these conditions. On the other hand, in the absence of GSH and Trx, APE1/Ref-1 C/S had negligible effect on p50 DNA binding even at the highest concentration examined (Figure 1C), consistent with the idea that redox activity of APE1/Ref-1 is involved in the activation of p50 DNA binding in the absence of other reducing molecules. These results revealed that APE1/Ref-1 has two distinct effects on p50 DNA binding. First, as reported previously, APE1/Ref-1 activates p50 DNA binding directly in a Cys-dependent manner, which requires high concentrations of APE1/Ref-1. Second, in the presence of GSH or Trx, APE1/Ref-1 also activates p50 DNA binding indirectly in a Cys-independent manner at low concentrations of APE1/Ref-1.
We previously showed that APE1/Ref-1 physically interacts with p50 (34). Since APE1/Ref-1 C/S stimulated p50 DNA binding as well as APE1/Ref-1 WT under certain conditions, we assumed that APE1/Ref-1 C/S may also interact with p50. As expected, GST pull-down assays showed that p50 binds to GST-fused APE1/Ref-1 C/S, but not to control GST (Figure 1D), indicating that APE1/Ref-1 interacts with p50 independently of its cysteine residues.
APE1/Ref-1 promotes GSH- or Trx-mediated reduction of p50
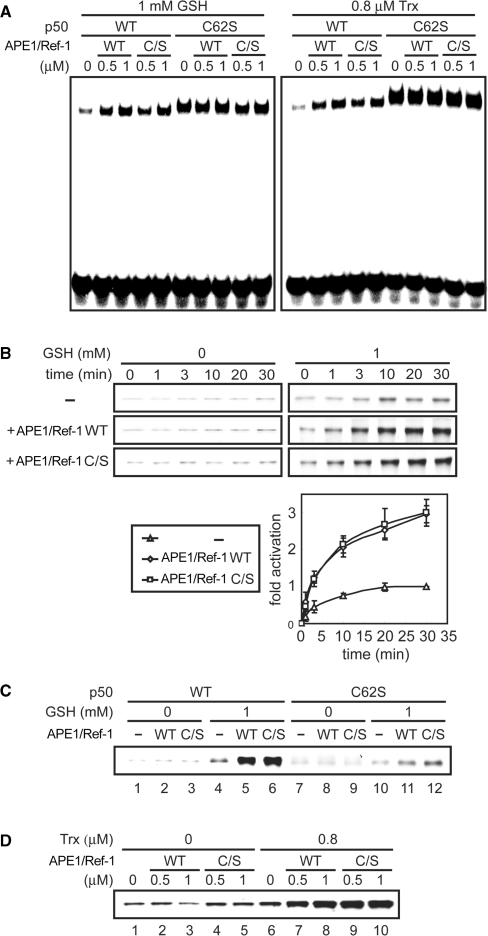
We investigated the cysteine-independent function of APE1/Ref-1 in more detail. The above findings may be interpreted as follows: APE1/Ref-1 and GSH or Trx may facilitate p50 DNA binding by a redox-independent mechanism. Alternatively, APE1/Ref-1 may promote GSH- or Trx-mediated reduction of p50. To discriminate these possibilities, we carried out two lines of experiments. First, we examined by EMSA whether the synergistic effect of APE1/Ref-1 and GSH or Trx is mediated by p50 Cys-62. For this, p50 C62S, which mimics the reduced form of p50 and binds to DNA in a redox-independent manner (21,23), was used. As shown in Figure 2A, APE1/Ref-1 WT or C/S had negligible effect on DNA binding by p50 C62S regardless of the presence of GSH or Trx, demonstrating that p50 Cys-62 is a target for the synergistic effect of APE1/Ref-1 and GSH or Trx. Second, we analyzed the redox status of p50 using a thiol-modifying fluorescent reagent, F5M, as described (23). Following redox reactions, free cysteine residues of p50 were fluorescently labeled and visualized by SDS–PAGE and fluorescence scanning. At the concentrations used here, APE1/Ref-1 WT or C/S had negligible effect on the redox status of p50 in the absence of GSH and Trx, but in the presence of either GSH or Trx, APE1/Ref-1 WT and C/S similarly enhanced p50 reduction (Figure 2B and D). On the other hand, p50 C62S was reduced only weakly under the same conditions (Figure 2C). These results indicate that independently of its cysteine residues, APE1/Ref-1 promotes GSH- or Trx-mediated reduction of p50. We termed the ability of APE1/Ref-1 to facilitate reduction of target transcription factors by other reducing molecules as redox chaperone activity.
Figure 2.
APE1/Ref-1 promotes GSH- or Trx-mediated reduction of p50. (A) The p50 Cys-62 is a target for the synergistic effect of APE1/Ref-1 and GSH or Trx. The p50 WT or C62S (0.05 μM) was incubated with the indicated concentrations of APE1/Ref-1, GSH and Trx/TrxR/NAPDH for 30 min, and then EMSA were performed with NF-κB probe. (B) The p50 (0.05 μM) was incubated with 0.5 μM APE1/Ref-1 WT or C/S with or without GSH for various times, and the redox state of p50 was visualized using F5M, as described in Materials and methods section. Fluorescence intensities of p50 were measured and plotted against time as fold change from the intensity of p50 incubated with GSH alone for 30 min. Plotted data are means ± SD from three independent experiments. (C) The p50 WT or C62S (0.05 μM) was incubated with 0.5 μM APE1/Ref-1 with or without GSH for 30 min, and then fluorescence assays were performed as in (B). (D) The p50 (0.05 μM) was incubated with the indicated concentrations of APE1/Ref-1 with or without Trx/TrxR/NAPDH for 30 min, and then fluorescence assays were performed as in (B).
Effects of APE1/Ref-1 and GSH on other transcription factors
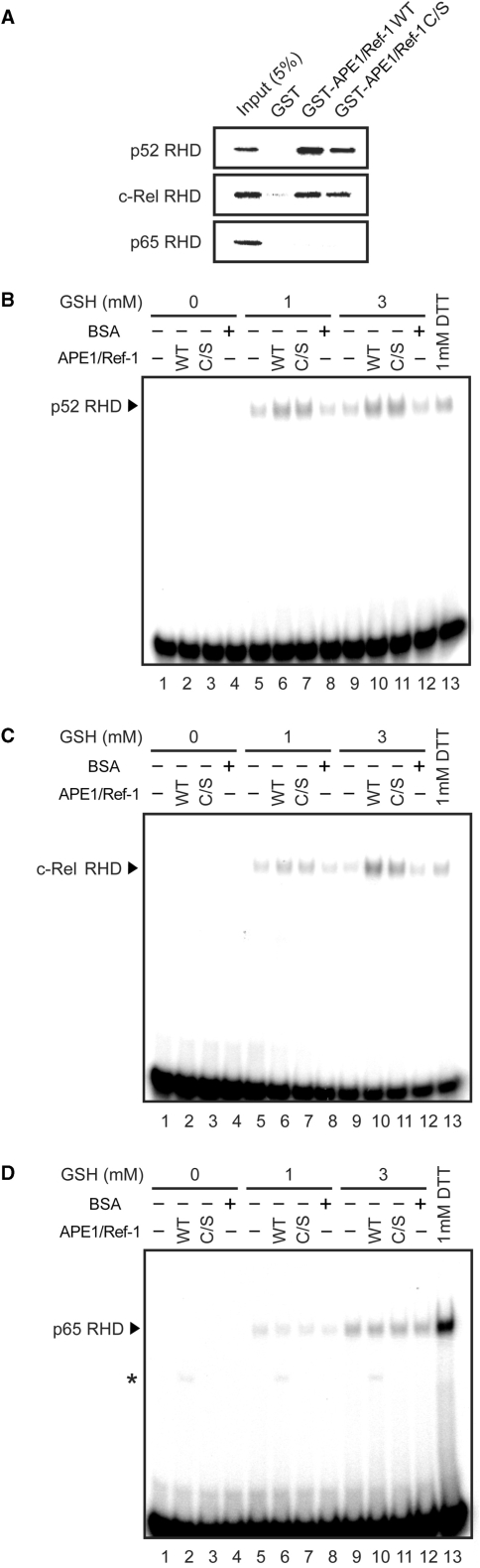
We extended the study on redox chaperone activity to other members of the Rel family, such as p52, p65 and c-Rel. They have a redox-sensitive cysteine residue in their respective DNA-binding domains (21). First, we examined whether APE1/Ref-1 WT and C/S bind to these Rel family members using GST pull-down assays. As shown in Figure 3A, p52 and c-Rel, but not p65, bound to both APE1/Ref-1 WT and C/S. Next, EMSA were performed to investigate whether APE1/Ref-1 affects their DNA-binding activity. At a low concentration of APE1/Ref-1 and in the presence of GSH, APE1/Ref-1 WT and C/S enhanced DNA-binding activity of p52 (Figure 3B) and c-Rel (Figure 3C), but not of p65 (Figure 3D). These results suggest that APE1/Ref-1 binds to and modulates a subset of transcription factors through its redox chaperone activity with some specificity. Apparent correlation between APE1/Ref-1 binding and redox chaperone activity suggests that selective binding to target transcription factors by APE1/Ref-1 may be a prerequisite for its redox chaperone activity.
Figure 3.
APE1/Ref-1 enhances DNA-binding activity of p52 and c-Rel, but not of p65. (A) APE1/Ref-1 physically interacts with p52 and c-Rel, but not with p65. GST pull-down assays were performed as described in Materials and methods section. (B–D) p52 (B), c-Rel (C) or p65 (D) was incubated with 0.5 μM APE1/Ref-1 WT or C/S, or control BSA together with the indicated concentrations of GSH for 30 min, and EMSA were performed with NF-κB probe. Asterisk indicates the APE1/Ref-1–DNAcomplex.
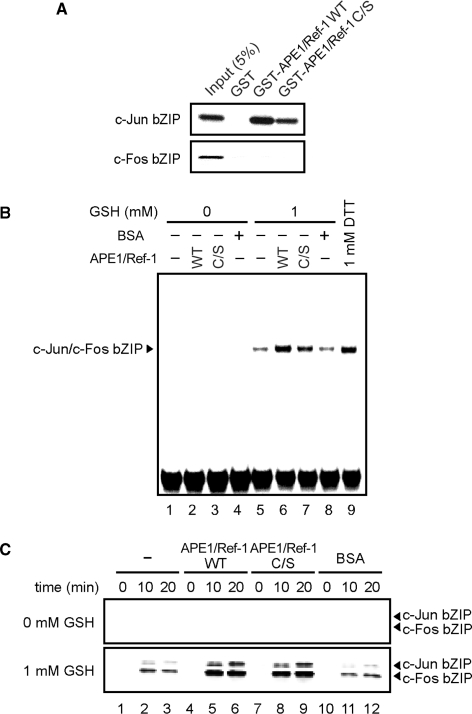
We also examined a potential role of redox chaperone activity in another redox-sensitive transcription factor, AP-1. AP-1 is a heterodimer of c-Fos and c-Jun, whose DNA-binding activity is known to be regulated by APE1/Ref-1 (6,7). APE1/Ref-1 targets Cys-272 of c-Fos and Cys-154 of c-Jun, conserved cysteine residues that are located in their respective basic leucine zipper (bZIP)-type DNA-binding domains (6). GST pull-down assays showed that APE1/Ref-1 WT and C/S bind to the c-Jun bZIP, but not to the c-Fos bZIP (Figure 4A). EMSA showed that in the presence of GSH, APE1/Ref-1 enhanced DNA-binding activity of the c-Jun/c-Fos bZIP heterodimer independently of the cysteine residues of APE1/Ref-1 (Figure 4B). These results suggest that DNA-binding activity of AP-1 is promoted not only by redox activity of APE1/Ref-1, as noted before (6,7), but also by its redox chaperone activity. This prompted us to examine the redox states of c-Fos and c-Jun by fluorescence assays. As expected, both the c-Jun bZIP and the c-Fos bZIP were weakly reduced by GSH, and the reduction was promoted by APE1/Ref-1 WT or C/S (Figure 4C). APE1/Ref-1 alone could not reduce the c-Jun bZIP and the c-Fos bZIP appreciably at the concentration used here (Figure 4C). These results suggest that APE1/Ref-1 binds to the c-Jun bZIP and thereby enhances the reduction of both the c-Jun bZIP and the c-Fos bZIP, leading to increased DNA binding by the c-Jun/c-Fos bZIP heterodimer.
Figure 4.
APE1/Ref-1 enhances DNA-binding activity of AP-1 independently of the cysteine residues of APE1/Ref-1. (A) APE1/Ref-1 physically interacts with the c-Jun bZIP, but not with the c-Fos bZIP. GST pull-down assays were performed as described in Materials and methods section. (B) APE1/Ref-1 WT or C/S enhances AP-1 DNA binding in the presence of GSH. A mixture (0.1 μM each) of the c-Jun bZIP and the c-Fos bZIP was incubated with 0.5 μM APE1/Ref-1 WT or C/S, or control BSA (lanes 4 and 8) with or without GSH for 30 min, and then EMSA were performed with AP-1 probe. (C) APE1/Ref-1 promotes GSH-mediated reduction of AP-1. A mixture (0.1 μM each) of the c-Jun bZIP and the c-Fos bZIP was incubated with 0.5 μM of APE1/Ref-1 WT or APE1/Ref-1 C/S, or control BSA with or without GSH for the indicated times, and the redox status of the bZIP proteins was visualized.
Redox activity, DNA-binding activity and AP endonuclease activity of APE1/Ref-1 are dispensable for its redox chaperone activity
Since APE1/Ref-1 is a multifunctional protein, we were interested in whether redox chaperone activity of APE1/Ref-1 is dependent on other activities it possesses. The ability of APE1/Ref-1 to promote p50 DNA binding in the absence of other reducing molecules (Figure 1A) has been attributed to its redox activity (20,23,34). As APE1/Ref-1 C/S did not promote p50 DNA binding appreciably even at the highest concentration used (Figure 1C), it is likely that APE1/Ref-1 C/S is deficient in redox activity. Besides, it is highly unlikely that a protein with no cysteine residue exerts redox activity.
APE1/Ref-1 also has DNA-binding activity (41). When APE1/Ref-1 WT is used in EMSA at high concentrations, a faster-migrating band is often detected (Figures 1A and 3D; indicated by asterisk). We examined the identity of this complex by using anti-APE1/Ref-1 and control antibodies and found that anti-APE1/Ref1 antibody selectively abolishes the formation of the faster-migrating complex (Figure 1A). Hence, this complex is most likely the APE1/Ref-1-DNA complex. APE1/Ref-1 C/S did not produce such a faster-migrating complex even at the highest concentrations examined (Figures 1C and 3D), indicating that APE1/Ref-1 C/S is deficient in DNA binding.
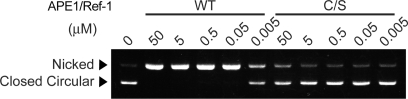
Next, AP endonuclease activity of APE1/Ref-1 was examined using supercoiled plasmid DNA containing AP sites as substrate. APE1/Ref-1 is known to cleave the phosphodiester bond immediately 5′ to AP sites and to create nicks on damaged DNA. As reported previously (7), APE1/Ref-1 WT converted the supercoiled DNA substrate to a relaxed form in a concentration-dependent manner (Figure 5). In contrast, APE1/Ref-1 C/S showed 10 000-fold less activity than APE1/Ref-1 WT (Figure 5). Collectively, APE1/Ref-1 C/S possesses redox chaperone activity comparable to that of wild-type (WT), but lacks redox activity, DNA-binding activity and AP endonuclease activity. These findings indicate that redox activity, DNA-binding activity and AP endonuclease activity of APE1/Ref-1 are dispensable for its redox chaperone activity.
Figure 5.
APE1/Ref-1 C/S lacks AP endonuclease activity. AP endonuclease activity of APE1/Ref-1 was examined by the conversion of supercoiled, depurinated pBluescript II SK(+) to a nicked form. Depurinated DNA (200 ng) in a 20 μl reaction volume was incubated with the indicated concentrations of APE1/Ref-1 for 15 min at 37°C, electrophoresed on a 0.8% agarose gel and visualized by ethidium bromide staining.
APE1/Ref-1 activates NF-κB-mediated transcription independently of its cysteine residues
A number of previous studies have implicated APE1/Ref-1 in the activation of NF-κB-mediated transcription in living cells (42,43). To investigate whether redox chaperone activity of APE1/Ref-1 is involved in the regulation of NF-κB in cells, we knocked down APE1/Ref-1 expression in human 293T cells using siRNA. Western blot analysis showed that the siRNA reduced the APE1/Ref-1 protein level by >90% 2–4 days after transfection, whereas it had little effect on the protein levels of actin and Trx (Figure 6A and C). During this period, no morphological or growth defects were observed in APE1/Ref-1-downregulated cells (data not shown). Then, we examined transcriptional activity of NF-κB using a reporter gene containing four NF-κB-binding sites. Transfection of Flag-tagged APE1/Ref-1 WT or C/S construct resulted in their expression at a level similar to that of endogenous APE/Ref-1 (Figure 6C) and also resulted in a 2-fold increase of TNF-α-induced expression of the reporter gene (Figure 6B). On the other hand, APE1/Ref-1 knockdown significantly attenuated TNF-α-induced expression of the reporter gene, and transfection of RNAi-resistant Flag-APE1/Ref-1 WT or C/S into APE1/Ref-1-downregulated cells restored TNF-α response to a control level (Figure 6B). TNF-α-induced activation of NF-κB was then directly measured by carrying out EMSA using nuclear extracts prepared from the transfected cells. We found that nuclear DNA-binding activity of NF-κB is correlated well with reporter gene activity; i.e. DNA binding was significantly upregulated by expression of Flag-APE1/Ref-1 and downregulated by APE1/Ref-1 siRNA, and there was no appreciable functional difference between APE1/Ref-1 WT and C/S (Figure 6D). These results demonstrate that APE1/Ref-1 activates NF-κB-mediated transcription independently of its cysteine residues, possibly through its redox chaperone activity in living cells.
DISCUSSION
A number of previous reports have demonstrated that APE1/Ref-1 activates DNA-binding activity of many redox-sensitive transcription factors by directly reducing their cysteine residues (6–17,20,23,34). In the present study, we discovered novel activity of APE1/Ref-1 termed redox chaperone activity, by which APE1/Ref-1 regulates DNA-binding activity of various transcription factors through promoting reduction of their critical cysteine residues by other reducing molecules such as GSH and Trx. Redox chaperone activity seems to be mediated by direct interactions between APE1/Ref-1 and target transcription factors, and does not require high concentrations of APE1/Ref-1 as its redox activity. From previous quantitative immunoblot analysis (23), the concentration of APE1/Ref-1 in the cell nucleus is estimated to be >1 μM, which is comparable to the concentrations needed to observe redox chaperone activity in vitro (Figures 1–4). On the other hand, intracellular concentrations of GSH and Trx are estimated to be around 1 mM and 1 μM, respectively (44), which are also comparable to the concentrations used in this study. It is therefore plausible that APE1/Ref-1 regulates redox-sensitive transcription factors through redox chaperone activity in living cells.
APE1/Ref-1 is, to our knowledge, the first example of a redox chaperone, and hence, this study adds a new dimension in our understanding of cellular redox regulation. Here, we propose three possible mechanisms for redox chaperone activity of APE1/Ref-1. First, APE1/Ref-1 may facilitate reduction of target transcription factors by bridging between the target transcription factors and reducing molecules such as GSH and Trx (recruitment model). Second, APE1/Ref-1 may facilitate reduction by inducing conformational change of target transcription factors such that redox-sensitive cysteine residues become more accessible to reducing molecules (conformational change model). Third, APE1/Ref-1 may stabilize the reduced states of target transcription factors by preventing oxidation of redox-sensitive cysteine residues, possibly through hydrogen bond formation with the thiol groups (oxidation barrier model). These models are not mutually exclusive. In support of the first model, it has been shown that APE1/Ref-1 physically interacts with Trx (45–47). However, the finding that APE1/Ref-1 facilitates reduction by structurally unrelated reducing molecules such as GSH and Trx may suggest that the specificity of redox reactions is determined solely by APE1/Ref-1, and that the type of reducing molecules is not important, the idea that is consistent with the second and third models.
Although redox activity of APE1/Ref-1 was found to be dispensable for its redox chaperone activity, we still think that these activities are mechanistically similar. This idea is more plausible than the idea that the two functionally related activities embedded in the single protein are exerted by distinct mechanisms. In our scenario, APE1/Ref-1 binding to target transcription factors enhances their ability to be reduced, and then reduction is carried out by other reducing molecules or APE1/Ref-1 itself; APE1/Ref-1 is regarded as a redox chaperone in the former case and as a redox factor in the latter case.
We showed that APE1/Ref-1 C/S possesses redox chaperone activity, but lacks redox activity, AP endonuclease activity and DNA-binding activity. From these findings, we discuss structure–function relationships of APE1/Ref-1. X-ray crystallography suggests that the seven cysteine residues of APE1/Ref-1 neither form any disulfide bond nor are integral to the tertiary structure (37). It is therefore plausible that the tertiary structure of APE1/Ref-1 C/S is similar to that of WT. For redox activity of APE1/Ref-1, the N-terminal region spanning amino acids 1–127 is shown to be necessary and sufficient for this activity (48). Here, we provided evidence that mutation of all the cysteine residues abolishes redox activity, the finding that is theoretically reasonable. Thus, Cys-65, Cys-93 and Cys-99, the three cysteine residues within the N-terminal region, may be responsible for redox activity. As for AP endonuclease activity, it has been shown that mutation of each of the seven cysteine residues has no discernable effect on this activity (49). Therefore, combined mutations of two or more cysteine residues are likely responsible for the defective AP endonuclease activity of APE1/Ref-1 C/S.
APE1/Ref-1 null mice are unable to develop beyond the point of implantation, underscoring its vital role in vivo (50). Others reported that moderate downregulation of APE1/Ref-1 with antisense cDNA in human cells increases sensitivity to oxidative stress and attenuates NF-κB activation (51). More recently, it was shown that potent downregulation of APE1/Ref-1 with siRNA arrests cell proliferation and causes apoptosis with accumulation of abasic DNA damage in several human cell lines (52). On the other hand, APE1/Ref-1 expression is induced by reactive oxygen species, such as the superoxide anion H2O2 and the hydroxyl radical (·OH) (53–56). Thus, APE1/Ref-1 plays a critical role in an antioxidative stress response pathway leading to activation of several transcription factors.
It remains elusive exactly how APE1/Ref-1 carries out its essential functions in vivo. Fung and Demple (52) showed that defects associated with APE1/Ref-1 downregulation in human cells, such as accumulation of DNA damage and apoptotic cell death, are largely rescued by ectopic expression of yeast Apn1, an AP endonuclease that shows little structural homology to APE1/Ref-1 and is thus thought to have no other APE1/Ref-1 activities. In contrast, we showed here that the attenuation of NF-κB-dependent reporter gene expression, triggered by APE1/Ref-1 downregulation in human cells, is reversed by overexpression of Ref1 C/S, the mutant that possesses redox chaperone activity but are defective in redox activity, DNA-binding activity and AP endonuclease activity. Thus, APE1/Ref-1 may play multiple roles in the antioxidative stress response pathway in vivo through its different biochemical activities.
In conclusion, we discovered a novel biochemical activity of APE1/Ref-1 termed redox chaperone activity that regulates DNA-binding activities of various transcription factors through promoting the reduction of their critical cysteine residues by other reducing molecules such as GSH and Trx. This finding provides new insight into the mechanism of cellular redox regulation. The physiological significance of this finding awaits further investigation.
ACKNOWLEDGEMENTS
We are grateful to Kosuke Kataoka for providing valuable reagents. This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Japan Science and Technology Agency, by a Grant from the Global COE Program from the Ministry of Education, Culture, Sports, Science and Technology, and by a grant from NEDO (to H.H.). K.A. is a JSPS Research Fellow. Funding to pay the Open Access publication charges for this article was provided by the Japan Science and Technology Agency.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 2.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 3.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 4.Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 5.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 6.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang RP, Adamson ED. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- 9.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 11.Lando D, Pongratz I, Poellinger L, Whitelaw ML. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 2000;275:4618–4627. doi: 10.1074/jbc.275.7.4618. [DOI] [PubMed] [Google Scholar]
- 12.Cao X, Kambe F, Ohmori S, Seo H. Oxidoreductive modification of two cysteine residues in paired domain by Ref-1 regulates DNA-binding activity of Pax-8. Biochem. Biophys. Res. Commun. 2002;297:288–293. doi: 10.1016/s0006-291x(02)02196-4. [DOI] [PubMed] [Google Scholar]
- 13.Tell G, Pellizzari L, Cimarosti D, Pucillo C, Damante G. Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun. 1998;252:178–183. doi: 10.1006/bbrc.1998.9548. [DOI] [PubMed] [Google Scholar]
- 14.Tell G, Scaloni A, Pellizzari L, Formisano S, Pucillo C, Damante G. Redox potential controls the structure and DNA binding activity of the paired domain. J. Biol. Chem. 1998;273:25062–25072. doi: 10.1074/jbc.273.39.25062. [DOI] [PubMed] [Google Scholar]
- 15.Tell G, Zecca A, Pellizzari L, Spessotto P, Colombatti A, Kelley MR, Damante G, Pucillo C. An ‘environment to nucleus’ signaling system operates in B lymphocytes: redox status modulates BSAP/Pax-5 activation through Ref-1 nuclear translocation. Nucleic Acids Res. 2000;28:1099–1105. doi: 10.1093/nar/28.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 18.Abate C, Patel L, Rauscher F.J., III, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 19.Matthews JR, Kaszubska W, Turcatti G, Wells TN, Hay RT. Role of cysteine62 in DNA recognition by the P50 subunit of NF-kappa B. Nucleic Acids Res. 1993;21:1727–1734. doi: 10.1093/nar/21.8.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitomo K, Nakayama K, Fujimoto K, Sun X, Seki S, Yamamoto K. Two different cellular redox systems regulate the DNA-binding activity of the p50 subunit of NF-kappa B in vitro. Gene. 1994;145:197–203. doi: 10.1016/0378-1119(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 21.Toledano MB, Ghosh D, Trinh F, Leonard WJ. N-terminal DNA-binding domains contribute to differential DNA-binding specificities of NF-kappa B p50 and p65. Mol. Cell Biol. 1993;13:852–860. doi: 10.1128/mcb.13.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishi T, Shimizu N, Hiramoto M, Sato I, Yamaguchi Y, Hasegawa M, Aizawa S, Tanaka H, Kataoka K, Watanabe H, et al. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J. Biol. Chem. 2002;277:44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 24.Droge W, Schulze-Osthoff K, Mihm S, Galter D, Schenk H, Eck HP, Roth S, Gmunder H. Functions of glutathione and glutathione disulfide in immunology and immunopathology. FASEB J. 1994;8:1131–1138. [PubMed] [Google Scholar]
- 25.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch. Biochem. Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 26.Dickinson DA, Forman HJ. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 2002;64:1019–1026. doi: 10.1016/s0006-2952(02)01172-3. [DOI] [PubMed] [Google Scholar]
- 27.Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 1999;27:922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 28.Kosower NS, Kosower EM. The glutathione status of cells. Int. Rev. Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 29.Meister A. Glutathione metabolism and its selective modification. J. Biol. Chem. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 30.Kobayashi-Miura M, Shioji K, Hoshino Y, Masutani H, Nakamura H, Yodoi J. Oxygen sensing and redox signaling: the role of thioredoxin in embryonic development and cardiac diseases. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H2040–H2050. doi: 10.1152/ajpheart.01316.2006. [DOI] [PubMed] [Google Scholar]
- 31.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl Acad. Sci. USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robson CN, Milne AM, Pappin DJ, Hickson ID. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res. 1991;19:1087–1092. doi: 10.1093/nar/19.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu N, Sugimoto K, Tang J, Nishi T, Sato I, Hiramoto M, Aizawa S, Hatakeyama M, Ohba R, Hatori H, et al. High-performance affinity beads for identifying drug receptors. Nat. Biotechnol. 2000;18:877–881. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- 35.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol. Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber E, Schaffner W. Long-range activation of transcription by SV40 enhancer is affected by ‘inhibitory’ or ‘permissive’ DNA sequences between enhancer and promoter. Somat. Cell Mol. Genet. 1989;15:591–603. doi: 10.1007/BF01534920. [DOI] [PubMed] [Google Scholar]
- 39.Hiramoto M, Shimizu N, Sugimoto K, Tang J, Kawakami Y, Ito M, Aizawa S, Tanaka H, Makino I, Handa H. Nuclear targeted suppression of NF-kappa B activity by the novel quinone derivative E3330. J. Immunol. 1998;160:810–819. [PubMed] [Google Scholar]
- 40.Kataoka K, Yoshitomo-Nakagawa K, Shioda S, Nishizawa M. A set of Hox proteins interact with the Maf oncoprotein to inhibit its DNA binding, transactivation, and transforming activities. J. Biol. Chem. 2001;276:819–826. doi: 10.1074/jbc.M007643200. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Prasad R, Beard WA, Kedar PS, Hou EW, Shock DD, Wilson SH. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J. Biol. Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan Z, Basi D, Li Q, Mariash A, Xia YF, Geng JG, Kao E, Hall JL. Loss of redox factor 1 decreases NF-kappaB activity and increases susceptibility of endothelial cells to apoptosis. Arterioscler. Thromb. Vasc. Biol. 2005;25:96–101. doi: 10.1161/01.ATV.0000150418.14698.75. [DOI] [PubMed] [Google Scholar]
- 43.Hall JL, Wang X, Van A, Zhao Y, Gibbons GH. Overexpression of Ref-1 inhibits hypoxia and tumor necrosis factor-induced endothelial cell apoptosis through nuclear factor-kappab-independent and -dependent pathways. Circ. Res. 2001;88:1247–1253. doi: 10.1161/hh1201.091796. [DOI] [PubMed] [Google Scholar]
- 44.Watson WH, Yang X, Choi YE, Jones DP, Kehrer JP. Thioredoxin and its role in toxicology. Toxicol. Sci. 2004;78:3–14. doi: 10.1093/toxsci/kfh050. [DOI] [PubMed] [Google Scholar]
- 45.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl Acad. Sci. USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin J, Clore GM, Kennedy WP, Kuszewski J, Gronenborn AM. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure. 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- 47.Wei SJ, Botero A, Hirota K, Bradbury CM, Markovina S, Laszlo A, Spitz DR, Goswami PC, Yodoi J, Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. [PubMed] [Google Scholar]
- 48.Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl Acad. Sci. USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantha AK, Oezguen N, Bhakat KK, Izumi T, Braun W, Mitra S. Unusual role of a cysteine residue in substrate binding and activity of human AP-endonuclease 1. J. Mol. Biol. 2008;379:28–37. doi: 10.1016/j.jmb.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daily D, Vlamis-Gardikas A, Offen D, Mittelman L, Melamed E, Holmgren A, Barzilai A. Glutaredoxin protects cerebellar granule neurons from dopamine-induced apoptosis by activating NF-kappa B via Ref-1. J. Biol. Chem. 2001;276:1335–1344. doi: 10.1074/jbc.M008121200. [DOI] [PubMed] [Google Scholar]
- 52.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 53.Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl Acad. Sci. USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang S, Misner B, Chiu R, Meyskens F.L., Jr Redox effector factor-1, combined with reactive oxygen species, plays an important role in the transformation of JB6 cells. Carcinogenesis. 2007;28:2382–2390. doi: 10.1093/carcin/bgm128. [DOI] [PubMed] [Google Scholar]
- 55.Yao KS, Xanthoudakis S, Curran T, O’Dwyer PJ. Activation of AP-1 and of a nuclear redox factor, Ref-1, in the response of HT29 colon cancer cells to hypoxia. Mol. Cell Biol. 1994;14:5997–6003. doi: 10.1128/mcb.14.9.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flaherty DM, Monick MM, Carter AB, Peterson MW, Hunninghake GW. Oxidant-mediated increases in redox factor-1 nuclear protein and activator protein-1 DNA binding in asbestos-treated macrophages. J. Immunol. 2002;168:5675–5681. doi: 10.4049/jimmunol.168.11.5675. [DOI] [PubMed] [Google Scholar]