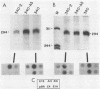
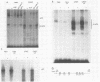
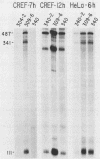
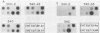Abstract
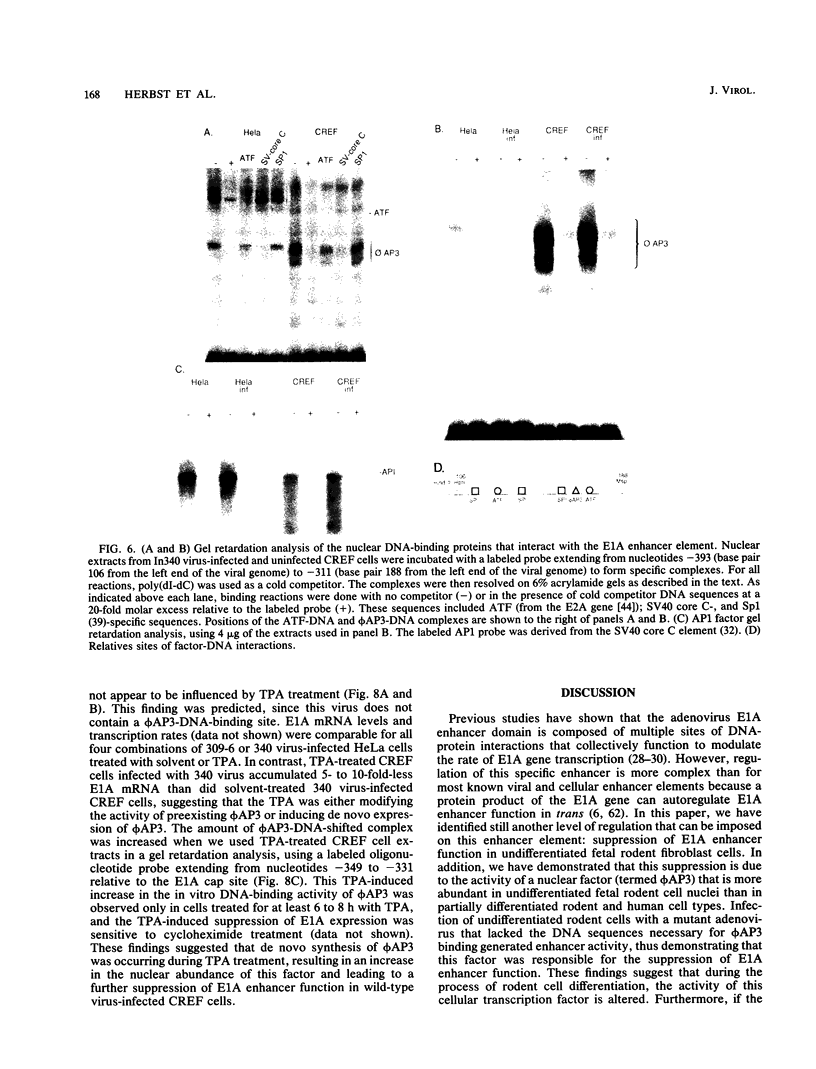
Most of the eucaryotic enhancer elements so far described consist of multiple DNA binding sites for proteins that act either synergistically or antagonistically to modulate the rate of transcription. In this report, we show that the activity of the adenovirus E1A enhancer element is suppressed in virus-infected undifferentiated rodent fetal fibroblast cells (CREF and F111 cells) and primary rat liver hepatocytes that have lost their fully differentiated phenotype (dedifferentiated). This contrasts with the results obtained for virus-infected differentiated or partially dedifferentiated rodent hepatocytes or hepatoma cell lines and human HeLa cells, in which deletion of the E1A enhancer domain greatly reduces the rate of E1A gene transcription. An in vitro quantitation of the nuclear proteins (from HeLa and CREF cells) that interact with and modulate the activity of the E1A enhancer revealed similar binding activities for the E2f and ATF proteins. However, an AP3-like (phi AP3) activity was present at a 10- to 20-fold higher concentration in CREF cells than in HeLa cells, and removal of this phi AP3-binding site on the viral genome resulted in an increase in the rate of E1A gene transcription in virus-infected CREF cells. Together, these results demonstrated that the factors which positively regulate enhancer function were present in CREF cells and that the phi AP3 factor was acting to suppress the activity of the E1A enhancer. Furthermore, the level of this factor was found to increase to even higher levels in CREF cells treated with 12-O-tetradecanoylphorbol-13-acetate, and this induction resulted in a further suppression in the rate of E1A gene transcription. On the basis of these observations, we propose that E1A expression is negatively regulated by the phi AP3 factor in undifferentiated rodent fetal fibroblast cells and that this could be an important mechanism that distinguishes between establishment of the differentiated cell versus transformed cell phenotypes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abmayr S. M., Workman J. L., Roeder R. G. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 1988 May;2(5):542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987 Jun 19;49(6):729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Babiss L. E. The cellular transcription factor E2f requires viral E1A and E4 gene products for increased DNA-binding activity and functions to stimulate adenovirus E2A gene expression. J Virol. 1989 Jun;63(6):2709–2717. doi: 10.1128/jvi.63.6.2709-2717.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett P., Clark L., Hay R. T. A cellular protein binds to a conserved sequence in the adenovirus type 2 enhancer. Nucleic Acids Res. 1987 Mar 25;15(6):2719–2735. doi: 10.1093/nar/15.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli E., Hen R., Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984 Dec 13;312(5995):608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Blumenfeld M., Yaniv M. A liver-specific factor essential for albumin transcription differs between differentiated and dedifferentiated rat hepatoma cells. Genes Dev. 1988 Aug;2(8):957–974. doi: 10.1101/gad.2.8.957. [DOI] [PubMed] [Google Scholar]
- Chiu R., Imagawa M., Imbra R. J., Bockoven J. R., Karin M. Multiple cis- and trans-acting elements mediate the transcriptional response to phorbol esters. Nature. 1987 Oct 15;329(6140):648–651. doi: 10.1038/329648a0. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Clayton D. F., Darnell J. E., Jr Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol Cell Biol. 1983 Sep;3(9):1552–1561. doi: 10.1128/mcb.3.9.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. F., Harrelson A. L., Darnell J. E., Jr Dependence of liver-specific transcription on tissue organization. Mol Cell Biol. 1985 Oct;5(10):2623–2632. doi: 10.1128/mcb.5.10.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Pirozzi A., Blance C., Cortese R. Negative control of liver-specific gene expression: cloned human retinol-binding protein gene is repressed in HeLa cells. EMBO J. 1987 Mar;6(3):631–636. doi: 10.1002/j.1460-2075.1987.tb04801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Xanthopoulos K. G., Darnell J. E., Jr A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, alpha 1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschatrette J., Moore E. E., Dubois M., Weiss M. C. Dedifferentiated variants of a rat hepatoma:reversion analysis. Cell. 1980 Apr;19(4):1043–1051. doi: 10.1016/0092-8674(80)90095-1. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987 Sep 11;50(6):863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Fisher P. B., Babiss L. E., Weinstein I. B., Ginsberg H. S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF). Proc Natl Acad Sci U S A. 1982 Jun;79(11):3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Hay N., Takimoto M., Bishop J. M. A FOS protein is present in a complex that binds a negative regulator of MYC. Genes Dev. 1989 Mar;3(3):293–303. doi: 10.1101/gad.3.3.293. [DOI] [PubMed] [Google Scholar]
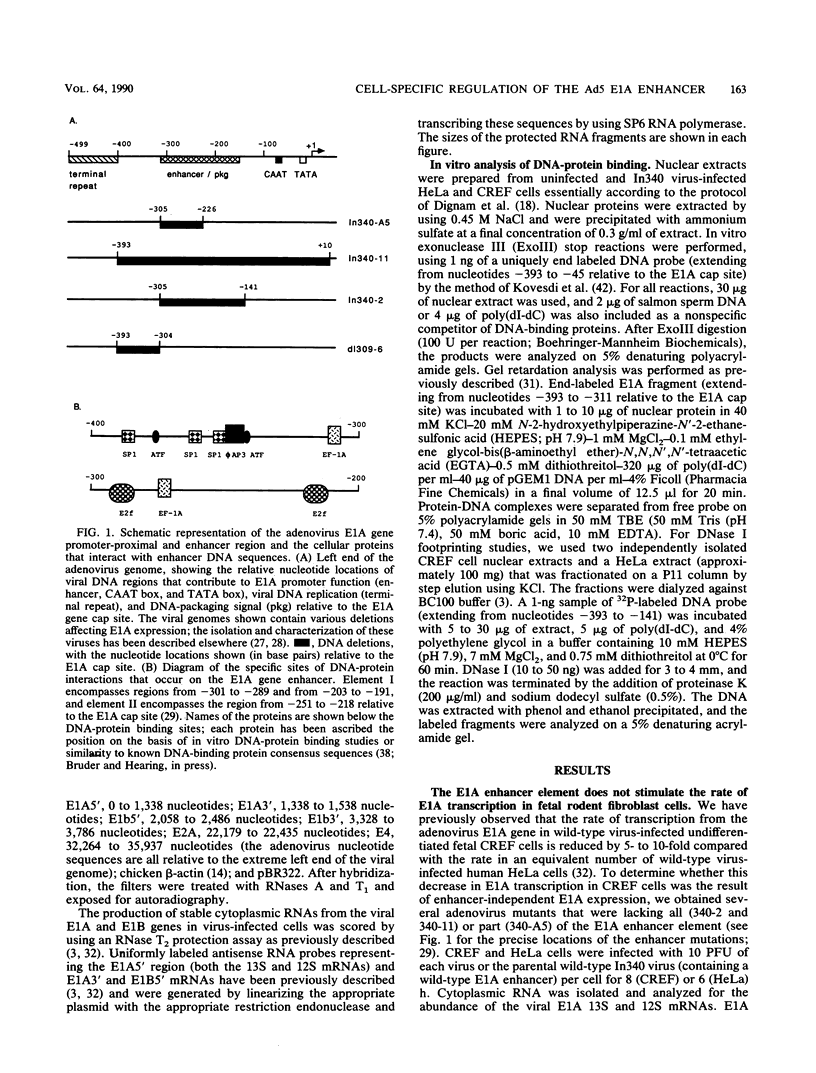
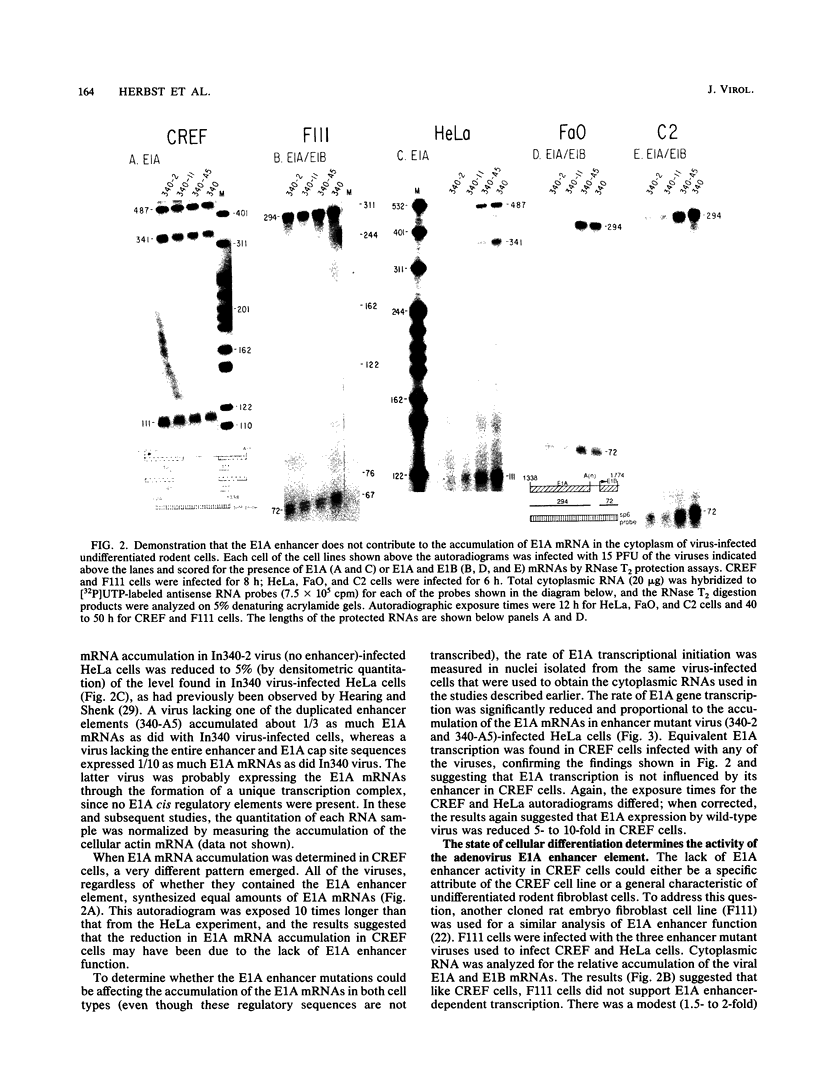
- Hearing P., Shenk T. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell. 1986 Apr 25;45(2):229–236. doi: 10.1016/0092-8674(86)90387-9. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hen R., Borrelli E., Sassone-Corsi P., Chambon P. An enhancer element is located 340 base pairs upstream from the adenovirus-2 E1A capsite. Nucleic Acids Res. 1983 Dec 20;11(24):8747–8760. doi: 10.1093/nar/11.24.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R. S., Friedman N., Darnell J. E., Jr, Babiss L. E. Positive and negative regulatory elements in the mouse albumin enhancer. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1553–1557. doi: 10.1073/pnas.86.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
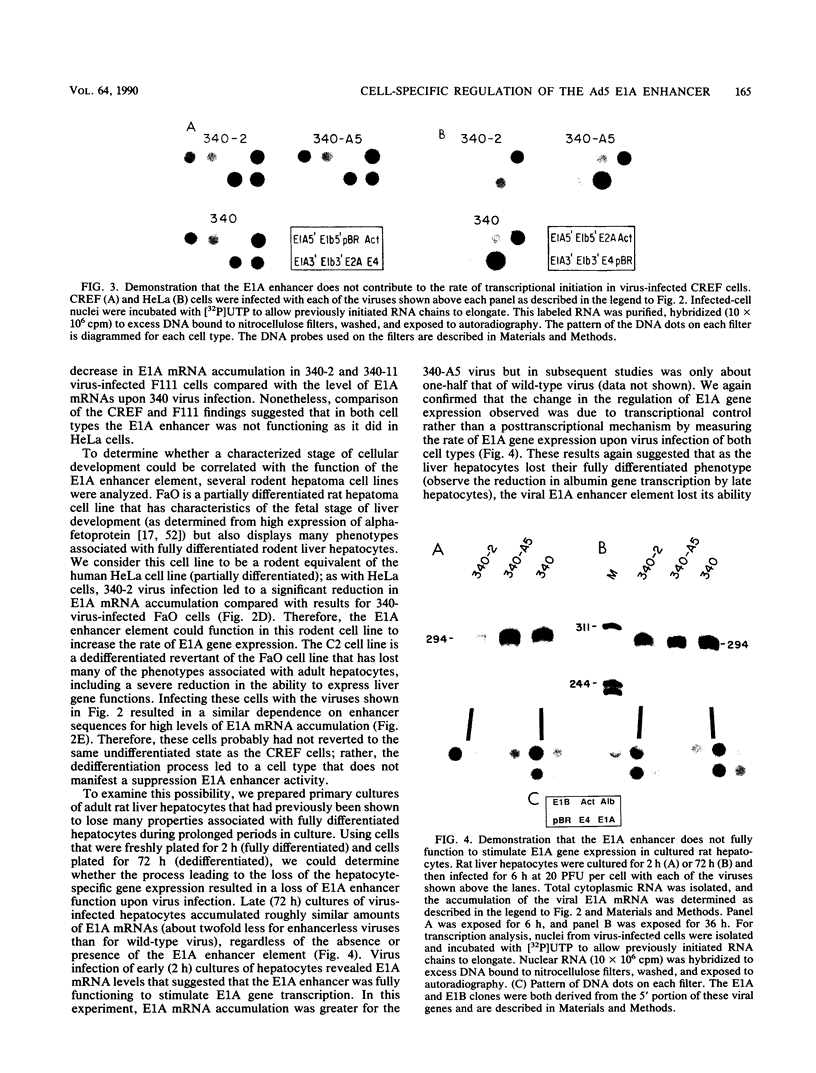
- Herbst R. S., Hermo H., Jr, Fisher P. B., Babiss L. E. Regulation of adenovirus and cellular gene expression and of cellular transformation by the E1B-encoded 175-amino-acid protein. J Virol. 1988 Dec;62(12):4634–4643. doi: 10.1128/jvi.62.12.4634-4643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Herr W., Gluzman Y. Duplications of a mutated simian virus 40 enhancer restore its activity. Nature. 1985 Feb 21;313(6004):711–714. doi: 10.1038/313711a0. [DOI] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Hai T., Lin Y. S., Green M. R., Roeder R. G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988 Sep 23;54(7):1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- Imler J. L., Ugarte E., Wasylyk C., Wasylyk B. v-jun is a transcriptional activator, but not in all cell-lines. Nucleic Acids Res. 1988 Apr 11;16(7):3005–3012. doi: 10.1093/nar/16.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kovesdi I., Reichel R., Nevins J. R. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986 Apr 25;45(2):219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Green M. R. A cellular transcription factor E4F1 interacts with an E1a-inducible enhancer and mediates constitutive enhancer function in vitro. EMBO J. 1987 May;6(5):1345–1353. doi: 10.1002/j.1460-2075.1987.tb02374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnenbach A., Huebner K., Croce C. M. DNA-transformed murine teratocarcinoma cells: regulation of expression of simian virus 40 tumor antigen in stem versus differentiated cells. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4875–4879. doi: 10.1073/pnas.77.8.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Montell C., Courtois G., Eng C., Berk A. Complete transformation by adenovirus 2 requires both E1A proteins. Cell. 1984 Apr;36(4):951–961. doi: 10.1016/0092-8674(84)90045-x. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- REUBER M. D. A transplantable bile-secreting hepatocellular carcinoma in the rat. J Natl Cancer Inst. 1961 Apr;26:891–899. [PubMed] [Google Scholar]
- Sassone-Corsi P., Lamph W. W., Kamps M., Verma I. M. fos-associated cellular p39 is related to nuclear transcription factor AP-1. Cell. 1988 Aug 12;54(4):553–560. doi: 10.1016/0092-8674(88)90077-3. [DOI] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- SivaRaman L., Thimmappaya B. Two promoter-specific host factors interact with adjacent sequences in an EIA-inducible adenovirus promoter. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6112–6116. doi: 10.1073/pnas.84.17.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Stuhlmann H., Jähner D., Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzendruber D. E., Lehman J. M. Neoplastic differentiation: interaction of simian virus 40 and polyoma virus with murine teratocarcinoma cells in vitro. J Cell Physiol. 1975 Apr;85(2 Pt 1):179–187. doi: 10.1002/jcp.1040850204. [DOI] [PubMed] [Google Scholar]
- Thalmeier K., Synovzik H., Mertz R., Winnacker E. L., Lipp M. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev. 1989 Apr;3(4):527–536. doi: 10.1101/gad.3.4.527. [DOI] [PubMed] [Google Scholar]
- Thiel J. F., Smith K. O. Fluorescent focus assay of viruses on cell monolayers in plastic Petri plates. Proc Soc Exp Biol Med. 1967 Jul;125(3):892–895. doi: 10.3181/00379727-125-32232. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Imler J. L., Chatton B., Schatz C., Wasylyk C. Negative and positive factors determine the activity of the polyoma virus enhancer alpha domain in undifferentiated and differentiated cell types. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7952–7956. doi: 10.1073/pnas.85.21.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Jelinek W., Darnell J. E., Jr The definition of a large viral transcription unit late in Ad2 infection of HeLa cells: mapping of nascent RNA molecules labeled in isolated nuclei. Cell. 1977 Apr;10(4):611–616. doi: 10.1016/0092-8674(77)90093-9. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]