Abstract
Direct visualization of DNA and proteins allows researchers to investigate DNA–protein interactions with great detail. Much progress has been made in this area as a result of increasingly sensitive single-molecule fluorescence techniques. At the same time, methods that control the conformation of DNA molecules have been improving constantly. The combination of both techniques has appealed to researchers ever since single-molecule measurements have become possible and indeed first implementations of such combined approaches have proven useful in the study of several DNA-binding proteins in real time. Here, we describe the technical state-of-the-art of various integrated manipulation-and-visualization methods. We first discuss methods that allow only little control over the DNA conformation, such as DNA combing. We then describe DNA flow-stretching approaches that allow more control, and end with the full control on position and extension obtained by manipulating DNA with optical tweezers. The advantages and limitations of the various techniques are discussed, as well as several examples of applications to biophysical or biochemical questions. We conclude with an outlook describing potential future technical developments in combining fluorescence microscopy with DNA micromanipulation technology.
INTRODUCTION
The most powerful and convincing approach to decipher protein–DNA interactions is to directly observe proteins binding to DNA under well-controlled conditions. To achieve this, scientists have developed ever more powerful methods and instrumentation. In most of these approaches, the DNA is manipulated in some way, while the DNA and/or proteins binding to it are observed with fluorescence microscopy. Fluorescence microscopy has become so sensitive that single fluorophores can be detected and interrogated routinely. Single-molecule fluorescence microscopy on DNA and DNA-binding proteins has been successfully applied to many biologically relevant systems (1). In most of these experiments, short DNA fragments are used, free in solution or attached to a surface. In organisms, however, DNA molecules are in general very long, flexible and dynamic. In order to observe the interaction of a protein with such more natural DNA substrates, fluorescent observation of the protein (or DNA) is not enough: the shape and position of the DNA has to be well-controlled. Over the years, many methods for manipulating single DNA molecules have been developed. Approaches such as magnetic and optical tweezers and flow stretching have led to a remarkable understanding of the complex mechanical properties of DNA. Furthermore, these methods have been applied to study the actions of proteins interacting with the DNA, indirectly by measuring changes in the physical properties of the DNA strand (length, stiffness, elasticity) (2). Such indirect detection has proven to be powerful, but in many cases the actual location and potential multimerization state of the proteins on the DNA are crucial to understand their functioning. Moreover, having mechanical cues backed up by simultaneous direct (fluorescent) observation of proteins interacting with DNA strengthens the quality of the data: both signatures have to match, providing an intrinsic experimental control: seeing is believing. These considerations have led researchers to combine (single-molecule) fluorescence microscopy with DNA manipulation tools. In this review we will discuss this development, from relatively simple (but powerful) methods that allow limited control of the DNA up to sophisticated instruments with high degrees of control of the DNA and capabilities of high-resolution detection and localization of DNA-bound proteins. A summary of the various techniques, including some of their advantages and disadvantages, is shown in Table 1.
Table 1.
Summary of pros and cons of the various combined visualization and manipulation techniques described
| Technique | Pros | Cons |
|---|---|---|
| Molecular Combing | Technically simple Many DNA molecules can be studied in parallel TIRF compatible Compatible with low protein concentrations (∼10 pM) | Multiple uncontrolled attachment points No control of force Slow buffer exchange |
| Surface-tethered DNA | Many DNA molecules can be studied in parallel TIRF compatible Single attachment point to surface Can be compatible with low protein concentrations (∼10 pM) | Little force control Slow buffer exchange Requires continuous smooth fluid flows |
| DNA fixed to one optically trapped bead | Fast buffer exchange Some force control No background fluorescence from surface | Requires continuous smooth fluid flows Minimum protein concentration ∼1 nM because of microfluidics system (46) Technically more challenging |
| DNA tethered between two optically trapped beads | Fast buffer exchange High degree of force control Direct measurement of force on DNA No background fluorescence from surface (using water immersion objective) | Technically challenging Minimum protein concentration ∼1 nM because of microfluidics system (46) |
MOLECULAR COMBING
Technique description
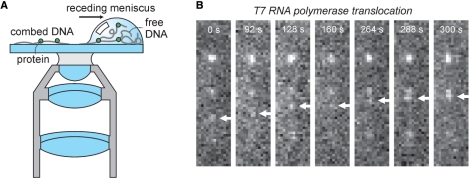
One of the least complicated techniques to manipulate single DNA molecules for visualization with fluorescence microscopy is molecular combing. In molecular combing, DNA molecules are (non-)specifically attached to a solid surface, extended and aligned by various means. The first report of molecular combing was by Bensimon et al. (3). They stretched long pieces of DNA (50 kbp up to 1 Mbp) on hydrophobic silanized glass surfaces using a receding air–water interface (Figure 1A). Surface tension extends the DNA and results in stretched DNA. In this way, DNA is irreversibly stuck to the surface and rehydration of the sample does not result in detachment of the DNA. The non-specific attachment of the DNA to the surface is probably the result of hydrophobic interaction with parts of the double-stranded DNA that have frayed and thus expose hydrophobic single-stranded DNA pieces. This combing procedure has found powerful uses in mapping and analyzing complete genomes with fluorescent in situ hybridization (FISH)—see for more details review of Lebofsky and Bensimon (4). Its major drawback is, however, that it overstretches the DNA (i.e. the DNA is longer than its contour length) (5) because of the high surface tension at the air–water interface (∼0.5 nN) (6). This major structural change most probably interferes with DNA–protein interaction.
Figure 1.
Immobilizing DNA on a surface by combing. (A) Cartoon of the combing procedure. DNA can be stretched and immobilized using hydrophobic silanized glass surfaces and a receding air–water interface. After rehydration of the sample the DNA stays firmly attached to the glass slide. Combing can also be achieved using fluid flow which results in lower stretching forces. (B) Kim and Larson visualized in real time the motion of T7 RNAP along combed DNA strands (9). The directional movement of the T7 RNAP elongation complex along a DNA molecule is observed using the incorporation of fluorescent UTP into RNA strand. Adapted by permission of Oxford University Press from Ref. (9).
In an elegant solution to this problem, Gueroui et al. (7) lowered the surface tension by addition of a monolayer of the fatty alcohol 1-dodecanol at the air–water interface. This resulted in combed DNA extended to its contour length. The low solubility of 1-decanol in water (∼1 p.p.m.) results in a negligibly low effect on the properties of the DNA molecules in solution. Another solution is to use fluid flow in a flow cell to stretch the DNA. Although this approach requires more sophisticated instrumentation, it provides much more control on the degree of DNA extension (8).
By using a different treatment of the surfaces, the extent and nature of the interactions of the DNA with the surface can be altered. On hydrophobic silanized surfaces DNA is stuck along its full length with a very high density of attachment points. When the glass is coated with hydrophobic polymers such as polymethylmetacrylate (PMMA), polydimethyl-siloxane (PDMS) or polystyrene, the combed DNA only attaches in a few places to the surface (7,8). When the pH is lowered to a value of 6 during combing, the tethering can be restricted to only the extremities of the DNA (8). The use of biotin labels on the DNA ends in combination with biotin binding glass (for example coated with streptavidin) provides another means to ensure that DNA only sticks with its end to the surface (9). When the DNA is only attached to the glass at a few positions and not overstretched, combing is an excellent means to study interactions between the DNA and DNA-binding proteins.
In molecular combing, DNA is often visualized with intercalating dyes such as YOYO-1 (3). YOYO-1 lengthens and stiffens the DNA, which can actually help to stretch the DNA to full length (10). On the other hand, structural modification of DNA by YOYO-1 might interfere with DNA-binding proteins and therefore other means of visualization have been developed, in most cases involving specific end-labeling of the DNA. Crut et al., for instance, used quantum dots to discern the ends of combed DNA (11), while Chan et al. generated fluorescent end-labels using PCR with Cy3- and Cy5-labeled primers (10). The latter study demonstrated that it is possible to use a mixture of Cy5-labeled nucleotides and unlabeled nucleotides in the PCR to lightly stain an entire DNA molecule (10), albeit constrained to a DNA length of ∼25 kb due to PCR limitations. For fluorescence imaging of combed DNA wide-field, epi-illuminated fluorescence microscopy is often used, using CCD cameras as detector. Alternatively, with the combed DNA attached to a glass–water interface, total internal reflection fluorescence (TIRF) microscopy can be used (12).
Applications
A good example of the combination of combed DNA and TIRF microscopy for the study of protein–DNA interactions is the work of Kim and Larson, who visualized in real time the diffusive motion of immunolabeled T7 RNA polymerase (RNAP) along DNA (9). They combed λ-phage DNA by flow, thus avoiding overstretching of the DNA, and attached it to a hydrophobic surface, resulting in multiple anchor points along the backbone. They did not observe any hindrance of RNAP's diffusive motion by the anchor points. RNAP was localized by fitting the fluorescence intensity profiles with 2D-Gaussians (13) with an accuracy of 60 nm (standard error). They determined RNAP's diffusion coefficient to be 1.2 ± 1.0 × 10−9 cm2/s (3.6 × 107 bp2/s) from the mean square displacement along the contour. They showed that the large spread in the diffusion constants is not due to the uncertainty of protein localization but to a surprising, intrinsic variability of individual RNAPs. This conclusion could only be reached because this approach allows the observation of individual proteins moving along DNA. Next, the researchers visualized RNAP binding, initiation and transcription. In the latter experiment, they used fluorescently labeled nucleotides that are incorporated in the nascent RNA produced during transcription (Figure 1B). During transcription, however, the moving RNAP halts at the location where the DNA is anchored to the surface and transcription stalls. The authors estimated the tension on the DNA from the rates of transcription they observed. This indirect way of determining the force requires the assumption that only force alters the transcription rate and that surface proximity or non-uniform NTP concentrations do not alter the rate. A more direct way of controlling the force on the DNA would be preferable.
SURFACE-TETHERED DNA EXTENDED IN FLOW
Technique description
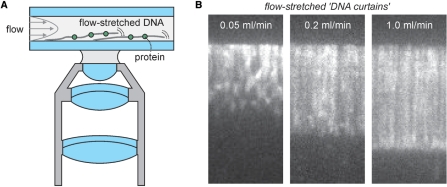
In order to improve the control over the tension on the DNA, other approaches than combing have been taken. A particularly straightforward one is the attachment of a DNA to a surface with one end and stretching it with shear flow (Figure 2A). For attachment, DNA (in most cases the 48 502-bp double-stranded DNA from λ-phage, with a contour length of about 16.5 µm) is biotinylated on one end by hybridizing one of its single-stranded overhangs to a short complementary strand modified with biotins. This biotinylated λ-DNA can then be attached to a surface coated with streptavidin or other biotin-binding protein. Entropic forces will keep the DNA in a relatively compact, random coil. However, these forces can be overcome by applying a shear flow with flow rates of the order tens of ml/hr (14,15). The drag force working on a DNA stretched in this way decreases along the length of the DNA, in the direction of the free end. This force can be made homogeneous along the DNA by attaching a micron-sized bead to the free end. The drag force the bead experiences significantly exceeds the drag on the DNA and will therefore be the major source of tension experienced by the DNA (16,17). A shear flow can be created by pumping a buffer solution through a flow cell using a syringe pump. Flow cells can either be commercially purchased (e.g. Integrated Biodiagnostics) or custom made using (soft) lithography or by gluing two glass plates on a patterned spacer (like parafilm or adhesive tape) (18). Recent advances in microfluidics instrumentation for single-molecule studies have been described in a review by Brewer and Bianco (19). In such thin flow chambers, the flow follows a parabolic Poiseuille velocity profile, with zero velocity at the surface. This depth-dependent velocity profile complicates determination of the local shear the DNA experiences. It has been estimated that a surface-tethered λ-DNA that is extended to 80% of its contour length using buffer flow (viscosity η ≈ 1 Pa s) is on average 0.2 µm away from the surface and experiences a flow velocity of 100 µm/s (14). As an alternative to the use of flow to stretch DNA, the use of magnets in combination with magnetic beads has been reported (20). Wide-field fluorescence imaging can be readily combined with these DNA-stretching approaches. A key advantage of the proximity of the surface is that TIRF microscopy can be applied to reduce background fluorescence. It should, however, be realized that the distance from the surface is not constant in time and along the DNA. This results in fluctuations in fluorescence intensity since the excitation intensity in the evanescent wave used in TIRF illumination decays exponentially with the distance from the interface, with a decay constant of typically 100–150 nm. The proximity of the surface, however, can also be problematic due to undesired interactions with the DNA or DNA-binding proteins. To reduce these effects, the glass surface can be coated with poly(ethylene)glycol (PEG) (14), or supported lipid bilayers (21). Like DNA combing, the surface-tethered DNA stretching approach has one key advantage over other DNA manipulation methods: it can be performed using many DNA molecules in parallel, which can be simultaneously observed in one field of view, e.g. to produce flow-stretchable ‘DNA curtains’ (Figure 2B) (21).
Figure 2.
Stretching of surface-tethered DNA using continuous flow. (A) Schematic of the assay. DNA is attached to the glass surface with one end. To overcome the entropic forces that keep the DNA compact a continuous solvent flow is applied, extending the DNA. Visualization of DNA or associated proteins can be realized using fluorescence microscopy. (B) Application of this assay to YOYO-stained λ-DNA (21), demonstrating how DNA stretching and extension depends on the flow rates. It furthermore highlights ability of this approach to allow the simultaneous observation of many DNA molecules in one field of view. Adapted with permission from Ref. (21). Copyright 2006 American Chemical Society.
Applications
This flow-stretching approach has been applied to study fluorescently labeled proteins diffusing along DNA. Xie and coworkers have investigated how individual base-excision DNA-repair proteins (human oxoguanine DNA glycolase 1, hOgg1) find damaged bases by fast diffusion along the DNA. They studied the pH, salt and flow dependence of the diffusive interaction and concluded that hOgg1, at near-physiological pH and salt concentrations, binds to the DNA for on average 0.025 s while sliding along it with a diffusion constant of 5 × 106 bp2/s, leading to a mean sliding length of 440 bp. The surface-tethered DNA flow-stretching approach was also used to study recombinase RAD51-diffusion along double-stranded DNA (22). The authors proposed that the diffusing complexes are multimers of RAD51 forming a ring around the DNA, since the complexes remained bound for many minutes while diffusing over substantial distances along λ-DNA. The reported diffusion constants range from 104–106 bp2/s. In their experiment, diffusion also appeared to take place when the flow was switched off after attaching the DNA with both ends to the surface (i.e. combing the DNA). In other studies (23,24), the formation and ATP-hydrolysis-dependent disassembly of RAD51 nucleoprotein filaments, which do not move along the DNA were visualized. Subsequent incubation with RAD51 labeled with different dyes yielded ‘harlequin’ filaments, indicating that RAD51-filaments nucleate on multiple sites on the DNA (23). A related approach was taken to study the ejection of DNA by bacteriophage T5 (25) and λ (26). In these studies, the phages were non-specifically attached to the glass surface of a flow chamber. DNA ejection was triggered by addition of the E. coli outer membrane proteins that serve as receptor for the phages. Ejected DNA was stretched in a buffer flow and visualized by addition of the intercalating dye YO-PRO-1. DNA ejection could thus be followed and quantified in real time. The T5 DNA was shown to be ejected in a stepwise manner, pausing at the five specific nicks on T5's genome (25). A maximum ejection rate of 75 kbp/s was observed. In the case of phage λ (26), the genome was ejected in a continuous manner in about 1.5 s, with a maximum speed of 60 kbp/s.
DNA FIXED TO AN OPTICALLY TRAPPED BEAD EXTENDED IN FLOW
Technique description
In order to reduce the complications of the nearby surface (unwanted DNA and protein interactions and the Poiseuille flow profile), an approach where the DNA is tethered to an optically trapped microsphere and extended in a flow away from the surface of the flow chamber can be used (Figure 3A). In this approach, the DNA is attached with similar chemistry to a polystyrene or silica sphere with a diameter of typically a micrometer. Such a sphere can be optically trapped in the focus of an intense near-infrared laser beam (2,27). The physical origin of optical trapping is the transfer of momentum from photons refracted or reflected by the sphere to the sphere itself. In the geometry of the tight focus of a laser beam generated by a high numerical aperture objective, the momenta of many photons add up leading to a 3-dimensional ‘potential well’, keeping the sphere in place (28). Although the high intensity of this trapping laser may lead to enhanced photobleaching (29), selective fluorescence detection of dyes very close to or even in the laser focus has been proven to be possible (29–32).
Figure 3.
Stretching of DNA held with one side in an optical trap using continuous flow. (A) Schematic of the assay. DNA is attached to a bead with one end, the bead is held in an optical trap. To overcome the entropic forces that keep the DNA compact a continuous solvent flow is applied extending the DNA. DNA or associated proteins can be visualized using fluorescence microscopy. (B) Application of this assay to the formation of RecA filaments (40). λ-DNA is incubated with fluorescent RecA and filament formation is monitored by fluorescence. Note the increase of the length of the DNA due to RecA binding. Adapted by permission from Macmillan Publishers Ltd: Nature (40), copyright 2006.
Applications
Chu and coworkers have used this assay extensively to study the mechanical properties of DNA as a prototypical semi-flexible polymer (33–35). In order to visualize the DNA they used intercalating dyes and wide-field fluorescence imaging. They did not drive the buffer flow required for extending the DNA with a pump, but rather moved the whole sample chamber with respect to the optical trap. This allowed them to overcome two shortcomings of stretching in buffer flow: in general it is difficult (i) to get a stable flow and (ii) to quickly change the flow rate. Movement of the sample, however, can only be performed over a limited range and thus for a limited time. Using this approach, they visualized and measured the relaxation of stretched DNA after abrupt release of the shear force. They found that the relaxation rate depends on the length of the DNA and can be described by a distribution of decay times with sharp peaks, ranging from a tenth of a second to tens of seconds, in agreement with theoretical predictions (34). In a subsequent publication (33) they studied the extension and dynamics of the DNA exposed to different drag velocities. They found that for DNA stretched in flow, the interaction of the DNA with the solvent is not constant along the chain, i.e. the DNA is not ‘free draining’. This finding illustrates that extending DNA in a flow without a bead being attached to the free end results in a complicated profile of forces acting along the DNA chain and, as a consequence, to non-uniform extension.
An extensive series of fluorescence experiments using optically trapped, flow-stretched DNA has been performed by Kowalczykowski and coworkers on RecBCD (36–39). RecBCD is a highly processive DNA helicase and nuclease, among others involved in homologous repair. They have directly visualized DNA unwinding by RecBCD by making use of the property of intercalating dyes to bind to double-stranded DNA and not to single-stranded DNA. They demonstrated that RecBCD unwinds DNA at a rate of 100–1000 bp/s and does so over lengths of tens of thousands of bases (36). In a next study using the same intercalating-dye approach, they showed that when the helicase encounters a specific sequence (‘χ’) it pauses for several seconds and subsequently proceeds unwinding the DNA at a substantially lower velocity. This observation raised the question how the two motor subunits of RecBCD, RecB and RecD, work together. They showed, by attaching a fluorescent bead to the RecD subunit, that after χ-recognition, the whole complex stays intact and that RecD does not fall off (37), as was hypothesized before. Later they measured the activity of RecBCD with a mutation in the RecD domain abolishing RecD's helicase activity (38). This mutant unwound DNA at an equal velocity before and after χ recognition. On the basis of this and their previous results they could propose a detailed model of how the two motor domains in RecBCD work together. The Kowalczykowski group has also used a similar assay to study the formation of RecA nucleoprotein filaments on single-stranded DNA (Figure 3B) (40). They directly visualized the nucleation of fluorescently labeled RecA recombinases and demonstrated that 4–5 subunits are necessary for a stable nucleus; after formation of a stable nucleus the filaments elongate on both ends at a rate of several nanometers per second.
DNA TETHERED BETWEEN TWO OPTICALLY TRAPPED BEADS
Technique description
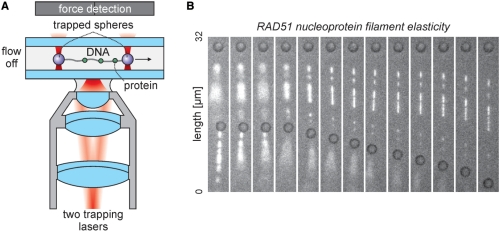
As discussed before, the inhomogeneous force distribution along flow-stretched DNA may pose problems to a quantitative analysis. Moreover, the approach requires the continuous application of flow: when flow is switched off, the DNA swiftly relaxes to a globular shape (34) (see also Figure 2B). This continuous flow precludes the application of such techniques to the study of many dynamic DNA–protein interactions, including the direct visualization of the diffusional target search of site-specific proteins along an extended DNA molecule. For the aforementioned hOgg1 system, this continuous flow apparently did not impose a preferential direction (14), yet for the sliding of RAD51 multimers, the diffusion on a one-side tethered DNA was governed by flow (22). Such drawbacks in flow-controlled experiments can be overcome by attaching the DNA from both ends, as in the molecular combing experiments. However, increased control is obtained because the DNA is manipulated from both ends by optical tweezers (Figure 4A) (alternatively, the DNA can be manipulated using an optical trap on one end and a micropipette (41)). In that case, control over the distance between the traps allows for fine adjustment of the extension of and the tension in the DNA. Another advantage of this method is that no special surface treatments of the microscope slides are required to specifically attach the DNA and at the same time block the binding of unwanted species. Most importantly, optical trapping permits accurate measurement of the forces applied to the DNA with a range of different optical detection methods (see (27) and references therein). This simultaneous fine control over the DNA tension and the quantitative detection of this tension enables one to directly probe the effect of force on biochemical or biophysical processes, such as the binding, locomotion or release of DNA binding enzymes, at the same time visualized using fluorescence.
Figure 4.
Enhanced control using two force-measuring optical traps. (A) Schematic of the dual-trap assay. Two traps can be generated from a single laser source by splitting into two orthogonal polarizations, which may be independently steered in the sample. After suspending a single DNA molecule in between two trapped beads, the DNA can be manipulated without the application of force. In addition, optical tweezers can be employed to quantitatively detect the forces exerted on the DNA. The fluorescence from DNA-staining dyes or fluorescently labeled DNA-binding proteins can be detected using a CCD camera. (B) The assay from panel a employed in our study of the elasticity of (fluorescently labeled) RAD51 nucleoprotein filaments formed on double-stranded DNA (50). One DNA molecule is suspended between two optically trapped beads (dark circles); a second molecule is tethered from the lower bead and freely diffusing once buffer flow is switched off (between 2nd and 4th frame). By increasing the distance between the traps, tension can be applied to the suspended DNA in a controlled manner. The differentiated extension of the fluorescent, RAD51-coated segments and the dark, uncoated segments can be directly seen. The increasing suppression of thermally excited diffusion of the DNA is readily observed.
Applications
In a first application of such dual-beam optical tweezers featuring DNA visualization, Chu and coworkers extended their aforementioned flow-stretched DNA control to observe the conformational dynamics of a DNA molecule stained with YOYO-1 without flow (42). This allowed for an expansion of the polymer's dynamics in linearly independent normal modes after video analysis. Doing so, they observed that higher-order fluctuations are suppressed when the DNA is extended, as expected from polymer dynamics theory. For these experiments, direct measurement of forces was not implemented and the extension of the DNA was used as a control parameter instead. Arai et al. used a similar system to illustrate the versatility of simultaneous single-molecule observation and manipulation, by tying a knot in individual DNA molecules (43). To suppress Brownian fluctuations, they performed their tricky single-molecule gymnastics in a high-viscosity solvent containing sucrose and actin filaments. By simultaneously measuring forces, they used this approach to investigate the dependence of the knot diameter on tension. Since the DNA knots were generated in a dense network of (unlabeled) actin and thus wrapped around actin filaments, the tension in the knot could not be increased enough to break the DNA.
The Yanagida group has been able to track individual E. coli RNA polymerase enzymes interacting with a DNA molecule suspended between two traps (44). To visualize the Cy3-labeled RNA polymerase, they used TIRF microscopy. For efficient TIRF excitation, they used cover slips with pedestals. The beads used to trap the DNA were held, using the tweezers, in the depressions facing the pedestals. In this way, the DNA could be kept at a distance of about 100 nm from the pedestal quartz surface, allowing for efficient TIRF excitation. The pedestals were chemically etched into the cover slip, and had been used in a similar experiment assaying the mechano-chemistry of individual myosins on a suspended actin track (45). Due to the enhanced enzyme affinity for the two asymmetrically positioned promoter sites along the DNA, the authors could readily determine the orientation of the captured DNA based on their knowledge of the promoter positions. Doing so, they showed that association and dissociation rates of RNA polymerase are different for GC-rich and for AT-rich regions. Moreover, they were able to observe polymerases undergoing linear diffusion along the DNA (‘sliding’), which had been anticipated for decades but at the time lacked solid and direct observation. Finally, decreased enzyme affinity for DNA was reported under higher tensions. Similar effects of tension on enzymatic activity have been reported for many different systems, including DNA polymerase and restriction enzymes (46,47). This TIRF approach is complicated by intensity fluctuations (due to movement of the DNA with respect to the interface) and the requirement to etch the cover slips. Wide-field epi-illuminated fluorescence microscopy would not suffer from these drawbacks.
The lab of Greve has used polarized fluorescence detection to get an estimate for the orientation of absorption dipole moments of intercalated YOYO dyes (48). They immobilized DNA between an optically trapped bead and one held in a micropipette. Here, the fluorescence detection was used mostly as a control in a range of experiments mainly focusing on the physics of YOYO intercalation and of DNA binding by RecA. Remarkably, these promising first experiments combining dual-tweezers and fluorescence microscopy in the late nineties have, to our knowledge, led to only a limited number of follow-up reports.
Recently, our group has demonstrated the feasibility of simultaneously performing controlled force-extension measurements and sensitive fluorescence detection in wide-field epi-fluorescence mode. The use of a water-immersion objective allowed for stable trapping even at a significant distance of the captured DNA from the glass surfaces of the sample chamber (41,49), which reduced background fluorescence without the need for any surface blocking. We applied this to the study of RAD51 filament mechanics on double-stranded DNA (50) (Figure 4B). By carrying out these experiments on partly RAD51-coated DNA molecules and using fluorescently labeled RAD51 mutants, we were able to dissect the inhomogeneous elasticity of the DNA into uncoated (dark), partly coated (intermittently fluorescent) and fully coated (contiguously fluorescent) segments (50). Figure 4B shows the controlled extension of such a RAD51-coated DNA molecule suspended between two trapped beads. A second DNA molecule is tethered on only one side from the lower bead and thus freely diffusing once buffer flow is switched off. Integrated analysis of the fluorescence images and synchronously recorded force-extension data revealed that on the suspended molecule only the uncoated or partly coated segments showed the well-known overstretching transition (5); fully coated segments, in which the DNA is held in a 150% extended configuration, could not be stretched further (50). Such dissection could only be performed using a combination of manipulation and visualization techniques.
RULES OF DISCIPLINE AND POINTS FOR ATTENTION
In the preceding paragraphs we have discussed the development and successful application of tools to concomitantly manipulate DNA and visualize associated proteins and/or the DNA itself using fluorescence approaches. Notwithstanding the great power of these approaches, we see two important limitations and/or challenges that need to be taken into consideration. First, in all approaches discussed, the DNA is extended from a random coil into a linear chain. This has obvious advantages for visualization and localization, but it should be taken into account that in the cell DNA is a random coil (often densely bound by proteins). It is well established that the mechanism by which many proteins (such as restriction enzymes or transcription factors) search for a specific target sequence not only comprises 3-dimensional diffusion in solution, but also 1-dimensional diffusion along the DNA chain (sliding) and jumping processes from one location on the DNA to one further down the chain, transiently close by due to the flexibility of the DNA. If the DNA is extended by flow or direct mechanical force, this latter process is strongly suppressed (51). In addition, certain restriction enzymes and other DNA-binding proteins require the formation of bends (46) or even loops in the DNA (52), which is obviously more difficult when the DNA is being pulled taut at the same time. The kinetic rates of many DNA enzymes (e.g. DNA polymerase and RNA polymerase (53,54)) are strongly dependent on the DNA tension. This, however, need not be a limitation but can actually be exploited to dissect the mechanism of complex reactions by varying the forces applied to the DNA (55).
A second point of concern when applying the described approaches is the potential effect of DNA or protein labeling. In order to visualize DNA or most proteins, fluorescent labeling is required. To label DNA, intercalating dyes such as YOYO-1 are frequently used. It is important to realize that these dyes may have substantial effects on the extension and mechanical properties of the DNA (depending on the labeling density) and might in this way influence the interaction of proteins with the DNA (56,57). An often used approach to label a protein of choice is to chemically attach reactive synthetic dyes (58). The preferred method is to generate a single-cystein variant of the protein and to specifically attach a thiol-reactive dye to that cystein. However, this approach can be time consuming, and great care has to be taken that protein activity has not been altered. A variation to this theme is to use semiconductor quantum dots as label (often streptavidin-coated quantum dots are linked to biotin groups attached to the protein) (59). Quantum dots are substantially brighter and more photostable than organic dyes. A disadvantage of quantum dots is their size, on the order of 10 nm, which might alter the protein's dynamics and physical properties. Furthermore, their fluorescence intensity tends to fluctuate under continuous illumination (‘blinking’). A third fluorescent labeling approach is to generate a fusion of the protein with green fluorescent protein (or one of its variants) (58). This is in general less tedious with respect to the molecular biology and no labeling reaction is required after purification of the fusion protein. Limitations are, however, the generally poorer fluorescence properties and the larger size of green fluorescent protein compared to synthetic dyes. Whichever way of labeling is chosen, care has to be taken to reduce the effect of photobleaching, not only to prolong measurement times, but also to prevent photodamage to DNA and proteins. For example, many researchers have noticed that the prolonged illumination of intercalating dyes bound to DNA can lead to breakage of the DNA (60). In general, photodamage can be reduced by working in oxygen-free conditions, the addition of antioxidants and by reducing the illumination intensities and duration (61,62).
OUTLOOK
The methods and applications presented here highlight the potential of combinations of DNA manipulation techniques with fluorescence imaging. We have summarized some of the advantages and disadvantages in Table 1. Over the years, approaches have evolved from relatively straightforward ones, allowing only limited control over the DNA with respect to location and tension (such as combing and flow stretching), to more advanced ones, allowing full control over these parameters using dual optical tweezers. It is to be expected that in particular the combination of dual optical tweezers and fluorescence microscopy will be further developed, allowing for more sophisticated DNA manipulation schemes, for instance, involving force feedback (63), or the simultaneous trapping of two DNA strands (18). At the same time, we and others are adding more advanced fluorescence methods to the tweezers, such as super-resolution methods (64), FRET (30,31), and polarization measurements (48). It is to be expected that soon combinations of fluorescence imaging and magnetic tweezers (28,65,66) will be reported, allowing simultaneously the visualization of and control over the DNA torsion, i.e. the degree of supercoiling. The strength of these combined imaging and manipulation approaches is such that we expect to be only at the beginning of a technical development that will have great impact on the field of protein–DNA interactions in the coming years.
ACKNOWLEDGEMENTS
This work is part of the research program of the ‘Stichting voor Fundamenteel Onderzoek der Materie (FOM)’, which is financially supported by the ‘Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)’. E.J.G.P. and G.J.L.W. are recipients of Vidi grants from NWO. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cornish PV, Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- 2.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 3.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 4.Lebofsky R, Bensimon A. Single DNA molecule analysis: applications of molecular combing. Brief. Funct. Genomic. Proteomic. 2003;1:385–396. doi: 10.1093/bfgp/1.4.385. [DOI] [PubMed] [Google Scholar]
- 5.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 6.Bensimon D, Simon AJ, Croquette V, Bensimon A. Stretching DNA with a receding meniscus - experiments and models. Phys. Rev. Lett. 1995;74:4754–4757. doi: 10.1103/PhysRevLett.74.4754. [DOI] [PubMed] [Google Scholar]
- 7.Gueroui Z, Place C, Freyssingeas E, Berge B. Observation by fluorescence microscopy of transcription on single combed DNA. Proc. Natl Acad. Sci. USA. 2002;99:6005–6010. doi: 10.1073/pnas.092561399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crut A, Lasne D, Allemand JF, Dahan M, Desbiolles P. Transverse fluctuations of single DNA molecules attached at both extremities to a surface. Phys. Rev. E. 2003;67:051910. doi: 10.1103/PhysRevE.67.051910. [DOI] [PubMed] [Google Scholar]
- 9.Kim JH, Larson RG. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Res. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan TF, Ha C, Phong A, Cai DM, Wan E, Leung L, Kwok PY, Xiao M. A simple DNA stretching method for fluorescence imaging of single DNA molecules. Nucleic Acids Res. 2006;34:e113. doi: 10.1093/nar/gkl593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crut A, Geron-Landre B, Bonnet I, Bonneau S, Desbiolles P, Escude C. Detection of single DNA molecules by multicolor quantum-dot end-labeling. Nucleic Acids Res. 2005;33:e98. doi: 10.1093/nar/gni097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 13.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle PS, Ladoux B, Viovy JL. Dynamics of a tethered polymer in shear flow. Phys. Rev. Lett. 2000;84:4769–4772. doi: 10.1103/PhysRevLett.84.4769. [DOI] [PubMed] [Google Scholar]
- 16.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 17.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 18.Noom MC, van den Broek B, van Mameren J, Wuite GJL. Visualizing single DNA-bound proteins using DNA as a scanning probe. Nat. Methods. 2007;4:1031–1036. doi: 10.1038/nmeth1126. [DOI] [PubMed] [Google Scholar]
- 19.Brewer LR, Bianco PR. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat. Methods. 2008;5:517–525. doi: 10.1038/nmeth.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 21.Granéli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DIVA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 22.Granéli A, Yeykal CC, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc. Natl Acad. Sci. USA. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJG, Wuite GJL, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Prasad TK, Yeykal CC, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J. Mol. Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 25.Mangenot S, Hochrein M, Radler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Curr. Biol. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 26.Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl Acad. Sci. USA. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman KC, Block SM. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustamante C, Macosko JC, Wuite GJL. Grabbing the cat by the tail: Manipulating molecules one by one. Nat. Rev. Mol. Cell Biol. 2000;1:130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- 29.van Dijk MA, Kapitein LC, van Mameren J, Schmidt CF, Peterman EJG. Combining optical trapping and single-molecule fluorescence spectroscopy: Enhanced photobleaching of fluorophores. J. Phys. Chem. B. 2004;108:6479–6484. doi: 10.1021/jp049805+. [DOI] [PubMed] [Google Scholar]
- 30.Lang MJ, Fordyce PM, Block SM. Combined optical trapping and single-molecule fluorescence. J. Biol. 2003;2:6. doi: 10.1186/1475-4924-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Interlaced optical force-fluorescence measurements for single molecule biophysics. Biophys. J. 2006;91:1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins TT, Smith DE, Larson RG, Chu S. Stretching of a single tethered polymer in a uniform-flow. Science. 1995;268:83–87. doi: 10.1126/science.7701345. [DOI] [PubMed] [Google Scholar]
- 34.Perkins TT, Quake SR, Smith DE, Chu S. Relaxation of a single DNA molecule observed by optical microscopy. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 35.Perkins TT, Smith DE, Chu S. Direct observation of tube-like motion of a single polymer-chain. Science. 1994;264:819–822. doi: 10.1126/science.8171335. [DOI] [PubMed] [Google Scholar]
- 36.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 37.Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after Chi recognition. Mol. Cell. 2005;17:745–750. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: The recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 40.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 41.Wuite GJL, Davenport RJ, Rappaport A, Bustamante C. An integrated laser trap/flow control video microscope for the study of single biomolecules. Biophys. J. 2000;79:1155–1167. doi: 10.1016/S0006-3495(00)76369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quake SR, Babcock H, Chu S. The dynamics of partially extended single molecules of DNA. Nature. 1997;388:151–154. doi: 10.1038/40588. [DOI] [PubMed] [Google Scholar]
- 43.Arai Y, Yasuda R, Akashi K, Harada Y, Miyata H, Kinosita K, Itoh H. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 44.Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 46.van den Broek B, Noom MC, Wuite GJL. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 48.Bennink ML, Scharer OD, Kanaar R, Sakata-Sogawa K, Schins JM, Kanger JS, de Grooth BG, Greve J. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry. 1999;36:200–208. doi: 10.1002/(sici)1097-0320(19990701)36:3<200::aid-cyto9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 49.Vermeulen KC, Wuite GJL, Stienen GJM, Schmidt CF. Optical trap stiffness in the presence and absence of spherical aberrations. Appl. Opt. 2006;45:1812–1819. doi: 10.1364/ao.45.001812. [DOI] [PubMed] [Google Scholar]
- 50.van Mameren J, Modesti M, Kanaar R, Wyman C, Wuite GJL, Peterman EJG. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophys. J. 2006;91:L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finzi L, Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 53.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 54.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 55.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 56.Berman HM, Young PR. The interaction of intercalating drugs with nucleic-acids. Annu. Rev. Biophys. Bioeng. 1981;10:87–114. doi: 10.1146/annurev.bb.10.060181.000511. [DOI] [PubMed] [Google Scholar]
- 57.Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 58.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 59.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 60.Åkerman B, Tuite E. Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res. 1996;24:1080–1090. doi: 10.1093/nar/24.6.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dittrich PS, Schwille P. Photobleaching and stabilization of fluorophores used for single-molecule analysis with one- and two-photon excitation. Appl. Phys. B. 2001;73:829–837. [Google Scholar]
- 62.Widengren J, Chmyrov A, Eggeling C, Lofdahl PA, Seidel CAM. Strategies to improve photostabilities in ultrasensitive fluorescence spectroscopy. J. Phys. Chem. A. 2007;111:429–440. doi: 10.1021/jp0646325. [DOI] [PubMed] [Google Scholar]
- 63.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Capitanio M, Maggi D, Vanzi F, Pavone FS. FIONA in the trap: the advantages of combining optical tweezers and fluorescence. J. Optics A. 2007;9:S157–S163. [Google Scholar]
- 65.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 66.van Oijen AM. Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity. Biopolymers. 2007;85:144–153. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]