Abstract
Today's proteome is the result of innumerous gene duplication, mutagenesis, drift and selection processes. Whereas random mutagenesis introduces predominantly only gradual changes in protein function, a case can be made that an abrupt switch in function caused by single amino acid substitutions will not only considerably further evolution but might constitute a prerequisite for the appearance of novel functionalities for which no promiscuous protein intermediates can be envisaged. Recently, tetracycline repressor (TetR) variants were identified in which binding of tetracycline triggers the repressor to associate with and not to dissociate from the operator DNA as in wild-type TetR. We investigated the origin of this activity reversal by limited proteolysis, CD spectroscopy and X-ray crystallography. We show that the TetR mutant Leu17Gly switches its function via a disorder–order mechanism that differs completely from the allosteric mechanism of wild-type TetR. Our study emphasizes how single point mutations can engender unexpected leaps in protein function thus enabling the appearance of new functionalities in proteins without the need for promiscuous intermediates.
INTRODUCTION
Evolution is considered to proceed through countless rounds of random mutagenesis and natural selection. To date, a wealth of data exist showing how point mutations gradually alter protein function in support of the notion that proteins with novel functions are the result of an evolutionary drift process that follows an initial gene duplication event (1,2). However, as a consequence of gene duplication, protein intermediates must occur that display redundant functions. Because such redundancies in protein function put into question any evolutionary advantages that such early gene duplication products might provide, these intermediates are considered to be under strong purifying selection with the conversion into a silenced pseudo gene as the likely outcome (3,4). In this context, an evolutionary step, in which random mutations would not lead to a gradual change but to an abrupt switch in function, might prove highly advantageous as a potential, but so far neglected silencing escape mechanism.
That functional leaps occurred during evolution seems obvious when considering evolutionary related proteins with functions that exclude each other and for which, therefore, no intermediates can be envisaged that would contain both functionalities. The bacterial repressors LacI (5) and PurR (6) provide an example for such a pair. They are considered paralogs and share 35% sequence identity (7). Their ability to bind to DNA is modulated via their interaction with low molecular weight effector molecules. However, whereas in case of PurR, binding of the effector hypoxanthine to PurR enables PurR to bind to DNA (6), in LacI, binding of allolactose abolishes its DNA-binding ability (5,8). Obviously, no single protein could display both activities. The frequency and consequently the probability for the occurrence of such functional leaps, greatly increases as the number of mutations that have to be introduced simultaneously decreases. Of particular interest are therefore proteins in which a single point mutation causes a dramatic change in function.
Alongside the bacterial regulator LacI, the tetracycline repressor (TetR) has been established as a paradigm of an effector-regulated DNA-binding protein (9–13). In TetR, the monomers in the homodimeric protein consist of two globular domains, namely a DNA-binding domain (residues 1–45) comprising a helix–turn–helix motif and an effector-binding domain (residues 46–208) that also contains the dimerization interface (13). As is the case for LacI, TetR binds tightly to its palindromic tetO operator DNA in the absence of a low-molecular weight effector molecule, thereby blocking the transcription of any downstream genes. Binding of tetracycline (TC) or anhydrotetracycline (ATC) to TetR causes the repressor to dissociate from the DNA and gene transcription can occur. In both TetR and LacI conformational changes, allosterically induced upon effector binding, have been shown to cause the loss of DNA-binding affinity (5,12).
Recently, it has been reported that the allosteric response of TetR to an effector molecule can be reversed by mutating one or more amino acids (14–16). Since these TetR variants now require TC to bind to tetO, they are called reverse tetracycline repressors (revTetR), and in these variants TC takes over the role of a corepressor and not of an inducer of gene transcription. When fused to an eukaryotic transcriptional activator domain, these variants can be used as molecular tools to specifically induce gene expression in vivo in eukaryotes (17,18).
The behavior of revTetR in the presence of TC is identical to that of PurR with respect to its corepressor hypoxanthine. Therefore, the pair TetR and revTetR constitute a functionally similar pair as LacI and PurR. However, in contrast to the latter two, which deviate by as much as 65% in sequence, in the revTetR variant revTetR-Leu17Gly, a single point mutation suffices to reverse its functional behavior. It should be noted that also for the Lac repressor, variants have been reported with a reversed phenotype (19,20). However, none of the mechanisms responsible for the reversal of phenotype in these variants have been elucidated to date.
Here, we report the mechanism by which the biological function is reversed in revTetR-Leu17Gly. We show that this is achieved by switching to an orthogonal mechanism and not by a mere reversal or extension of the mechanism that causes the induction of wild-type TetR upon TC binding. Our study emphasizes how drastically a single mutation can alter the mechanism by which a protein exerts its function.
MATERIALS AND METHODS
Protein expression and purification
Escherichia coli strain RB791 was transformed with pWH610 containing the respective revtetR alleles (14). Cells were grown in LB medium in the presence of 100 μg/ml ampicillin at 37°C to an OD600 of 0.8, before the temperature was lowered to 22°C and protein expression induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). After 3 h, the cells were harvested upon centrifugation, resuspended in 20 mM phosphate buffer (pH 6.2), 50 mM NaCl, 5 mM EDTA, 1 mM PMSF and disrupted by sonication. The revTetR variants were purified from the supernatant using three subsequent ion exchange chromatography steps and one gel filtration step as described for wild-typeTetR (21). To increase protein yields, the pH of the first cation exchange step (SP-Sepharose, Amersham Biosciences, Uppsala, Sweden) was decreased to pH 6.2 and increased to pH 8.9 for the following two anion exchange chromatography steps (ResourceQ and MonoQ, Amersham Biosiences). The protein was concentrated to 16 mg/ml in 50 mM Tris–HCl (pH 8.0), 0.2 M NaCl after gel filtration and stored at −20°C. Typically, 1 l of bacterial culture yielded about 0.5 mg of pure protein.
Limited proteolysis
RevTetR1, revTetR2, revTetR4 and wild-type TetR were incubated at concentrations of 25 μM at 20°C with 0.015 U/ml of the nonspecific serine protease subtilisin (EC number 3.4.21.62, from Fluka, Neu-Ulm, Germany) in 50 mM Tris–HCl, 0.2 M NaCl (pH 8.0), and in the presence or absence of 2 mM ATC, MgCl2 or both. EDTA was added at a concentration of 15 mM to some samples, in order to chelate excessive Mg2+. After about 5 min, samples were drawn and the digestion stopped by adding equal volumes of SDS sample buffer. Samples were immediately boiled and subsequently examined by SDS gel electrophoresis using 15% (w/v) SDS gels.
Edman sequencing
The proteins and fragments thereof were electrophoretically separated by SDS–PAGE, transferred to an Immobilon-polyvinylidene difluoride membrane (Westran-S-PVDF, Whatman/Schleicher & Schuell BioScience, Keene, NH, USA) and stained with 0.2% (w/v) Ponceau red S (Sigma-Aldrich, Munich, Germany). They were then excised and sequenced N-terminally by Edman degradation using a pulsed-liquid sequencer (Applied Biosystems, Inc., Foster City, CA, USA model 477A/120A).
Circular dichroism spectroscopy
Circular dichroism (CD) measurements were performed at 15°C using a Jasco J-810 spectropolarimeter (Jasco, Tokyo, Japan) and a cuvette with 0.1 cm path length. All experiments were performed in 50 mM sodium phosphate buffer (pH 8.0) at a protein concentration of 5 µM. ATC was added to a final concentration of 1.5 mM. No magnesium ions were present in any of the buffers. Spectra were registered between 180 and 260 nm and corrected for the contributions from the phosphate buffer and ATC. Spectra were accumulated eight times with a band width of 2.0 nm and a sensitivity of 100 mdeg. The scan speed was 20 nm/min, the time response 1 s and data pitch 0.1 nm. CD spectra were analyzed using the deconvolution program CDSSTR from the DichroWeb server (22).
Protein crystallization
Prior to crystallization, revTetR1 was mixed with ATC (from Acros, Geel, Belgium) to obtain a protein solution consisting of 12 mg/ml protein, 50 mM Tris·HCl (pH 8.0), 0.2 M NaCl and 2 mM ATC (Molar ratio ATC to revTetR1 equal 4 to 1). Crystals were grown by the hanging drop vapor diffusion method over a reservoir consisting of 1 ml of 0.1 M Tris–HCl (pH 8.7), 0.3 M MgCl2, 20% PEG 4000 and 3% dioxane. Drops were formed by mixing 1 μl of the revTetR1–[Mg-ATC]+ complex with equal amounts of the reservoir solution. X-ray quality crystals grew within 16 days at 4°C to an average size of about 100 × 100 × 80 μm3. Crystals were flash-frozen in liquid nitrogen after a stepwise transfer into a cryoprotectant solution consisting of 80% (v/v) mother liquor and 20% (v/v) ethylene glycol. Despite repeated efforts, no crystals suitable for X-ray analysis could be obtained for corepressor free revTetR1 or for the ternary complex of revTetR1 in complex with ATC and operator DNA.
Crystal structure determination
X-ray data were collected in house to a resolution of 1.74 Å at 100 K and a wavelength of 1.542 Å using a MicroMax™-007 HF microfocus X-ray generator (Rigaku-MSC, The Woodlands, TX, USA) and a Mar345 image plate detector (MAR Research, Hamburg, Germany). The data recorded during a low and high resolution pass were indexed and integrated with XDS and scaled with XSCALE (23). The crystals have a solvent of 47.8% and contain one molecule per asymmetric unit. The structure was solved by molecular replacement with PHASER (24) using the induced wild-type TetR structure (PDB ID: 2TCT) (9) as a search model. Initial model bias was reduced using the Prime and Switch phasing protocol in program RESOLVE (25). Rigid-body refinement of the top solution was followed by cycles of manual rebuilding with program COOT (26) and restrained atom position, and individual B-factor refinement with program REFMAC5 (27). Mg2+ ions and water molecules were added towards the end of the refinement. During the final stages of refinement, a translation, libration and screw-rotation (TLS refinement) (28) protocol was used and the model divided into four TLS groups (residues 6–45; 46–91; 92–161; 162–205) as suggested by the TLS motion determination server (http://skuld.bmsc.washington.edu/~tlsmd/) (29,30). TLSANL (31) was used to analyze the resulting TLS tensors. A summary of the crystallographic data collection and refinement statistics is presented in Table 1. For comparison, an identical TLS refinement protocol was used to derive TLS parameters for the deposited structures of tetO-bound wild-type TetR (PDB ID: 1QPI) (12), TC-bound wild-type TetR (1DU7) (12) and ligand-free wild-type TetR (1A6I) (11). 1DU7 was used instead of 2TCT as a model for TC-bound wild-type TetR, because no structure factors are available from the data bank for 2TCT.
Table 1.
Crystallographic data collection and refinement statistics
| Data collection | revTetR1 in complex with [Mg-ATC]+ |
|---|---|
| Space group cell dimensions | P21212 |
| a, b, c (Å) | a = 70.65, b = 54.46, c = 56.85 |
| α, β, γ (°) | α = β = γ = 90 |
| Resolution (Å) | 19.66–1.74 (1.80–1.74)a |
| Rintb (%) | 4.1 (46.4) |
| Rmeasc (%) | 5.2 (62.6) |
| Rmrgd-Fc (%) | 5.8 (38.5) |
| I/σI | 26.2 (3.8) |
| Wilson B-value (Å2) | 35.3 |
| Completeness (%) | 98.6 (96.9) |
| Total number of reflections (unique reflec.) | 239 734 (22 843) |
| Refinement statistics | |
| Rwork/Rfree/Rtotal (%)d | 17.55/22.50/18.01 |
| Number of nonhydrogen atoms | 1821 |
| Number of residues | 200 |
| Number of solvent molecules | 166 |
| Ligands present in the model | 1 × anhydrotetracyclin, 2 × Mg2+ |
| Overall mean B-value (Å2) | 41.3 |
| Mean B-value DNA-binding domain/effector-binding domain (Å2) | 73.6, 33.7 |
| Mean B-value ATC/Mg2+/ solvent molecules (Å2) | 24.8, 36.7, 48.1 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.12 |
| Ramachandran statistics (%)e | 97.2/2.8 |
aValues in this column reported in parentheses are for the highest resolution shell.
bRint = Σ|Ij − <I> |/ΣIj, where Ij is the intensity measurement for reflection j, and < I > is the mean intensity of symmetry-related reflections.
cRmeas is the multiplicity weighted merging R-factor, and Rmrgd-F is an indicator for the quality of the reduced data (43).
dRwork/Rfree = 100 (Σh | Fo(h) − Fc(h) |/Σh Fo(h)), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated from 9.7% of the data, that were randomly removed before the refinement was started.
eThe Ramachandran statistics were obtained with program PROCHECK (44). Reported is the percentage of residues in the most favored and additionally favored areas of the Ramachandran plot.
Structure analysis
Structure superpositions were calculated with LSQKAB (32) and the rotation axes visualized with CALC-AX (Joachim Meyer, University of Freiburg, personal communication). For the latter, the structures were superimposed first based on a set of 93 residues that immediately surround the effector-binding site and in a second step based on 28 residues from the DNA-binding domain. The rotation and translation matrices describing the second superposition were then analyzed to calculate the position and orientation of the rotation axis. All structure depictions were generated using program PYMOL (33). The coordinates and structure factors have been deposited with the RCSB Protein Data Bank, the accession code is 2VKV.
RESULTS
Quantification of the induction efficiencies and effector-binding affinities in revTetR1, revTetR2 and revTetR4
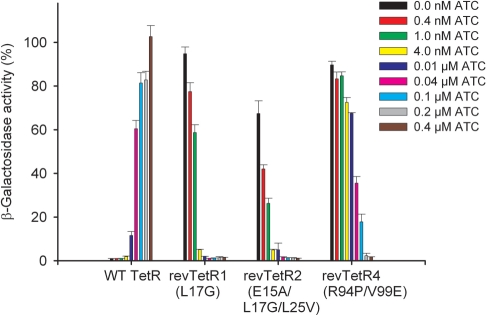
Of the known TetR variants with reverse phenotype (14–16,34), the following E. coli TetR-BD (16) mutants were studied here: revTetR1 containing the mutation Leu17Gly, the triple mutant revTetR2 (Glu15Ala/Leu17Gly/Leu25Val) and the double mutant revTetR4 (Arg94Pro/Val99Glu). All these variants exhibit a dose-dependent corepression response with ATC (Figure 1). In wild-type TetR, induction can be observed starting from 0.01 µM ATC, and close to full induction is reached with 0.1 µM. The reverse variants, revTetR1 and revTetR2, appear much more sensitive since full corepression is reached with as little as 0.01 µM ATC. RevTetR4 appears less sensitive; here, 0.2 µM ATC are required for full repression. Thus, there are slight differences in the revTetR variants with respect to their sensitivity towards ATC in vivo. Effector binding was also investigated by determining ATC-binding constants in vitro using fluorescence titration with ATC in complex with Mg2+ ([Mg-ATC]+, Table 2). These experiments show that of all revTetRs, revTetR1 displays the highest affinity for [Mg-ATC]+ but, that at the same time, all the revTetR variants exhibit strongly decreased affinities for ATC in comparison to wild-type TetR. The reason for this is not immediately obvious (see below), since none of the mutated residues reside in the effector-binding site (12).
Figure 1.
Corepression of different revTetR variants with ATC in comparison to the induction of wild-type TetR (WT TetR) analyzed using a β-galactosidase-activity reporter assay as described (16). Corepression was measured at least twice and was analyzed in vivo in media containing increasing ATC concentrations, namely 0, 0.0004, 0.001, 0.004, 0.01, 0.04, 0.1, 0.2 μM and 0.4 mM ATC. The β-galactosidase activity in the absence of TetR was set to 100% and corresponds to 8300 ± 300 units.
Table 2.
RevTetR effector and tetO operon-binding affinities
| ATC-binding constantsa |
tetO-Binding constantsb |
|||
|---|---|---|---|---|
| +[Mg-ATC]+ KA [·107 M−1]c | +[Mg-ATC]+ KA [·107 M−1]c | +ATC KA [·107 M−1]c | −ATC KA [·107 M−1]c | |
| Wild-type TetR | ≥100d | 0.04e | 0.02 | 560e |
| revTetR1 | 4.5 | 320 | 190 | NDf |
| revTetR2 | 2.4 | 140 | 7.8 | ND |
| revTetR4 | 0.9 | 64 | 2.4 | ND |
aATC-binding constants (association constants, KA) have been determined in vitro using fluorescence titration by a direct titration of the TetR variants with [Mg-ATC]+ (45,46).
btetO-Binding constants have been determined by SPR measurements, as described (15).
cStandard deviations typically range from 10% to 40%.
dThe affinity to wild-type TetR is too large to be determined by this method.
eValues taken from ref. (15).
fND = not detectable. The binding constant was below 105 M−1, which is too low for quantification.
The revTetR corepressor complexes bind DNA with high affinity
Surface plasmon resonance (SPR) experiments were performed to quantify the tetO DNA-binding affinities of the different revTetR variants alone and in the presence of ATC or [Mg-ATC]+. DNA binding of revTetR in the absence of effector could not be quantified because injection of 50 µM revTetR did not lead to saturation and gave only very low signals. Thus, we assume weak binding constants below 105/M for the free revTetR variants to tetO. The binding of wild-type TetR to tetO is reduced to a similar extent by ATC and [Mg-ATC]+, showing residual binding with an association constant (KA) of 0.02 × 107/M and 0.04 × 107/M, respectively (Table 2). From this, we conclude that the non-DNA-binding conformations of the revTetR variants exhibit lower residual tetO binding than effector-bound wild-type TetR. TetO binding in the presence of [Mg-ATC]+ was quantified for revTetR1, revTetR2 and revTetR4 (Table 2). The reverse variants bind to tetO in the presence of [Mg-ATC]+ with affinities that are comparable to the binding of wild-type TetR to tetO in the absence of the effector. This is most obvious for revTetR1, where almost identical values (320 versus 560 · 107/M) are obtained (Table 2).
Effector-free revTetRs are susceptible to proteolytic degradation
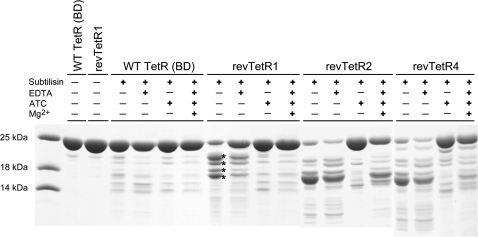
In order to gain insight into the structural integrity of the revTetR variants, we investigated these variants with limited proteolysis in the presence and absence of ATC and [Mg-ATC]+ (Figure 2). In the absence of the corepressor ATC and upon exposure to subtilisin, the different variants yielded similar degradation patterns. These are characterized in case of revTetR1 by the appearance of four major bands in SDS gels. The susceptibility of the revTetR variants to proteolytic cleavage is significantly reduced when ATC and [Mg-ATC]+ is added. In contrast, wild-type TetR is largely protected against proteolysis both in the presence and absence of ATC and [Mg-ATC]+ (Figure 2). Limited proteolysis experiments with trypsin yielded similar results (data not shown).
Figure 2.
Limited proteolysis analysis of wild-type TetR and revTetR variants with subtilisin. Whereas in wild-type TetR, the presence or absence of the effector does not alter the proteolytic susceptibility of TetR, the revTetR variants are only protected against proteolysis in the presence of the effector ATC or [Mg-ATC]+. The four asterisks mark protein bands that were N-terminally sequenced.
In case of revTetR1, the observed fragments were N-terminally sequenced. The positions at which cleavage occurred could be unambiguously identified as K29, G35, W43 and N47 (residues C-terminal to the scissile peptide bond). SDS–PAGE-derived estimates of the apparent masses of these fragments are in agreement with the assumption that these fragments result from a single cleavage and that no further truncation occurred towards their C-terminus. Because the aforementioned residues all map to the DNA-binding domains of TetR, our result shows that in the absence of any effector the DNA-binding heads in the revTetR variants are prone to proteolytic cleavage indicating that they are structurally flexible and conformationally less rigid than in effector-free wild-type TetR.
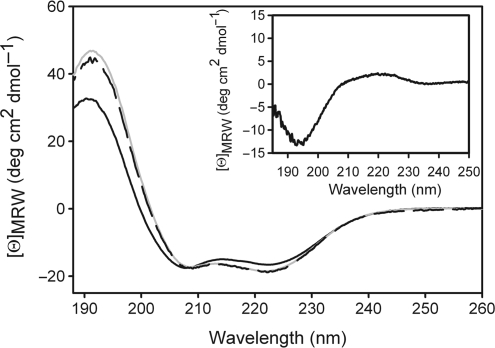
Effector binding increases the α-helical content of revTetR1
Far UV-CD spectroscopy was employed to study any changes in secondary structure induced in revTetR1 upon ATC binding. The CD spectra of revTetR1 with and without corepressor are typical for an all α-helical protein (Figure 3). Close inspection reveals, however, that a 5% increase in helicity [according to the deconvolution analysis with CDSSTR (22)] can be observed in revTetR1 when ATC is added. Only upon addition of ATC, the spectrum of revTetR1 resembles that of wild-type TetR. This increase in helicity can be explained either by about 8–10 amino acids undergoing a random coil to helix transition or, likewise, by a significant higher number of residues undergoing a near-helix to helix structure transition. In agreement with this, the difference spectrum calculated by subtracting the spectrum of revTetR1 with ATC from that of revTetR1 without ATC corresponds to that expected for a polypeptide chain in a random coil conformation with a minimum at 195 nm and positive ellipticities in the region around 212 nm (Figure 3). These results hint that revTetR1 is partially unfolded in the absence of ATC and becomes fully structured upon ligand binding as apparent from the increase in the α-helical CD signal.
Figure 3.
Gain of secondary structure upon corepressor binding to revTetR1 monitored by CD. CD spectra of purified revTetR1 (5 µM) were recorded at 15°C before (continuous line) and after addition of ATC (broken line). Addition of ATC to revTetR1 leads to a CD spectrum that is identical to that of wild-type TetR (shown in gray). Please note that both spectra were recorded in the absence of any magnesium ions. The inset displays the difference spectrum obtained by subtracting the spectrum of ATC-bound from ATC-free revTetR1. The difference spectrum displays many characteristics of an unfolded polypeptide chain.
The crystal structure of ATC-bound revTetR1
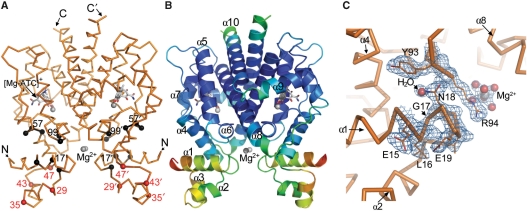
The structure of revTetR1 in complex with [Mg-ATC]+ has been determined at a resolution of 1.7 Å (Figure 4 and Table 1). RevTetR1 forms a dimer and its predominantly α-helical structure is closely similar to that observed for wild-type TetR (9,12,13). The structure reveals the presence of the effector molecule [Mg-ATC]+ in the effector-binding site. The effector forms highly similar interactions with the protein than those seen in wild-type TetR in complex with Mg2+-bound TC ([Mg-TC]+) (PDB code: 2TCT) (9,35). Close inspection of these interactions does not explain why [Mg-ATC]+ exhibits such a strongly decreased affinity for revTetR1 when compared to wild-type TetR (Table 2), since the differences in the effector-binding modes are small. The structure of the revTetR1 monomer contains one additional Mg+2 ion (Figure 4C) that is coordinated by six water molecules. This cation was identified as magnesium based on the residual density observed at this position when modeled as water, its octahedral coordination sphere and the average ligand cation distance of 2.14 Å (±0.15). Furthermore, 150 mM MgCl2 were present in the crystallization setup, increasing the likelihood that any divalent metal-binding site in the crystals is occupied by magnesium. The two Mg2+ ions in the revTetR1 dimer are only indirectly bound to the protein because only the attached water molecules are hydrogenbridged to residues E23, R94, D95, K98 and E150 of revTetR1.
Figure 4.
Crystal structure of the revTetR1 dimer. (A) Together with the Cα-backbone (orange), the effector molecule [Mg-ATC]+ and two additionally bound Mg2+ ions (gray) are depicted. The Cα positions of residues mutated in revTetR1, revTetR2 and revTetR4 as well as in additional TetR-variants with reverse phenotype (mutated residues 56–58) (16) are marked with black spheres. Residues at which proteolytic cleavage occurs in effector-free revTetR1 are marked in red. (B) Cartoon representation of revTetR1 colored according to the individual crystallographic thermal displacement factors (B-values) and explaining the naming of the helices in TetR (α1 to α10). In red, yellow, green and blue are displayed residues with high, medium high, medium low and low B-values, respectively. (C) The σA-weighted 2Fo−Fc electron density map around glycine 17 in revTetR1 contoured at a 1.0 σ cut-off. At the position of the missing Leu17 side-chain a water molecule binds within the interdomain interface of revTetR1.
The mutated glycine residue at position 17 could be ascertained readily because it lacks the density of the former leucine side-chain in wild-type TetR (Figure 4C). Superimposition of the region that surrounds Gly17 in revTetR1 onto the corresponding region around Leu17 in TC-bound and DNA-bound wild-type TetR (PDB code 1QPI) (12), shows that the Leu17Gly mutation does not alter the backbone conformation in any of the surrounding residues (Figure 5A). Since Leu17 is embedded in a network of hydrophobic residues in wild-type TetR, the removal of its side-chain significantly reduces the number of interside-chain interactions formed across the interface between the DNA-binding and the effector-binding domain in revTetR1 in comparison to wild-type TetR. Remarkably, the residues that induce the reverse phenotype in the different revTetR variants all cluster around Leu17 and participate in this hydrophobic interface to a very similar extent than Leu17 (Figure 5A).
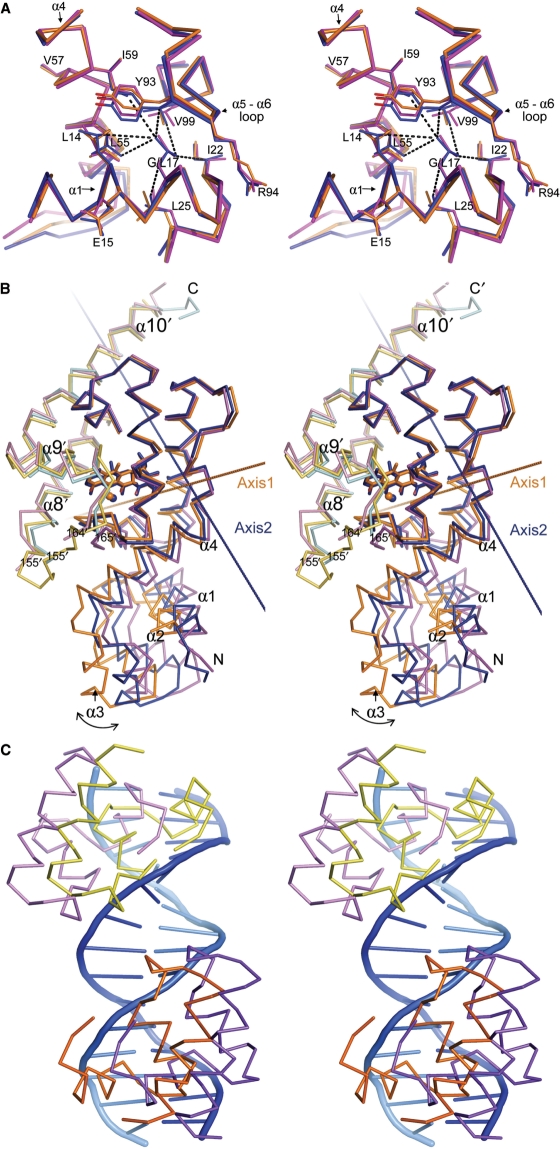
Figure 5.
Stereo representation of selected structural features in the revTetR1-[Mg-ATC]+ structure in comparison to wild-type TetR. (A) Superimposition of the region around Gly17 in revTetR1 (orange) and Leu17 in wild-type TetR (blue, PDB ID: 2TCT) (9) and tetO-bound TetR (purple, PDB ID: 1QPI) (12). Substituting Leu17 against glycine in revTetR1 does not alter the backbone conformation in any of the surrounding residues. It does, however, cause a significant reduction in the number of interside-chain interactions formed between residues from helices α1 and α4, and the α5 to α6 loop segment. Strikingly, all the residues that give rise to the reverse phenotype in the different revTetRs cluster around Leu17. (B) Comparison of the domain orientations in the revTetR1-[Mg-ATC]+ complex (orange/yellow for monomer one/two of the protein dimer), the wild-type TetR–[Mg-TC]+ complex (blue/cyan) and the tetO-bound wild-type TetR dimer (purple/pink). The structures were superimposed by matching 93 residues that immediately surround the effector-binding site. A rotation of 14.4° around axis 1 is required to orient the DNA-binding domain of revTetR1 onto that of tetO-bound wild-type TetR. Similarly, a rotation of 9.2° around axis 2 orients the DNA-binding domain of the wild-type TetR–[Mg-TC]+ complex onto that of tetO-bound wild-type TetR. The two rotation axes are clearly oriented differently but intersect in proximity of residues His63 and Ser67. This is in agreement with previous reports that indicated that in TetR these residues form a molecular hinge (12). (C) The two DNA-binding domains as observed in the revTetR1 dimer (orange/yellow) superimposed onto the corresponding domains in DNA-bound wild-type TetR (purple/pink). The superposition shows that, prior to binding, the DNA-binding domains in revTetR1 must be reoriented.
Although the interactions in the effector-binding sites are very similar in ATC-bound revTetR1 and TC-bound wild-type TetR (9), the orientations of the DNA-binding domains differ significantly. After superposition of the effector-binding domains, the DNA-binding domain in revTetR1 differs from that of wild-type TetR in complex with TC and effector-free wild-type TetR bound to DNA by r.m.s.d. of 4.0 and 7.2 Å, respectively (Table 3). To investigate this further, we determined to what extend the DNA-binding domain in revTetR1 has to be shifted and rotated, in order to match the orientation seen in the DNA-bound wild-type TetR structure. When starting from the above pair-wise superpositions, we calculated that a rotation of 14.4° (plus a translation of 0.6 Å) is required to move the DNA-binding domain of revTetR1 on top of the DNA-binding domain of tetO-bound wild-type TetR (Figure 5B). For comparison, a rotation of only 9.2° (plus a translation of 0.5 Å) is required in order to move the DNA-binding domain of TetR-[Mg-TC]+ on top of the DNA-binding domain of DNA-bound TetR (Figure 5B). The orientation of the DNA-binding domain in revTetR1 is also not identical to that in wild-type TetR in complex with [Mg-TC]+ either, since a rotation of 11.0° (plus a translation of 0.5 Å) is required to superimpose the DNA-binding domain of revTetR1–[Mg-ATC]+ onto the DNA-binding domain of TetR-[Mg-TC]+ (data not shown). In summary, these considerations show that the DNA-binding domain in revTetR1 is oriented differently than in tetO-bound wild-type TetR. As a consequence, the two DNA-binding domains present in revTetR1 must adjust their orientation, in order to interact with DNA (Figure 5C).
Table 3.
Structural comparison between revTetR and wild-type TetR
| RevTetR1 in complex with [Mg-ATC]+ versus wild-type TetR in complex with [Mg-TC]+ (PDB ID: 2TCT) (9,35) | RevTetR1 in complex with [Mg-ATC]+ versus tetO-bound wild-type TetR (PDB ID: 1QPI) (12) | |
|---|---|---|
| Comparison based on the superposition of the effector-binding domainsa,b | ||
| R.m.s.d. of the effector-binding domains (Å)b | 1.04 | 1.84 |
| R.m.s.d. of the DNA-binding domains (Å) | 4.01 | 7.24 |
| Comparison based on the superposition of the DNA-binding domains | ||
| R.m.s.d. of the effector-binding domains (Å) | 4.32 | 7.20 |
| R.m.s.d. of the DNA-binding domains (Å) | 1.32 | 2.80 |
aIn these calculations, the dimer revTetR1 in complex with [Mg-ATC]+ was compared to the corresponding wild-type TetR dimer.
bFor both the superpositions and the calculation of the r.m.s.ds backbone atoms only were compared.
Because the SPR measurements and the β-galactosidase-activity reporter assay unequivocally showed that revTetR1 in complex with [Mg-ATC]+ interacts tightly with DNA (Table 2), we investigated whether we could detect any hints for an unusual flexibility in the revTetR1 structure. Such an increased flexibility could explain why the two DNA-binding domains present in the revTetR1 dimer are able to readily adjust their orientation in order to bind to DNA, a behavior that is not possible in wild-type TetR. An initial hint for this is obtained from the high static disorder that we observe in the DNA-binding domain of revTetR1 and which is reflected by a high average isotropic thermal displacement factor (B-value). While the effector domain and the dimer interface are characterized by an average B-value of 34 Å2, the adjacent DNA-binding domain displays an average B-value of 74 Å2 (Figure 4B). In wild-type TetR, the DNA-binding domains display considerably lower B-values (9). A direct comparison of the TetR structures must however be cautioned. Since wild-type TetR crystallizes in a different space group, the differences in B-values might solely reflect differences in the number of packing contacts in which these domains participate in the different crystals.
TLS analysis reveals increased domain motion flexibility in revTetR1
To further investigate the hypothesis that the DNA-binding domains in ATC-bound revTetR1 display an increased flexibility in comparison to the structures of TC-bound wild-type TetR, DNA-bound wild-type TetR and ligand-free wild-type TetR (PDB code: 1A6I) (11), we subjected all four structures to a detailed anisotropic motion analysis. Because individual atomic anisotropic displacement parameters cannot be refined at the resolution at which the structure of revTetR1 was determined, we took recourse to the TLS parameterization model, instead. In this model, collective variables are used to describe the displacements of atom groups as pseudo-rigid bodies in terms of translation, liberation and screw-rotation motions (28).
For this analysis, the peptide chain was divided into four different segments comprising residues 6–45, 46–91, 92–161 and 162–205. The refined TLS parameters indicate that of all the structures that were compared, the DNA-binding domains of revTetR1-[Mg-ATC]+ (residues 6–45) exhibit the highest flexibility with libration representing the dominant motion (Supplementary Table S1A–C). The graphical representation of the nonintersecting screw tensors and the depiction of the 50% thermal ellipsoids for the revTetR1–[Mg-ATC]+ complex, in comparison to wild-type TetR (Supplementary Figure S1) indicate that the DNA-binding heads in revTetR1 in complex with [Mg-ATC]+ are highly flexible, and that the motions the DNA-binding heads are able to undergo, might be extended enough to allow them to adopt a DNA-binding competent orientation.
DISCUSSION
Our studies enable us to propose a mechanism by which the single TetR point mutant Leu17Gly (revTetR1) switches the function of TetR (Figure 6). This mechanism is in full agreement with the observed binding data (Table 2), namely it explains why ligand-free revTetR1 cannot bind DNA and why complex formation with the effector ATC is required for revTetR1 to interact with DNA. The behavior of revTetR1 is diametrically opposite to that of wild-type TetR, since wild-type TetR binds to the operator DNA only in the absence of the effector and loses its DNA-binding affinity upon effector binding. We suggest that the mechanism described below also holds true for other revTetR variants.
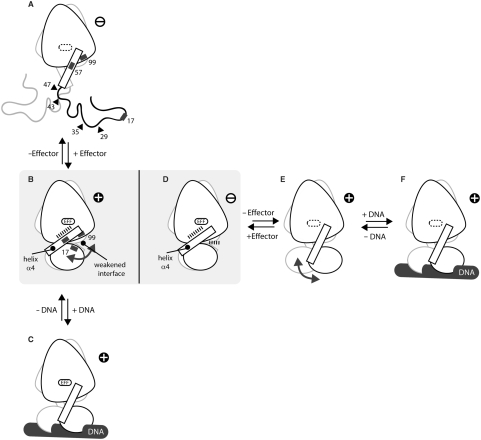
Figure 6.
An orthogonal and not inverse mechanism is responsible for the effector-mediated corepression in revTetR in comparison to the induction of wild-type TetR. Whereas effector binding induces a disorder–order transition in revTetR1, which then enables effector-bound revTetR1 to interact with DNA (A–C), effector binding to wild-type TetR induces a defined conformational change in TetR that locks the protein in a conformation that is not able to interact with DNA any more (D–F). The crystal structures of revTetR1 in complex with ATC (B) and that of wild-type TetR in complex with TC (PDB ID: 2TCT) (9) (D) resemble each other. However, whereas in revTetR1 the mutations that are responsible for the reverse phenotype (black rectangles in A and B) weaken the interface between the DNA- and the effector-binding domain and thereby allow the orientation of the DNA-binding domains to freely adjust to the DNA upon binding (indicated by a double-headed arrow), in wild-type TetR the DNA-binding domains are locked in a non-DNA-binding orientation (D) because of a pendulum-like motion of helix α4 that is triggered by the binding of the effector (E–D). The structures of effector-free wild-type TetR (E) and DNA-bound effector-free wild-type TetR (F) have been sketched according to PDB-ID codes 1A6I (11) and 1QPI (12), respectively. No crystal structure is yet available for (C). The structure of (A) is inferred from CD measurements and limited proteolysis experiments. The identified cleavage sites are marked with black triangles. All DNA-binding competent structures are marked with ‘+’, those not able to bind to the operator DNA with ‘−’.
The CD measurements and the limited proteolysis experiments show that in the absence of effector molecules, the DNA-binding heads in the revTetR variants are partially unfolded and as a consequence are prone to proteolysis. Upon addition of ATC, all revTetR variants studied here become as resistant to proteolysis as effector-free wild-type TetR. This gain in proteolysis resistance upon complex formation with ATC is paralleled by a disorder to order transition and an about 5% gain in helical secondary structure, as shown for revTetR1. Only upon addition of ATC, the CD spectrum and hence the helical content of revTetR1 resembles that of wild-type TetR. Taken together, these observations hint that effector-free unbound revTetR1 cannot bind to its operator DNA because its DNA-binding heads are partially unfolded. Addition of the effector molecule is required to induce proper folding of the DNA-binding domains (Figure 6A and B). In contrast, in wild-type TetR the DNA-binding heads are already properly folded and enable wild-type TetR to bind to DNA in the absence of any effector molecule (Figure 6E and F).
Interestingly we observe that the binding affinities of the revTetR variants for ATC are significantly lower than for wild-type TetR (Table 2). Comparing the effector-bound revTetR1 structure to those available for effector-bound wild-type TetR reveals no obvious differences in the geometry and number of atomic interactions between the effector and the protein in the effector-binding site. The reduced binding affinity can however be explained if one assumes that in case of the revTetR variants, folding of the DNA-binding heads is thermodynamically unfavorable and therefore a fraction of the free energy gained upon binding of the effector is used to induce the folding of the DNA-binding domains.
In an important study by Reichheld and Davidson (36), it has previously been noticed that, in TetR, the effector-binding affinity and the stability of the DNA-binding domains are intercorrelated. In mutants where the DNA-binding heads are partially truncated or destabilized through the introduction of point mutations, the effector-binding affinity is significantly reduced. Conversely, Reichheld and Davidson also observed that effector binding to these variants lead to a general stabilization of the DNA-binding domain with a similar protection against proteolysis and increase in secondary structure as we now observe for the revTetR variants (36). They suggested that helix α4 plays a key role in propagating the stabilization to the DNA-binding heads. Small readjustments in helix α4 upon effector binding would allow for proper folding of the DNA-binding head against helix α4. The author suggested that a similar effector-induced folding of the DNA-binding heads might also be central in TetR variants with reverse phenotype. However, none of the mutants they studied did actually display the reverse phenotype and the changes they observed in the CD spectra also significantly deviate from those seen in revTetR1 (36), (Figure 3).
Unexpectedly, the crystal structure of revTetR1 in complex with ATC reveals that upon addition of ATC, the DNA-binding domains do not adopt a DNA binding competent orientation. The DNA-binding domains seem to be oriented even further apart than observed in wild-type TetR in complex with TC (Figure 5 and Table 3) (9,12). Because ATC-bound revTetR1 interacts with DNA with high affinity (Table 2), it is obvious that the DNA-binding domains must be able to readily adjust their orientation in the presence of DNA. We propose that this becomes possible because the removal of the side-chain of Leu17 in revTetR1 significantly reduces the number of interactions between the DNA- and the effector-binding domain (Figure 5), rendering the orientation of the DNA-binding domains much more flexible. The importance of the reduction of interdomain interactions and concomitant increase in domain flexibility is further substantiated by the observation that when replacing Leu17 by all possible amino acids, only a leucine to glycine substitution and, to some extent, a leucine to serine substitution lead to the reverse phenotype (14).
Albeit to a significant lesser extent, certain flexibility in the domain orientations is also observed in wild-type TetR, since in the absence of any ligand, the DNA-binding domains are also oriented slightly different than in the DNA-bound complex (11). It is possible that in case of revTetR1, the presence of high concentrations of Mg2+ in the crystallization buffer lead to the selection in the crystal of a single conformation from the multiple orientations sampled in solution. The two hydrated Mg2+ ions that are bound at the center of the domain interfaces (Figure 4) possibly stabilize this conformation. The increased flexibility of the DNA-binding domains in revTetR1 is supported by the TLS-analysis. Of all TetR crystal structures, the DNA-binding domains seem to be the most flexible in the revTetR1 structure in complex with ATC (Supplementary Figure S1 and Supplementary Table S1A–C).
A possible drawback of the increased domain flexibility is that in revTetR1 DNA binding is accompanied by a higher entropy loss. This is mirrored by the observation that, although none of the mutated residues in the revTetR variants immediately contact the operator DNA, the DNA-binding affinities are slightly reduced when compared to effector-free wild-type TetR (Table 2). The ability to freely readjust the orientation of the DNA-binding domains appears to be a hallmark of the reverse behavior of all revTetRs. In the revTetRs studied here as well as in additional variants, where for example residues such as 55, 56 and 57 were substituted (16), the mutated residues map without exception into or in immediate proximity of the interdomain interface formed by α-helices α1, α4 and α6 between the DNA- and the effector-binding domain (Figure 5A). This hints that these variants share a common mechanism which can be summarized as an effector-induced disorder–order transition of the DNA-binding heads in combination with a weakened interdomain interface that allows the DNA-binding heads to readily adjust to the operator site (Figure 6B and C). It is very likely that this mechanism also extends to revTetR variant Gly96Glu Leu205Ser for which initially a different mechanism has been suggested (15). This variant displays an identical binding behavior with respect to corepressor and tetO binding, and residue Gly96 is in immediate proximity to residue Leu17 (Cα–Cα distance = 9.1 Å) and located in the same interdomain interface. When examined carefully, the CD spectrum reported by Kamionka et al. (15) for the effector-free protein also displays a reduced α-helical content.
This mechanism is significantly different from the mechanism by which wild-type TetR regulates gene transcription. Wild-type TetR binds tightly to the palindromic DNA operator sequence tetO in the absence of effector molecules (Figure 6 and Table 2) (10). Binding of the effector [Mg-TC]+ induces a defined conformational change in TetR, which locks the DNA-binding domains in an orientation that is not compatible with DNA-binding any more. At the center of this conformational change is a pendulum-like motion of helix α4, which links the effector-binding domain to the DNA-binding domain in TetR (12). During this rotation, His64 acts as a C-terminal pivot and is anchored tightly to [Mg-TC]+. The tight fixation of helix α4 increases the distance between the midpoints of the DNA recognition helices α3 and α3’ by 3 Å, thereby abolishing DNA binding in the induced complex (Figure 6D–F) (13).
Although the mechanism that we observe for revTetR differs considerably from that of wild-type TetR, it is not unparalleled. In fact, it constitutes the second most common mechanism by which repressors exert their function. Bacterial repressor proteins can be classified in general into two categories regarding the mechanism of interaction with their ligands. In the first category, the effector binds to a preformed oligomer and alters its affinity for DNA by triggering a conformational change in the protein. In the second, the small molecule effector acts by modulating the assembly properties and structural organization of the protein. Whereas TetR, LacI and the tryptophan repressor from E. coli belong to the first class (8,12,37), the tyrosine, arginine and biotin repressors belong to the second (38–41). Interestingly, the mechanism that is used to regulate gene expression does not depend on whether the effector molecule takes over the role of a corepressor or an inducer, since in the tryptophan repressor the ligand tryptophan acts as a corepressor. Overall, the mechanism that we propose for revTetR closely resembles the disorder to order transition that has been reported for the transcriptional regulator TraR (42) and the gain of function upon ligand binding reported of the E. coli biotin repressor. The biotin repressor is allosterically activated through the binding of the corepressor bio-5′-AMP and similarly to revTetR becomes then resistant to limited proteolysis (39,41).
This is the first study exploring the structure and mechanism of a reverse repressor variant. We showed that the reversal of function observed in revTetR1 is not based on a mere reversal of the mechanism that regulates wild-type TetR but that the single mutation Leu17Gly enables revTetR1 to switch mechanisms namely from a mechanism in wild-type TetR, that is based on an defined conformational change to an effector-induced disorder–order transition mechanism in revTetR. By doing so, it switches between the two most commonly observed mechanisms used by repressor proteins to regulate gene transcription in bacteria. The single point mutant revTetR1 might therefore represent a missing link that not only highlights the close relationship between these mechanisms but also demonstrates how these distinct mechanisms could easily have evolved from a common ancestor. Our study further emphasizes how single point mutations can engender unexpected leaps in protein function, a property so far predominantly attributed to the simultaneous appearance of correlated mutations. It is quite likely that such functional leaps have played important roles during the evolution of today's proteome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank C. Kisker and H. Schindelin from University of Würzburg for providing access to the spectropolarimeter. We thank C. Jäger from the Computer Chemistry Center, Erlangen, for generating energy-minimized coordinates for the ligand ATC. Financial support for this work was obtained from the Volkswagen foundation, Hannover, Gemany, the BIGSS graduate school financed by the state of Bavaria, the DFG-SFB473 from the Deutsche Forschungsgemeinschaft, Bonn, Germany and the Fonds der Chemischen Industrie (Frankfurt, Germany).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 2.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergthorsson U, Andersson DI, Roth JR. Ohnos dilemma: evolution of new genes under continuous selection. Proc. Natl Acad. Sci. USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 5.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher MA, Choi KY, Lu F, Zalkin H, Brennan RG. Mechanism of corepressor-mediated specific DNA binding by the purine repressor. Cell. 1995;83:147–155. doi: 10.1016/0092-8674(95)90243-0. [DOI] [PubMed] [Google Scholar]
- 7.Swint-Kruse L, Larson C, Pettitt BM, Matthews KS. Fine-tuning function: correlation of hinge domain interactions with functional distinctions between LacI and PurR. Protein Sci. 2002;11:778–794. doi: 10.1110/ps.4050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell CE, Lewis M. A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 9.Kisker C, Hinrichs W, Tovar K, Hillen W, Saenger W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 1995;247:260–280. doi: 10.1006/jmbi.1994.0138. [DOI] [PubMed] [Google Scholar]
- 10.Lederer T, Takahashi M, Hillen W. Thermodynamic analysis of tetracycline-mediated induction of Tet repressor by a quantitative methylation protection assay. Anal. Biochem. 1995;232:190–196. doi: 10.1006/abio.1995.0006. [DOI] [PubMed] [Google Scholar]
- 11.Orth P, Cordes F, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Conformational changes of the Tet repressor induced by tetracycline trapping. J. Mol. Biol. 1998;279:439–447. doi: 10.1006/jmbi.1998.1775. [DOI] [PubMed] [Google Scholar]
- 12.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 13.Saenger W, Orth P, Kisker C, Hillen W, Hinrichs W. The tetracycline repressor-A paradigm for a biological switch. Angew. Chem. Int. Ed. Engl. 2000;39:2042–2052. doi: 10.1002/1521-3773(20000616)39:12<2042::aid-anie2042>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 14.Henssler EM, Bertram R, Wisshak S, Hillen W. Tet repressor mutants with altered effector binding and allostery. FEBS J. 2005;272:4487–4496. doi: 10.1111/j.1742-4658.2005.04868.x. [DOI] [PubMed] [Google Scholar]
- 15.Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholz O, Henssler EM, Bail J, Schubert P, Bogdanska-Urbaniak J, Sopp S, Reich M, Wisshak S, Kostner M, Bertram R, et al. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol. Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 17.Berens C, Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 18.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 19.Kleina LG, Miller JH. Genetic studies of the lac repressor. XIII. Extensive amino acid replacements generated by the use of natural and synthetic nonsense suppressors. J. Mol. Biol. 1990;212:295–318. doi: 10.1016/0022-2836(90)90126-7. [DOI] [PubMed] [Google Scholar]
- 20.Suckow J, Markiewicz P, Kleina LG, Miller J, Kisters-Woike B, Muller-Hill B. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J. Mol. Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 21.Luckner SR, Klotzsche M, Berens C, Hillen W, Muller YA. How an agonist peptide mimics the antibiotic tetracycline to induce tet-repressor. J. Mol. Biol. 2007;368:780–790. doi: 10.1016/j.jmb.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 29.Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr. D Biol. Crystallogr. 2005;61:465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 31.Howlin B, Butler SA, Moss DS, Harris GW, Driessen HPC. TLSANL: TLS parameter-analysis program for segmented anisotropic refinement of macromolecular structures. J. Appl. Crystallogr. 1993;26:622–624. [Google Scholar]
- 32.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.DeLano W. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2003. [Google Scholar]
- 34.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichheld SE, Davidson AR. Two-way interdomain signal transduction in tetracycline repressor. J. Mol. Biol. 2006;361:382–389. doi: 10.1016/j.jmb.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 37.Zhang RG, Joachimiak A, Lawson CL, Schevitz RW, Otwinowski Z, Sigler PB. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987;327:591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- 38.Bailey MF, Davidson BE, Minton AP, Sawyer WH, Howlett GJ. The effect of self-association on the interaction of the Escherichia coli regulatory protein TyrR with DNA. J. Mol. Biol. 1996;263:671–684. doi: 10.1006/jmbi.1996.0607. [DOI] [PubMed] [Google Scholar]
- 39.Kwon K, Streaker ED, Ruparelia S, Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J. Mol. Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 40.Van Duyne GD, Ghosh G, Maas WK, Sigler PB. Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 1996;256:377–391. doi: 10.1006/jmbi.1996.0093. [DOI] [PubMed] [Google Scholar]
- 41.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc. Natl Acad. Sci. USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl Acad. Sci. USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 45.Takahashi M, Altschmied L, Hillen W. Kinetic and equilibrium characterization of the Tet repressor-tetracycline complex by fluorescence measurements. Evidence for divalent metal ion requirement and energy transfer. J. Mol. Biol. 1986;187:341–348. doi: 10.1016/0022-2836(86)90437-7. [DOI] [PubMed] [Google Scholar]
- 46.Scholz O, Schubert P, Kintrup M, Hillen W. Tet repressor induction without Mg2+ Biochemistry. 2000;39:10914–10920. doi: 10.1021/bi001018p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.