Abstract
We assign a function for a small protein, YacG encoded by Escherichia coli genome. The NMR structure of YacG shows the presence of an unusual zinc-finger motif. YacG was predicted to be a part of DNA gyrase interactome based on protein–protein interaction network. We demonstrate that YacG inhibits all the catalytic activities of DNA gyrase by preventing its DNA binding. Topoisomerase I and IV activities remain unaltered in the presence of YacG and its action appears to be restricted only to DNA gyrase. The inhibition of the enzyme activity is due to the binding of YacG to carboxyl terminal domain of GyrB. Overexpression of YacG results in growth inhibition and alteration in DNA topology due to uncontrolled inhibition of gyrase.
INTRODUCTION
The functional assignment of proteins identified in genome sequencing projects is a major challenging task in the postgenomic era. However, even for the most intensively studied eubacteria, Escherichia coli, a number of the open reading frames (ORFs) have unknown functions owing to their unrelatedness to other functionally well-characterized proteins. While loss of function mutational analysis has been successful in identifying protein functions in several cases, it fails miserably in determining the function of ‘non essential’ genes. Further, with the advent of structural genomics, there has been a rapid increase in the repertoire of proteins whose structures are known but functions are still elusive. Predicting function from structure is a relatively young and exciting field but has its own limitations in defining the functions for proteins with novel and unusual structural motifs (1).
Interaction network of protein complexes involved in diverse biological processes in E. coli was uncovered and validated by sequential rounds of tagging and purification. In this network, the interacting partners of protein complexes were purified by affinity chromatography and identified by mass spectrometry. A zinc-finger protein, YacG of yet unknown function, was shown to be a member of DNA gyrase interactome in the network (2). The NMR structure of YacG revealed the presence of a unique zinc-finger motif in this protein and an unstructured tail (3). No function could be assigned to this protein from any studies so far (2–4).
Amongst the enzymes responsible for maintaining the topological state of DNA in cells, DNA gyrase is unique in its ability to catalyze negative supercoiling of DNA in an ATP-dependent fashion (5). A steady state level of negative supercoiling is required for chromosome condensation, transcription initiation and enzyme complex movement during replication and transcription (6). The optimal level of intracellular activity of DNA gyrase is very crucial and strictly controlled for cell division, functioning and survival. In the crowded cellular confines, uncontrolled DNA gyrase activity could be disastrous to the cell. It is now apparent that certain endogenous proteins play a role in regulating DNA gyrase activity. For example, in case of thioredoxins (TrxA, TrxC) from Rhodobacter sp., a change in oxygen tension influences their redox-state, resulting in altered gyrase activity (7). Recently, few other cellular proteins have been identified and shown to be inhibitors of DNA gyrase. These include GyrI (8,9), MurI (10,11) and MfpA (12). In the present study, we describe the function for YacG in specific inhibition of DNA gyrase. Our studies reveal that YacG inhibits the enzyme activity with a mechanism of action distinct from that of well characterized group of gyrase inhibitors, viz. quinolones and coumarins (13).
MATERIALS AND METHODS
Bacterial strains and plasmids
E. coli strain BL26 (DE3) was used for overexpression of YacG from pET15b construct. pTrc99C-yacG was generated and used for assessing the in vivo effect of YacG overexpression in DH10B strain of E. coli. Overexpression plasmids for E. coli topoisomerase IV, DNA gyrase and topoisomerase I constructs were employed for purifying ParC, ParE, GyrA, GyrB and topoisomerase I, respectively.
Enzyme and substrate preparation
E. coli DNA gyrase subunits, GyrA and GyrB were purified as described previously (14). E. coli topoisomerase I and topoisomerase IV subunits, ParC and ParE were purified as described earlier (15). Supercoiled pUC18 and pBR322 were prepared by standard DNA purification protocols (16).
Enzyme assays
Supercoiling assays were performed in supercoiling buffer [35 mM Tris–HCl pH-7.5, 5 mM MgCl2, 25 mM potassium glutamate, 2 mM spermidine, 2 mM ATP, 50 mg/l bovine serum albumin (BSA) and 90 mg/l yeast tRNA in 5% (v/v) glycerol] as described (10). DNA gyrase was incubated with different concentrations of BSA or YacG prior to the addition of DNA substrate and incubated at 37°C for 15 min. The relaxation reactions were carried out with supercoiled DNA substrate and 100 nM enzyme in the buffer devoid of ATP at 37°C for 60 min. The decatenation reactions were carried out with 300 ng kinetoplast DNA and 100 nM DNA gyrase as described earlier (10). Relaxation assays with E. coli topoisomerase I and topoisomerase IV were performed as described (10,17). Electrophoretic mobility shift assays (EMSAs) were carried out as before (10). 1 μM GyrA was used to assess the intrinsic DNA-binding activity of the subunit alone and EMSAs were carried out at 37°C as described (18). ATPase assays were performed as described earlier (10). The reactions (20 μl each) were carried out in supercoiling buffer containing 2 mM ATP and 0.02 μCi of [γ-32P] ATP, at 37°C for 30 min. The reactions were terminated by adding chloroform and 2 μl of the aqueous layer was resolved on polyethyleneimine–cellulose thin layer chromatography plate. The spots corresponding to ATP and Pi were quantitated using PhosporImager. Sonicated salmon sperm DNA (150 μg/ml) was used wherever indicated.
Surface plasmon resonance (SPR)
YacG was immobilized on a CM5 sensor surface via amine coupling in acetate buffer (pH3.0). The surface was blocked with ethanolamine hydrochloride. The interaction was assessed in a buffer containing 35 mM Tris–HCl pH8.0, 1 mM EDTA and 50 mM NaCl. Varying amounts of GyrA and GyrB were passed over the immobilized YacG in each of the channel and the subsequent changes in the resonance units were recorded in a BIAcore 2000 system (Pharmacia) as described earlier (10). All the proteins were dialyzed against the running buffer prior to the experiment and the surface was regenerated using 10 mM NaOH.
Protease probing
2 μg each of GyrA and GyrB were subjected to partial tryptic digestion in absence or presence of YacG. Reactions were carried out in supercoiling buffer at 37°C for 10 min with 1μg/ml trypsin (Sigma). Samples were quenched by adding an equal volume of SDS loading buffer [125 mM Tris–HCl (pH 6.8), 20% (v/v) glycerol, 4% (w/v) SDS, 10% (v/v) 2-mercaptoethanol and 0.004% (w/v) bromophenol blue] and boiling for 5 min. The products were analyzed by SDS-PAGE. The fragments were eluted from the gel and subjected to tryptic mass fingerprinting analysis.
Ni-NTA pull-down assays
2 μg GyrB was preincubated with 4 μg of YacG in the supercoiling buffer for 30 min at 37°C followed by the addition of Ni-NTA sepharose beads. After 2 h of incubation at 4°C in a rotary shaker, followed by centrifugation, the supernatant was removed and the pellet was washed thrice with the buffer (20 mM Tris–HCl, pH-7.5, 50 mM NaCl, 0.1% NP40). Both supernatant and pellet fractions were then resolved on 12% SDS–PAGE and the proteins were visualized by western blotting with anti-GyrB polyclonal antibodies and anti-His tag monoclonal antibodies to probe for GyrB and YacG, respectively.
Cell growth and plasmid topology measurements
DH10B cells transformed with either pTrc99C vector or pTrc-yacG construct were grown in M9 media [Na2HPO4 6 g/l, KH2PO4 3 g/l, NaCl 0.5 g/l, NH4Cl 1 g/l, CaCl2 11 mg/l, supplemented with vitamin B1 (1 μg/ml) and 0.4% glucose] in the presence of 0.1 mM IPTG at 37°C and growth was monitored spectrophotometrically. The plasmids (pTrc99C vector or pTrc99C-yacG) were extracted by alkaline lysis method from the cultures grown till mid-logarithmic phase and the plasmid topology was visualized on agarose gel containing chloroquine (4 μg/ml) as described (19).
RESULTS
YacG inhibits DNA gyrase activity
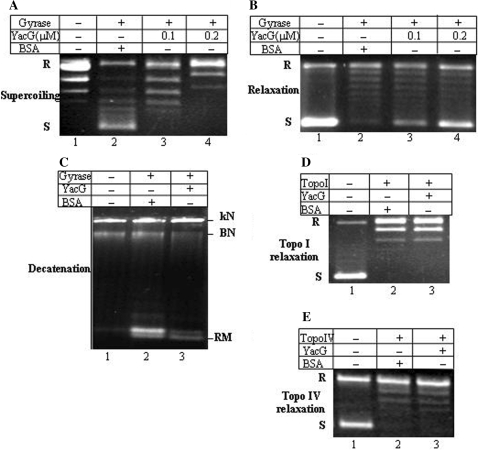
We asked whether the interaction of YacG with DNA gyrase in the interactome described has any functional significance. Specifically, what is the effect of YacG on the catalytic activity of the enzyme? For this, YacG was purified to apparent homogeneity (Supplementary Figure S1). The data presented in Figure 1A show that YacG inhibited supercoiling activity of DNA gyrase. BSA (Figure 1A, lane 2) or some other proteins tested (not shown) did not inhibit the enzyme activity. YacG also inhibited other reactions catalyzed by DNA gyrase viz. ATP-independent relaxation activity in a dose-dependent manner (Figure 1B) and also the decatenation activity (Figure 1C).
Figure 1.
YacG is a specific inhibitor of DNA gyrase. E. coli DNA gyrase activities are influenced by YacG (A–C). Supercoiling (A) and relaxation (B) activities of the enzyme. 10 and 100 nM enzyme was used for supercoiling and relaxation assays, respectively; lane 1, relaxed or supercoiled pUC18; lane 2, gyrase and 0.2 μM BSA; lanes 3 and 4, gyrase, 0.1 and 0.2 μM YacG, respectively; (C) decatenation activity. 100 nM enzyme was used for the reactions, lane 1, kinetoplast DNA; lane 2, DNA gyrase and 0.2 μM BSA; lane 3, DNA gyrase and 0.2 μM YacG. (D) Effect on E. coli topoisomerase I activity. 20 nM enzyme was used, lane 1, supercoiled pUC18; lane 2, topoisomerase I and 1 μM BSA; lane 3, topoisomerase I and 1 μM YacG. (E) Effect of YacG on topoisomerase IV from E. coli. 40 nM enzyme was used for ATP-dependent relaxation assay. Lane 1, supercoiled pUC18; lane 2, topoisomerase IV and 1 μM BSA; lane 3, topoisomerase IV and 1 μM YacG; S and R represent supercoiled and relaxed plasmid DNA; kN, kinetoplast; kDNA network; BN, broken network; RM, released minicircles. All the assays were repeated at least thrice. The representative figures have been presented.
YacG action is specific to DNA gyrase
Bacterial topoisomerase I relaxes negatively supercoiled DNA in an ATP-independent manner. DNA relaxation activity of E. coli topoisomerase I was not affected in the presence of YacG, even at a very high concentration (Figure 1D). Topoisomerase IV is the other eubacterial type II topoisomerase, which catalyses ATP-dependent relaxation of negatively supercoiled DNA (20). Topoisomerase IV activity from E. coli was also unaltered in the presence of YacG (Figure 1E). These studies indicate that the action of YacG is restricted to DNA gyrase amongst the major cellular topoisomerases.
Mechanism of inhibition by YacG
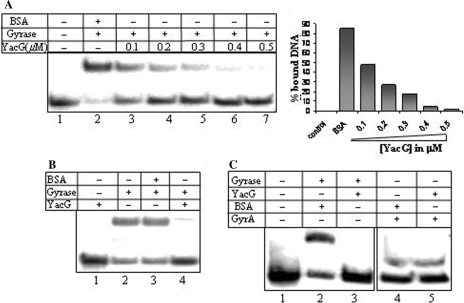
Since YacG inhibited all the three catalytic activities of DNA gyrase, it seems to target a step common to these reactions. During its catalytic cycle, gyrase remodels topology of DNA by passing one double-stranded DNA segment through a transient, enzyme-mediated break in another (21). Thus the reaction involves a series of orchestrated steps initiated by the binding of the tetrameric (A2B2 complex) holoenzyme to DNA followed by cleavage, strand passage and religation. ATP hydrolysis is required for the enzyme turnover to begin the next round of supercoiling. To elucidate the mode of action by YacG, we tested its effect on the first step of the gyrase reaction, i.e. DNA binding by EMSAs. The amount of retarded gyrase–DNA noncovalent complex was significantly reduced in the presence of YacG, with a concomitant increase in the free DNA species (Figure 2A). YacG did not bind DNA on its own, but could destabilize the preformed gyrase–DNA complex (Figure 2B). GyrA subunit of the enzyme is primarily responsible for the DNA binding while GyrB possesses the ATPase activity. However, the intrinsic DNA binding by GyrA subunit alone was unhindered in the presence of YacG, upon prior incubation of GyrA and YacG (Figure 2C, lanes 4 and 5). YacG did not abolish inter-subunit (GyrA–GyrB) interaction (not shown). These studies reveal that YacG inhibits all the catalytic activities of DNA gyrase by preventing the holoenzyme–DNA interaction. As a consequence, the subsequent steps in the gyrase reaction cycle viz. DNA cleavage are also affected (not shown).
Figure 2.
YacG prevents gyrase–DNA interaction. (A) Noncovalent complex formation by gyrase holoenzyme. EMSAs were carried out with 100 nM enzyme and radiolabeled 240 bp strong gyrase site (SGS) at 4°C; lane 1, free SGS; lane 2, DNA gyrase and 0.5 μM BSA; lane 3–7, gyrase and increasing concentrations (0.1–0.5 μM) of YacG. (B) YacG can destabilize preformed gyrase–DNA complex, lane 1, DNA in the presence of YacG; lane 2, gyrase and DNA for 30 min; lanes 3 and 4, gyrase and DNA preincubated for 15 min followed by addition of 0.5 μM BSA and YacG, respectively. (C) Effect on intrinsic DNA-binding activity of GyrA subunit. 1 μM GyrA subunit or 100 nM DNA gyrase incubated with radiolabelled SGS, in the presence or absence of YacG at 37°C; lane 1, free SGS; lane 2, DNA gyrase and 0.5 μM BSA; lane 3, gyrase and 0.5 μM YacG; lane 4, 1 μM GyrA and 5 μM BSA; lane 5, 1 μM GyrA and 5 μM YacG.
Direct interaction of YacG with GyrB
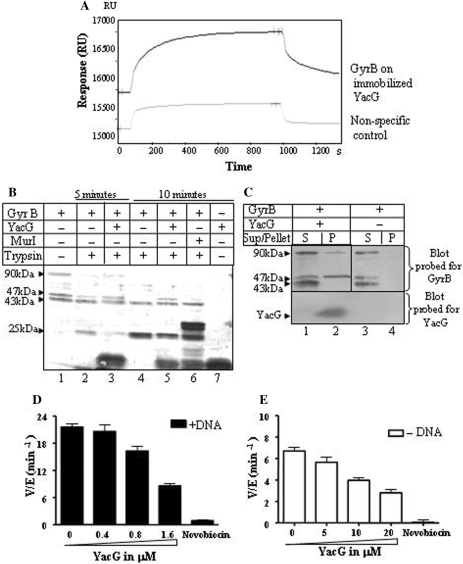
To understand how YacG specifically affects only gyrase and to determine which subunit is involved in the interaction, we examined the direct binding between the proteins. We carried out SPR, pull-down assays, proteolytic protection and gel filtration for this purpose. SPR analysis revealed a significant increase in the response units upon binding of GyrB to immobilized YacG thereby indicating a direct interaction between the two proteins (Figure 3A). Based on SPR data, we could obtain an apparent dissociation constant (Kd) value of 350 nM. No interaction could be detected between YacG and GyrA (not shown). Limited proteolysis experiments described in Figure 3B further supported these findings. GyrB harbors two stable domains, a 43 kDa amino terminal domain (NTD) and a 47 kDa carboxyl terminal domain (CTD). Proteolysis of GyrB produces primarily two fragments, a 43 kDa NTD, and 25 kDa polypeptide generated from the 47 kDa CTD (22). Upon trypsin digestion of GyrB in the presence of YacG, 47 kDa CTD was protected, with a concomitant reduction in the intensity of the 25 kDa fragment (Figure 3B, lanes 3 and 5). Protection was not observed in the presence of MurI, another endogenous gyrase interacting protein, which does not bind to GyrB (Figure 3B, lane 6) (10). As expected, the cleavage pattern of GyrA was unaltered in the presence of YacG (not shown). These data suggest that YacG binding alters protease sensitivity and protects the CTD of GyrB. Since the full length GyrB and the 47 kDa CTD fragment of GyrB interacted with YacG, these proteins were also precipitated along with YacG in the pellet fraction in the pull-down assays (Figure 3C, lane 2). Similar pull-down assays with gyrase holoenzyme (A2B2 complex) revealed that YacG did not hamper the interaction between GyrA and GyrB subunits (data not shown). Gel filtration analysis also revealed a specific interaction between YacG and GyrB subunit of gyrase (Supplementary Figure S2).
Figure 3.
Interaction between YacG and DNA gyrase subunits and its effect on ATPase activity. (A) SPR refractometry. Black colored line represents response for E. coli GyrB passed over YacG immobilized on CM5 sensor surface, grey colored line represents response for nonspecific control. Interaction was assessed in a buffer as described in Materials and Methods section. Passing 1 μM GyrB over immobilized YacG resulted in increase of 92 RU above nonspecific control. (B): Effect of YacG on trypsin-mediated proteolytic signature of GyrB subunit. 2 μg of GyrB was subjected to partial tryptic digestion: lane 1, undigested GyrB; lanes 2 and 4, GyrB and trypsin; lanes 3 and 5, GyrB, trypsin and YacG; lane 6, GyrB, trypsin and MurI; lane 7, undigested YacG. Duration of digestion: 5 min in lanes 2 and 3, 10 min in lanes 4–6; GyrB preparation (lane 1) used here was a mixture of full length GyrB and two stable fragments (47 kDa CTD and 43 kDa NTD). All these fragments were verified by mass spectrometry. (C) Western blots for the pull-down assay with Ni-NTA sepharose beads. The assays were carried out as described in the Materials and Methods section. Top panel: blot probed with anti-GyrB polyclonal antibodies. Lanes 1 and 2, supernatant and pellet fractions of the mixture of GyrB and YacG pulled down by Ni-NTA sepharose beads; lanes 3 and 4, supernatant and pellet fractions of the GyrB alone pulled down by Ni-NTA sepharose beads; bottom panel, same blot probed for YacG with anti-His tag antibodies. (D and E) Effect of YacG on ATPase activity of DNA gyrase. (D) DNA-stimulated ATPase activity. Reactions were performed with 400 nM each of GyrA and GyrB subunits and 150 μg ml−1 DNA, lane 1, gyrase; lanes 2–4, gyrase with 0.4, 0.8 and 1.6 μM YacG, respectively; lane 5, gyrase and 1 μg/ml novobiocin. (E) Intrinsic ATPase activity. 1.4 μM GyrB subunit was used, DNA and GyrA were omitted; lane 1, GyrB; lanes 2–4, GyrB with 5, 10, 20 μM YacG, respectively; lane 5, GyrB and 1 μg/ml novobiocin. 2 mM ATP present in all the reactions. The average of three independent experiments is depicted graphically.
GyrB subunit of the enzyme exhibits intrinsic ATPase activity, which is enhanced in the presence of GyrA and DNA (23). Binding of YacG to GyrB may alter the ATPase activity of GyrB. We observed that YacG inhibited DNA-stimulated ATPase activity of DNA gyrase (Figure 3D). Since YacG prevents DNA binding by gyrase, these results corroborate its mechanism of action. However, YacG also inhibited intrinsic ATPase activity of GyrB, albeit to a much lesser extent (Figure 3E). This latter effect indicates that the direct physical interaction between YacG and GyrB probably influences the ATP binding function of GyrB to a certain extent. YacG also inhibited the DNA independent ATPase activity of gyrase heterotetramer to a similar extent (not shown).
Growth profile upon YacG overexpression
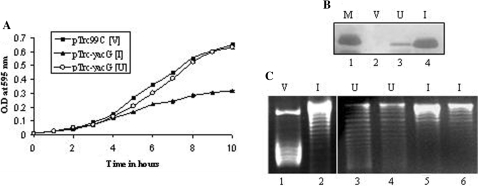
Since DNA gyrase is indispensable for all DNA transaction processes, one would expect the endogenous inhibitors to act only transiently on the enzyme in the normal intracellular milieu. Hence, inhibitory effect may be difficult to visualize in the highly regulated intracellular state. To extend our in vitro studies to in vivo scenario, we evaluated the growth pattern of the cells and the topological status of the DNA upon YacG overexpression. In the overexpressed state, continuous inhibition of DNA gyrase would hamper vital cellular processes and cause a growth disadvantage. The results presented in Figure 4A show that YacG overexpression (Figure 4B) results in growth inhibition. In order to confirm that the observed growth retardation was indeed due to uncontrolled inhibition of gyrase, we monitored the topological status of the plasmids extracted from these bacteria. The DNA was in a less supercoiled state in YacG expressing cells in contrast to the plasmid extracted from the cells harboring the vector alone (Figure 4C). These results indicate that the interaction of gyrase with YacG leads to the reduction in the negatively supercoiled status of the DNA and also a consequent inhibitory effect on cell growth.
Figure 4.
YacG overexpression leads to growth inhibition and relaxed DNA topology. (A) Growth profiles of DH10B strain of E. coli transformed with either pTrc99C vector (filled square) or pTrc99C-yacG construct, grown in the absence (open circle) or presence (filled triangle) of 0.1 mM IPTG. (B) Expression of YacG from pTrc99C-yacG construct in DH10B cells. Western blot with anti-His tag antibody to probe for His6-YacG. Lane 1, purified YacG marker (M); lane 2, induced cell extract from DH10B cells harboring pTrc99C vector; lane 3, extract from pTrc99C-yacG harboring cells grown in the absence of IPTG (U); lane 4, extract from pTrc99C-yacG harboring cells after induction with 0.1 mM IPTG (I). (C) Visualization of plasmid topology on agarose gel containing chloroquine (4 μg/ml). DH10B cells transformed with either pTrc99C vector (V) or pTrc99C-yacG construct were grown till mid-logarithmic phase either in the absence (U) or presence (I) of 0.1 mM IPTG and then plasmids were extracted by alkaline lysis method. Lane 1, pTrc99C vector from cells induced with 0.1 mM IPTG (V); lanes 2, 5 and 6, pTrc99C-yacG from cells induced with 0.1 mM IPTG (I); lanes 3 and 4, pTrc99C-yacG from cells grown in the absence of IPTG (U). Plasmid sizes are approximately in the range of 4.2 kb.
DISCUSSION
In the present study, we describe the function for a small zinc-finger protein, YacG. We demonstrate that YacG is an endogenous inhibitor of DNA gyrase. It physically interacts with the GyrB subunit and sequesters the enzyme away from its substrate DNA. The action of YacG is specific for gyrase. Other major cellular topoisomerases such as DNA topoisomerase I and topoisomerase IV are not inhibited by YacG.
The NMR structure of YacG revealed the presence of a novel sequence motif in the zinc-finger family of proteins (3). The unique consensus motif for the cysteine residues (-C-X2-C-X15-C-X3-C-) is conserved in all YacG homologues but absent in any other group of proteins. In this motif, zinc is coordinated by four cysteines residues, and the sequence pattern is similar to the characteristic ‘rubredoxin knuckle’ (24). It has a similar architecture and Zn2+ coordination to N-terminal Zn finger of GATA-1 (NF). Based on this structural similarity, YacG was thought to be involved in transcription either through protein–protein interactions (e.g. NF) or by DNA binding (C-terminal Zn finger of GATA-1). However, the critical residues important for these activities are not conserved in YacG (3) and the physiological function of the protein remained elusive until the present study.
Identification of topoisomerase interacting proteins is an emerging area. YacG is a new member in the growing list of DNA gyrase modulatory proteins. From the detailed characterization presented here, it is evident that its mode of action has similarity to other chromosomally encoded gyrase inhibitors like GyrI, MurI and MfpA (8–12). All these inhibitors essentially influence the enzyme activity by sequestering the enzyme away from DNA. None of them cause cytotoxicity, which usually arises as a result of DNA damage caused by accumulation of gyrase–DNA covalent intermediate. Comparative analysis of these proteinaceous inhibitors, however, does not reveal a common motif or structural fold required for their ability to inhibit DNA gyrase. Further, some of these endogenous proteins have their own primary physiological function. For instance, in addition to its gyrase inhibitory property MurI is an integral component of cell wall biosynthesis machinery (11). Thus, it appears that in at least few of the cases, their gyrase interaction function is of more recent origin.
At present, the physiological role of many of these inhibitors is not very apparent. In the intracellular milieu, one would expect the levels of gyrase and YacG to be vastly different. Gyrase is an integral house keeping function present at about 1000–3000 molecules/cell (25). In contrast, YacG is likely to be present at very low levels. The apparent Kd observed in the present studies would indicate inhibition of only few gyrase molecules in vivo without having any adverse effects to the cell under normal cellular growth. However, under conditions when YacG is upregulated, its modulation of DNA gyrase activity might lead to important physiological consequences. The activity of DNA gyrase needs to be regulated at various stages of cell growth as uncontrolled gyrase activity can be disastrous for the cell. YacG and other such proteins could bind transiently to DNA gyrase to sequester it away under situations when gyrase activity needs to be checked and kept under control. Sequestration of DNA gyrase by these modulators might serve as a ‘check point’ coordinating the cell division and DNA replication.
Finally, complete genome sequences of diverse bacteria have reinvigorated an effort to assign function to all the genes in any given organism. Until functions are assigned to the unknown ORFs, any organism's capabilities cannot be completely described. Thus, in this postgenome sequencing era, an immediate challenge would be to make best use of the large amount of high throughput data to determine functions for the currently uncharacterized proteins. Several approaches have been developed for determining the protein function using the information derived from sequence similarity, phylogenetic profiles and protein–protein interactions. Assignment of function for YacG described in this study is an example for integration of data obtained from both proteomic and reductionist approach.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank J. C. Wang, A. Maxwell, H. Hiasa, C. Arrowsmith for overexpressing constructs of E. coli topoisomerase I, DNA gyrase, topoisomerase IV, YacG, respectively and H. K. Majumder for kinetoplast DNA. We acknowledge the phosphorimager, Biacore and proteomics facilities of Indian Institute of Science, supported by the Department of Biotechnology, Government of India. Vandana Kumari is acknowledged for technical assistance. S.S. is the recipient of senior research fellowship from Council of Scientific and Industrial Research, Government of India. This work was supported by the Centre for Excellence grant from Department of Biotechnology, Government of India. Funding to pay the Open Access publication charges for this article was provided by COE grant from Department of Biotechnology, Government of India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg I, Jambon M, Godzik A. New avenues in protein function prediction. Protein Sci. 2006;15:1527–1529. doi: 10.1110/ps.062158406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 3.Ramelot TA, Cort JR, Yee AA, Semesi A, Edwards AM, Arrowsmith CH, Kennedy MA. NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins. Proteins. 2002;49:289–293. doi: 10.1002/prot.10214. [DOI] [PubMed] [Google Scholar]
- 4.Merlin C, McAteer S, Masters M. Tools for characterization of Escherichia coli genes of unknown function. J. Bacteriol. 2002;184:4573–4581. doi: 10.1128/JB.184.16.4573-4581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nöllmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Li K, Pasternak C, Hartig E, Haberzettl K, Maxwell A, Klug G. Thioredoxin can influence gene expression by affecting gyrase activity. Nucleic Acids Res. 2004;32:4563–4575. doi: 10.1093/nar/gkh794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakanishi A, Oshida T, Matsushita T, Imajoh-Ohmi S, Ohnuki T. Identification of DNA gyrase inhibitor (GyrI) in Escherichia coli. J. Biol. Chem. 1998;273:1933–1938. doi: 10.1074/jbc.273.4.1933. [DOI] [PubMed] [Google Scholar]
- 9.Chatterji M, Nagaraja V. GyrI: a counter-defensive strategy against proteinaceous inhibitors of DNA gyrase. EMBO Rep. 2002;3:261–267. doi: 10.1093/embo-reports/kvf038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengupta S, Shah M, Nagaraja V. Glutamate racemase from Mycobacterium tuberculosis inhibits DNA gyrase by affecting its DNA-binding. Nucleic Acids Res. 2006;34:5567–5576. doi: 10.1093/nar/gkl704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashiuchi M, Kuwana E, Yamamoto T, Komatsu K, Soda K, Misono H. Glutamate racemase is an endogenous DNA gyrase inhibitor. J. Biol. Chem. 2002;277:39070–39073. doi: 10.1074/jbc.C200253200. [DOI] [PubMed] [Google Scholar]
- 12.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 13.Maxwell A. DNA gyrase as a drug target. Biochem. Soc. Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell A, Howells AJ. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 1999;94:135–144. doi: 10.1385/1-59259-259-7:135. [DOI] [PubMed] [Google Scholar]
- 15.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 18.Huang YY, Deng JY, Gu J, Zhang ZP, Maxwell A, Bi LJ, Chen YY, Zhou YF, Yu ZN, Zhang XE. The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA) Nucleic Acids Res. 2006;34:5650–5659. doi: 10.1093/nar/gkl695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterji M, Sengupta S, Nagaraja V. Chromosomally encoded gyrase inhibitor GyrI protects Escherichia coli against DNA-damaging agents. Arch. Microbiol. 2003;180:339–346. doi: 10.1007/s00203-003-0598-4. [DOI] [PubMed] [Google Scholar]
- 20.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 21.Schoeffler A, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 22.Kampranis SC, Gormley NA, Tranter R, Orphanides G, Maxwell A. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry. 1999;38:1967–1976. doi: 10.1021/bi982320p. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell A, Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 1984;259:14472–14480. [PubMed] [Google Scholar]
- 24.Perry A, Lian LY, Scrutton NS. Two-iron rubredoxin of Pseudomonas oleovorans: production, stability and characterization of the individual iron-binding domains by optical, CD and NMR spectroscopies. Biochem. J. 2001;354:89–98. doi: 10.1042/0264-6021:3540089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton M, Armitage M, Maxwell A, Dosanjh B, Howells AJ, Norris V, Sigee DC. Immunogold localization of GyrA and GyrB proteins in Escherichia coli. Microbiology. 1994;140:2371–2382. doi: 10.1099/13500872-140-9-2371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.