Abstract
p73, the p53 homologue, exists as a transactivation-domain-proficient TAp73 or deficient deltaN(DN)p73 form. Expectedly, the oncogenic DNp73 that is capable of inactivating both TAp73 and p53 function, is over-expressed in cancers. However, the role of TAp73, which exhibits tumour-suppressive properties in gain or loss of function models, in human cancers where it is hyper-expressed is unclear. We demonstrate here that both TAp73 and DNp73 are able to specifically transactivate the expression of the anti-apoptotic member of the caspase family, caspase-2S. Neither p53 nor TAp63 has this property, and only the p73β form, but not the p73α form, has this competency. Caspase-2 promoter analysis revealed that a non-canonical, 18 bp GC-rich Sp-1-binding site-containing region is essential for p73β-mediated activation. However, mutating the Sp-1-binding site or silencing Sp-1 expression did not affect p73β's transactivation ability. In vitro DNA binding and in vivo chromatin immunoprecipitation assays indicated that p73β is capable of directly binding to this region, and consistently, DNA binding p73 mutant was unable to transactivate caspase-2S. Finally, DNp73β over-expression in neuroblastoma cells led to resistance to cell death, and concomitantly to elevated levels of caspase-2S. Silencing p73 expression in these cells led to reduction of caspase-2S expression and increased cell death. Together, the data identifies caspase-2S as a novel transcriptional target common to both TAp73 and DNp73, and raises the possibility that TAp73 may be over-expressed in cancers to promote survival.
INTRODUCTION
p73 is a member of the p53 family of transcription factors, existing as numerous NH2- and COOH-terminal isoforms (1,2) The NH2-terminal variant, known as the deltaNp73 (DNp73), is generated from an internal intronic promoter and lacks the NH2-terminal transactivation (TA) domain, and hence, has been suggested to bind to and counter the tumour-suppressive properties of the TA proficient full-length TAp73 forms (3,4). However, some reports have suggested that DNp73 have some ability to transactivate target genes due to the presence of a second TA domain, which includes the PxxP motif (5). The COOH-terminal variants arise due to alternate splicing resulting in multiple isoforms that exhibit varying degrees of TApotential (6,7). The longest isoform, the TAp73α, generally shows weaker activity than TAp73β and TAp73γ that exhibit stronger TA potential (7,8). Hitherto, it has been classically thought that the TAp73 forms primarily function as tumour suppressors, albeit weaker than p53 itself, whereas the DNp73 forms act as oncogenes, as has been demonstrated by genetic, over-expression and other in vitro studies (3,9,10).
However, clinical reports analysing p73 expression profile have highlighted a complicating scenario. Not only are the DNp73 forms over-expressed as expected, but also the TAp73 forms are over-expressed in a multitude of human cancers (6,11–17). It was shown that one-third of tumours that over-express DNp73 forms also exhibited concomitant up-regulation of the antagonistic TAp73 (12). Although co-over-expression of DNp73 with TAp73 may nullify the tumour-suppressive properties of the latter in human tumours, it is still unclear why there is a need for TAp73 forms to be over-expressed at all. Recent data from others and us have provided evidence for a role for TAp73 in supporting cellular growth, and hence, in tumour development. Ectopic expression of TAp73 was shown to support cellular survival under defined conditions, and conversely, absence of p73 led to reduced proliferation, through the regulation of AP-1 activity (18). Consistently, TAp73 expression was also found to lead to the activation of the promoter of gastrin, a peptide hormone that is important in determining the progression of a number of human malignancies, and a strong correlation was noted between gastrin and TAp73 levels in gastric cancers (19). In addition, over-expression of both TAp73 and DNp73 seen in upper gastrointestinal carcinomas correlated with TCF-dependent transcriptional activation and the up-regulation of β-catenin in gastrointestinal cells, implying a tumour-promoting role for combined expression of TAp73 and DNp73 (20). Finally, TAp73 was seen to negate p53-mediated suppression of human telomerase expression, suggesting a contributory role for TAp73 in carcinogenesis (21). These data therefore suggest that TAp73 forms may support cellular survival, besides their classical roles as tumour suppressors, though the exact context in which these properties are exhibited is unclear.
Nevertheless, as with p53, p73 forms have also been shown to regulate apoptosis, a critical process suppressing tumourigenesis. TAp73 has been shown to induce the expression of genes like puma and scotin amongst others (22), and absence of the anti-apoptotic DNp73 was shown to lead to massive apoptosis in the developing mouse brain (23). However, whether the core component of the apoptotic machinery—the proteolytic system involving a family of proteases known as caspases (24)—is regulated by p73 members is unclear. There are 14 members in the caspase family, which can be generally grouped into two main groups according to their functions: those involved in cytokine processing (caspase-1, -4, -5, -11 to -14) and those in apoptosis (caspase-2, -3, -6 to -10) (25). Of the apoptotic caspases studied, the function and regulation of caspase-2, -8 and -9 have been the best characterized. Of these, caspase-2 is interesting as it exists as two distinct isoforms with opposing functions: the long caspase-2L form induces cell death, while the short caspase-2S isoform inhibits cell death upon over-expression (26,27). The dominant caspase-2L form is expressed in most tissues, whereas caspase-2S is preferentially expressed in brain and skeletal muscles (27). The two mRNAs differ at their 5′-end, suggesting the existence of distinct transcriptional start sites (28). The 5′ RT–RACE and RNase protection assays showed that the main transcription start site of caspase-2S differs from the transcription start site of caspase-2L. Caspase-2S transcription initiates within intron 1 of the caspase-2 gene and the presence of a TATA box in caspase-2S promoter suggest that under specific conditions, caspase-2S expression can be up-regulated (28). In addition, caspase-2S isoform is produced by the insertion of a 61-bp exon at the 3′-end of the caspase-2 pre-mRNA, which introduces a premature stop codon (27).
Since TAp73 appears to regulate both apoptosis and also support cellular survival, we explored the possibility that it would differentially regulate caspase expression. Interestingly, we found that both TA73β and DN73β isoforms were able to induce the expression of caspase-2S, but not of the other caspases tested. This induction is dependent on a unique 18-bp p73β-recognition sequence on the caspase-2S promoter to which both TA73β and DN73β bind directly in vivo and in vitro. Consistently, over-expression of DN73β in neuroblastoma cells led to elevated levels of caspase-2S expression and these cells were resistant to cell death induced by several means. The data together identify caspase-2S as a novel target of both TA73β and DN73β isoforms, suggesting a role for them in promoting cellular survival.
MATERIALS AND METHODS
Cells, plasmids and transfections
The p53 null H1299 cells and Saos-2 cells inducibly expressing TAp73β have been described (18). SH-SY5Y neuroblastoma cells were transfected with pcDNA or DNp73β and selected on G418 to generate stable transfectants. Saos2-TAp73β inducible cells were maintained in DMEM supplemented with 10% tetracycline free FBS, and TAp73β was induced by addition of 2 μM doxocycline prior to harvesting at the indicated time points.
The 2 × 105 cells (in 6-well dishes) were used for transfection experiments using Lipofectamine PLUS-Reagent according to manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). H1299 cells were transiently transfected with various plasmids, and collected 36 or 24 h later for RT–PCR or luciferase analysis, respectively. siRNA was transfected at 3 μg per well using RNAifect (Qiagen, Valencia, CA, USA), as per manufacturer's protocols.
The pCDNA3-based expression plasmids for p53, TAp63α, TAp63β, TAp73α, TAp73β, TAp73β-292, DNp73β and DNp73β-292 have been described (21). TAp63α and TAp63β expression plasmids are gifts from Dr Giovanni Blandino (Regina Elena Cancer Institute, Rome) and Dr Massimo Broggini (Mario Negri Institute, Milan). The Sp-1 cDNA was a gift from Dr Robert Tjian (University of California, Berkerly, USA) (29). PCM2, Del1, Del2, Del3 and Del4 caspase-2 promoter-luciferase constructs have been described (28). Site-directed mutagenesis was performed as per manufacturer's instructions (Stratagene, La Jolla, CA, USA) using the Del4 promoter to generate Del4 truncations Del4.1, Del4.2 and Del4.3 by PCR cloning, using primers as follows: Del4.1-for: 5′AAAAGGTACCAGCCTGACTCCGCGCAAGG3′, Del4.2-for: 5′AAAAGGTACCTCCTTATGAGGGAAACTATAA3′, Del4.3-for: 5′AAAAGGTACCGTCTCTCGTGGGAAAAGACTGGC3′ and a common reverse primer Del4-rev: 5′ACTTAGATCGCAGATCTCGAGTCGATAC3′. Sp-1 siRNA was purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA), and p73 and the control siRNA were purchased from Qiagen. The sequence of p73 siRNA is as follows: 5′- - - AAGGCAATAATCTCTCGCAGT - - -3′, and targets both TAp73 and DNp73 forms.
Cell death assays
SH-SY5Y cells stably expressing pCDNA or DNp73β were seeded in triplicates in 6-well plates and serum-starved in serum-free DMEM for indicated time periods before harvesting for sub-G1 analysis. Cells were fixed in 70% ethanol overnight, washed twice with cold PBS, treated with RNase A for 20 min before addition of 5 μg/ml PI and analysed by BD Biosciences FACScalibur (Mountain View, CA, USA). Similarly, cells were treated with 20 μM cisplatin for 24 h or serum-starved for 48 h, and analysed for cell viability by their ability to exclude propidium iodide, by flow cytometry. For siRNA experiments, siRNA was transfected 24 h prior to serum-starvation for a further 48 h before analysis of cell death.
Luciferase assays
H1299 cells were transiently transfected with 0.2 μg of the various plasmids along with indicated caspase-2 promoter–reporter constructs and β-galactosidase construct to normalize for transfection efficiency. Luciferase assays were performed as described (21).
RNA analysis
Total RNA was prepared from cells using TRIzol reagent (Invitrogen) as per manufacturer's instructions. Semi-quantitative RT–PCR was performed using TAp73 (32 cycles), TAp63 (30), caspase-2L (33), caspase-2S (33), caspase-8 (30), caspase-9 (30), MDM2 (30), DNp73 (34) and gapdh (22) primers, under the following conditions: 94°C for 3 min, followed by cycling at 94°C for 50 s, 52°C for 50 s and 72°C for 1 min. Primers used are as follows—cas2L-for: 5′GCG GCG CCG AGC GCG GGG TCT TGG3′, cas2L-rev: 5′GTG GGA GGG TGT CCT GGG AAC3′, cas2S-for: 5′GAT GTG GAC CAC AGT ACT CTA G3′, cas2S-rev: 5′TCA TAG AGC AAG AGA GGC GGT G3′, cas8-for: 5′CAA GAA CCC ATC AAG GAT GCC TTG3′, cas8-rev: 5′CCA AAG TCT GTG ATT CAC TAT CC3′, cas9-for: 5′TGA TCG AGG ACA TCC AGC GG3′, cas9-rev: 5′GAA GCG ACG CCG CAA CTT CTC AC3′, mdm2-for: 5′ATG TGC AAT ACC AAC ATG TCT GTA CCT3′, mdm2-rev: 5′AGG GGA AAT AAG TTA GCA CAA TCA TTT GA3′, TAp73-for: 5′TCT GGA ACC AGA CAG CAC CT3′, TAp73-rev: 5′GTG CTG GAC TGC TGG AAA GT3′, DNp73-for: 5′CGC CTA CCA TGC TGT ACG TC3′, DNp73-rev: 5′GTG CTG GAC TGC TGG AAA GT3′, TAp63-for: 5′ACC TGA GTG ACC CCA TGT G 3″, TAp63-rev: 5′CGG GTG ATG GAG AGA GAG CA3″, gapdh-for: 5′ACC CCT TCA TTG ACC TCA AC3′, gapdh-rev: 5′CAG CGC CAG TAG AGG CAG3′.
Immunoblot analysis
Cell lysates prepared in lysis buffer containing 0.5% Nonidet P-40 or luciferase extracts were separated on SDS–polyacrylamide gels and western blotted using anti-p73 (ER15, Oncogene, Cambridge, MA, USA), anti-actin (Sigma, St Louis, MO, USA), anti-Sp-1, anti-TAp63 and anti-p53 (Santa Cruz) antibodies.
Chromatin immunoprecipitation assay
Cells were fixed with 1% formaldehyde for 15 min at room temperature (RT), stopped with glycine to a final concentration of 125 mM for 15min. Treated cells were then washed twice with PBS and once with KM buffer (pH 6.8, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 5 mM dithiothreitol, 10% glycerol, 10 mM MOPS), at 10 min interval. Cells were then lysed for 30 min using 4 ml KM buffer + 1% NP40 + protease inhibitors at 4°C and further incubated in 2.7 ml 5 M NaCl for 60 min at 4°C. Lysates were collected with TE buffer and sonicated with VCX130PB (Jencons, Bridgeville, PA, USA) five times for 10 s each, with about 10 min intervals on ice, centrifuged at 13 000 r.p.m. at 4°C for 30 min and supernatant was collected. A total of 400 μl of supernatant was used for immunoprecipitation with 10 μl of anti-p73 (ER15) or anti-HA (Santa Cruz) antibodies for 2 h at 4°C. 1 μl of Dynabeads® Protein A and G (Invitrogen) were added for a further 2 h. Immune complexes were then washed with 2 × RIPA buffer, 1 × HS buffer (0.1% SDS, 1% Triton-X, 2 mM EDTA, 20 mM Tris–HCl, pH 8, 500 mM NaCl), 1 × LS buffer (0.1% SDS, 1% Triton-X, 2 mM EDTA, 20 mM Tris–HCl, pH 8, 150 mM NaCl), 1 × 0.25 M LiCl buffer, 1 × 0.5 M LiCl buffer at 37°C for 10 min, followed by 2× RIPA buffer, 2× TE buffer. DNA/protein complex was eluted with 100 μl of TE buffer containing 1% SDS and was then reverse cross-linked by 200 mM NaCl at 65°C for 4 h, followed by proteinase K treatment (500 μg/ml) at 55°C for 2 h. DNA fragments were isolated using Qiagen PCR purification kit. PCR was performed using Taq polymerase (Qiagen) in a 50 μl solution as follows: 5 μl of DNA, 1 μl each of 10 pmol/μl of primers, 1 μl of 10 mM dNTPs, 5 μl of 10 × buffer and 0.4 μl of Taq polymerase. PCR conditions are as follows: Caspase-2S promoter containing the 18bp site → 95°C—3 min, 50°C—1 min, 72°C—30 s, 39 × using caspase-2SRE-for: 5′GGA CGC CCG CCC GAG CCG CTC3′ and caspase-2SRE-rev: 5′AGT CTT TTC CCA CGA GAG AGA CAA GGC C3′ (100 bp); non-specific site on Caspase-2S promoter using non-specific-for: 5′GGAATTGTGTGCTGCGGCTG3′ and non-specific-rev: 5′CGCAGAGCTCTAGCGGCGGC3′ (400 bp).
GST protein purification and in vitro DNA/protein binding assay
TAp73α, TAp73β, DNp73β and p53 were cloned into pGEX4T-1 (Pharmacia Biotech, GE, Princeton, NJ, USA) to generate GST-73α, GST-73β, GSTDNp73β and GST-p53 fusion proteins. These plasmids were transformed into BL21 Escherichia coli and cultured in 200 ml LB broth for 4–5 h, until OD600 = 0.5. 1 mM of IPTG was then added to the cultures and further cultured at 37°C for 4 h. The cultures were harvested and washed in PBS, and then lysed by sonication at rating 3, 30 s pulse, 30 s interval, 10× and the lysate was collected by centrifugation. The lysates were incubated with gluthatione beads for 2 h at 4°C. The beads were then washed several times in PBS and bound GST fusion proteins eluted with glutathione elution buffer (10 mM Tris, pH 8, 5 mM glutathione). A 18 bp TAp73β recognition site on caspase-2S promoter containing oligonucleotides, 5′GAC GCC CGC CCG AGC CGC TCC GAG3′, was synthesized with 5′ biotin label on both strands. The biotin-labelled recognition sequence was then attached to avidin-conjugated sepharose beads (Invitrogen). Purified GST proteins diluted in RIPA buffer (0.1% SDS, 1% Triton-X, 2 mM EDTA, 20 mM Tris–HCl, pH 8, 150 mM NaCl) were then incubated with the recognition sequence attached beads for 2 h at 4°C. After incubation, the mix was washed six times with RIPA buffer. After the last wash, 30 μl of protein loading buffer was added to the beads and boiled for 5 min before loading onto SDS–acrylamide gel for separation.
RESULTS
TAp73β, but not TAp73α, p53 or TAp63, induces caspase-2S expression
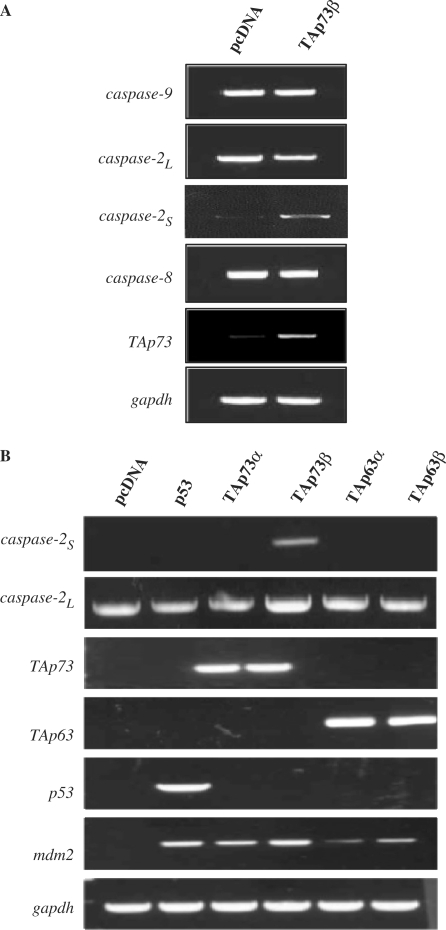
We first evaluated if p73 can transcriptionally regulate any of the initiator caspases. TAp73β was ectopically expressed in p53 null H1299 cells and the levels of caspase-2, -8 and -9 mRNA were determined by semi-quantitative RT–PCR. Although none of the full-length initiator caspases were up-regulated by TAp73β over-expression, the short isoform of the caspase-2, caspase-2S, was significantly up-regulated (Figure 1A). Unfortunately, we were unable to detect endogenous caspase-2S protein levels, as it is generated from a short-lived mRNA and hence, not easily detectable under these conditions (data not shown) (30). Nonetheless, we evaluated if this increase in caspase-2S was specific to TAp73β over-expression by expressing the other p53 family members. Surprisingly, we found that only TAp73β, but not p53, TAp73α, TAp63α or TAp63β was able to induce the expression of caspase-2S (Figure 1B). It is noteworthy that TAp73α and both TAp63α or TAp63β were unable to activate caspase-2S, though they were capable of activating Mdm2, suggesting that this induction is specific to the TAp73β form of TAp73.
Figure 1.
TAp73β, but not p53, TAp73α, TAp63α or TAp63β, induce caspase-2S expression. (A) TAp73β expression induces the up-regulation of caspase-2S but not other caspase transcripts. The pcDNA and TAp73 expression constructs were transfected into H1299 cells for 36 h before RNA extraction and cDNA synthesis. Semi-quantitative RT–PCR was performed for caspase-2L, caspase-8, caspase-9 and caspase-2S transcripts. (B) The pcDNA, p53, TAp63α, TAp63β, TAp73α and TAp73β expression constructs were transfected into H1299 cells similarly and expression of caspase-2L and caspase-2S transcripts were analysed. Mdm2 was used as a positive control and the expression of the transfected p53 family members is shown.
Both TAp73β and DNp73β activate the caspase-2S promoter
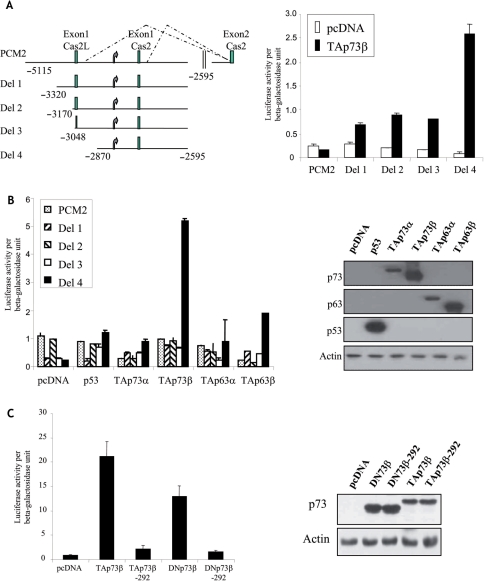
As TAp73β expression led to an increase in the steady-state levels of caspase-2S mRNA, we ascertained if this was due to transcriptional activation of the caspase-2S promoter. To this end, we utilized several caspase-2 promoter-luciferase reporter constructs described in our earlier study, which revealed that caspase-2S can be transcribed from an alternate promoter within intron 1 of the caspase-2 gene (28) (Figure 2A, left panel). Expectedly, TAp73β expression led to a significant increase in the reporter activity from the construct containing only caspase-2S promoter located within intron 1 (Del 4), but not from the construct containing full-length caspase-2L promoter (PCM2) (Figure 2A, right panel). TAp73β was also unable to effectively activate luciferase activity from the sequentially deleted promoter constructs containing the caspase-2L exon 1 and intron 1 (Del 1, Del 2 and Del 3) (28), despite all of them containing the caspase-2S promoter located in intron 1 (Figure 2A, right panel). This suggests that there may be negative regulatory elements in exon 1, preventing TAp73β-mediated caspase-2 activation. Nonetheless, these data confirms our initial finding that TAp73β induces caspase-2s expression by transcriptionally activating its promoter.
Figure 2.
Activation of caspase-2S promoter by TAp73β and DNp73β. (A) Left panel shows the schematic of the caspase-2S promoter-luciferase constructs used in the study. Curved arrows represent the TATA box. Exon 1 of caspase-2S and caspase-2L are shown. These constructs were transfected together with TAp73β and β-gal plasmid into H1299 cells for 24 h. The cultures were then lysed and used for luciferase assays (right panel). (B) Activation of Del 4 reporter construct is specific only to TAp73β. The indicated reporter constructs were transfected together with either of the following: p53, TAp63α, TAp63β, TAp73α and TAp73β, together with the β-gal plasmid into H1299 cells for 24 h, prior to analysis of luciferase activity (left panel). Right panel shows immunoblot analysis of the expression of the transfected plasmids. (C) DNA-binding ability of p73β is essential for induction of Del4 promoter. Del4 reporter constructs were transfected together with either TAp73β or TAp73β-292, or DNp73β or DNp73β-292 together with β-gal plasmid into H1299 cells, prior to analysis of luciferase activity (left panel). Right panel shows immunoblot analysis of the expression of the transfected plasmids. All luciferase assays were repeated at least thrice, each time in duplicates. The graphs are representation of average ± SED.
Analysis using the other p53 family members confirmed our earlier RT–PCR data that only TAp73β, but not p53, TAp63α or TAp63β is able to effectively activate the caspase-2 promoter (Del4), though all proteins were approximately equally expressed (Figure 2B). Next, we evaluated if the DNA-binding ability and TA domain of TAp73β are required to activate the Del4 reporter construct by utilizing the TAp73β-R292H, which has a point mutation in the DNA-binding domain and hence, defective in its ability to specifically bind DNA, and the DNp73β that lacks the TA domain. This analysis revealed that the DNA-binding ability of TAp73β is essential to activate caspase-2S promoter, as the TAp73β-R292H mutant was almost completely defective in transactivation, though being expressed efficiently (Figure 2C). However, unexpectedly, we found that DNp73β without the TA domain was able to transactivate the caspase-2S promoter, and mutation in the DNA-binding domain of DNp73β abrogated this effect (Figure 2C). These results together indicate that TAp73β and DNp73β are specifically capable of transcriptionally activating the caspase-2S promoter, though the NH2-terminal TA domain is dispensable for this process.
A unique element in the caspase-2S promoter is required for activation by p73β
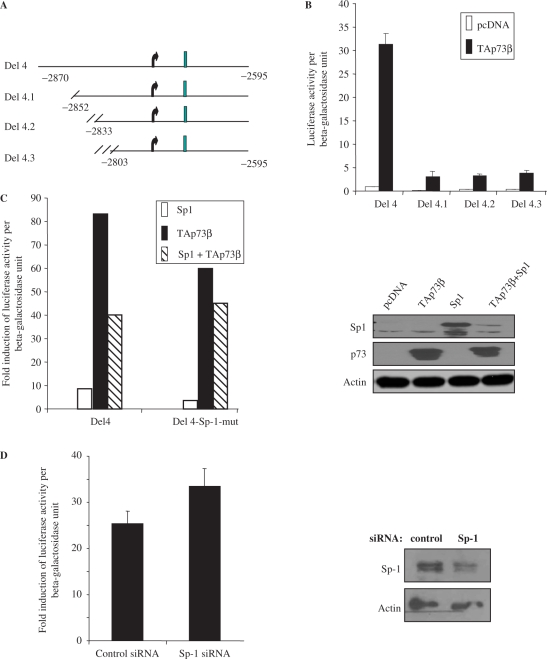
We next characterized the caspase-2S promoter to define the minimal DNA sequence required for activation, by making sequential deletions of the Del 4 construct (Figure 3A). We made the assumption that the essential site should lie upstream of the TATA box (Supplementary Figure 1A, indicated in bold), and hence, made deletions starting from the 5′-end of Del4 towards the 3′-end till prior to the TATA box (Figures 3A and Supplementary Figure 1A). These constructs, termed Del 4.1, 4.2 and 4.3, were co-transfected together with TAp73β to analyse their activity. Surprisingly, TAp73β was unable to activate any of these deletions constructs, when compared to the parent Del4 promoter (Figure 3B), suggesting that the site that is required for TAp73β to activate the caspase-2S promoter probably lies upstream of Del 4.1. The fragment between Del 4 and Del 4.1 is only about 18-bp long and contains a GC-rich box to which the Sp-1 transcription factor could potentially bind (31) (Supplementary Figure 1B). To test if the Sp-1-binding site is required for TAp73β-mediated activation, we generated a mutant construct in which the Sp-1 site was mutated by site-directed mutagenesis (Supplementary Figure 1B). Luciferase assays indicted that though there was a slight decrease in total activity, TAp73β was still capable of consistently activating the Del4-Sp-1 mutant construct, and the fold of activation of the promoter–luciferase activity was significant for both the wild-type and Sp-1-mutant promoters (Del4 versus Sp-1-mut: 83.0 versus 62.0) (Figure 3C, left panel). To evaluate if Sp-1 would affect TAp73β-mediated caspase-2S promoter activation, we co-transfected Sp-1 cDNA with TAp73β cDNA. Expression of Sp-1 alone led to a marginal activation of the caspase-2S promoter, and this effect was abrogated when the Sp-1 site was mutated (Del4 versus Sp-1-mut: 8.0 versus 3.0) (Figure 3C, left panel). Co-expression of TAp73β with Sp-1 resulted in a decrease in the TA potential compared to when TAp73β was expressed alone (Del4 versus Sp-1-mut: 40.0 versus 44.0) (Figure 3C, left panel). Nonetheless, the combined effect of TAp73β and Sp-1 was not affected by mutation of the Sp-1-binding site. Immunoblot analysis indicated that both the levels of Sp-1 and TAp73 were consistently reduced when co-expressed, suggesting other inter-regulation between them (Figure 3C, right panel). These data together suggest that the Sp-1-binding site may not be crucial for p73 to activate caspase-2S, and that Sp-1 may not have a significant role in regulating TAp73β-mediated caspase-2S promoter activation.
Figure 3.
Characterization of DNA elements required for activation of caspase-2S promoter by p73. (A) Schematic shows the deletion constructs made sequentially using Del 4. (B) The sequentially truncated Del4 constructs were transfected together with TAp73β and β-gal into H1299 cells for 24 h, prior to luciferase analysis. (C) Del4 and Del4-Sp-1 mutant reporter constructs were transfected together with TAp73β or Sp-1, alone or in combination, into H1299 cells for luciferase analysis. The fold induction of luciferase activity was derived by dividing the values obtained with the respective constructs with that of pcDNA-transfected samples. Right panel shows immunoblot analysis of the expression of the transfected plasmids. (D) Knockdown of Sp-1 does not significantly affect the activation of Del4 promoter by TAp73β. Control or Sp-1 siRNA were transfected into H1299 for 24 h prior to transfection of the Del4, TAp73β and β-gal constructs for luciferase analysis. Fold induction of luciferase activity is shown. Right panel shows efficiency of Sp-1 knockdown by immunoblotting. All luciferase assays were repeated at least thrice, each time in duplicates. The graphs are representation of average ± SED.
To further determine if Sp-1 is required for TAp73β-dependent activation of the caspase-2S promoter, we silenced its expression using Sp-1-specific siRNA 24 h prior to luciferase assays using the Del4 construct. As seen in Figure 3D, silencing of Sp-1 expression did not affect the induction of Del4 activity by TAp73β, and the ratio of activation was comparable between control and Sp-1 siRNA treatment (control versus Sp-1 siRNA: 26.5 versus 33.5). These results therefore together suggest that the 18-bp element within the GC-rich box in caspase-2S promoter is required for TAp73β-mediated expression, which is probably independent of Sp-1.
TAp73β and DNp73β bind to the unique site in vitro and in vivo
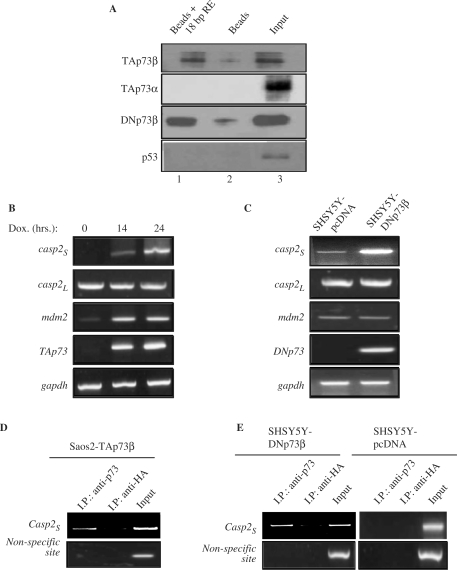
Since both TAp73β and DNp73β activated the caspase-2S promoter through their DNA-binding domains, we tested if they can directly bind to the 18-bp element identified as being important for caspase-2S promoter activation by TAp73β. In the first instance, in vitro DNA-binding assays were performed using bacterially purified GST-TAp73β, GST-TAp73α, GST-DN73β and GST-p53, and a biotin-labeled 24-bp oligonucleotide encompassing the 18-bp element. Incubation of GST proteins and oligonucleotides together with avidin-conjugated agarose beads, followed by washing to remove excess unbound proteins and subsequent immunoblotting revealed that only TAp73β and DN73β, but not TAp73α or p53, could bind to the beads containing the 18-bp DNA sequence (Figure 4A, compare lanes 1 to 3). This binding, though weak, was reproducible. To confirm this result, we attempted to detect endogenous binding of TAp73β and DNp73β to the caspase-2S promoter in vivo, using chromatin immunoprecipitation (ChIP) assays. We used two cell lines for this purpose: the Saos2-TAp73β inducible cells and the human SH-SY5Y neuroblastoma cell line stably expressing DNp73β. The caspase-2S mRNA expression was up-regulated upon TAp73β induction in Saos2-TAp73β cells and was higher in SH-SY5Y cells stably expressing DNp73β compared to their pcDNA-expressing control counterparts (Figure 4B and C). There were no differences in the levels of caspase-2L in both cases (Figure 4B and C). Immunoprecipitation with anti-p73 or the irrelevant anti-HA antibodies was followed by PCR amplification of the region flanking the 18-bp site on the caspase-2S promoter (indicated as Casp-2S) or an irrelevant site away from this region. As shown in Figure 4D, strong and specific binding of TAp73 was noted on the region surrounding the 18-bp element but not on the non-specific site in Saos2-TAp73β cells. Both these sites were amplifiable from crude lysates prior to immunoprecipitation, indicating the presence of these DNA fragments in the lysates (input). Similarly, ChIP analysis using the SH-SY5Y cells indicated a PCR product amplified specifically in DNp73β-expressing cells and not in pcDNA cells with the anti-p73 antibody, which was also not significantly present in anti-HA immunoprecipitates (Figure 4E, upper panel). No PCR product was amplified from the non-specific site on the caspase-2S promoter (Figure 4E, lower panel). These data together suggest that TAp73β and DNp73β can bind to the caspase-2S promoter containing the 18-bp site identified by the luciferase assays, both in vitro and in vivo, and highlight that the lack of the TA domain does not affect the binding of DNp73β to DNA sequences.
Figure 4.
The p73 binds to the 18-bp sequence element on the caspase-2S promoter in vitro and in vivo. (A) In vitro DNA-GSTp73 binding assay. Purified GST-TAp73α GST-TAp73β GST-DNp73β or GST-p53 proteins were incubated with biotinylated 18 bp caspase-2S promoter containing sequence elements before further incubation with avidin-conjugated beads. GST-p73/p53 without biotinylated DNA but with beads alone was used as negative controls. The beads were washed and separated onto SDS–acrylamide gel for immunoblot analysis with the indicated antibodies. Lane1: GST-protein + 18-bp element + beads. Lane 2: GST-protein + beads. Lane 3: GST-protein lysate. (B–C) Up-regulation of caspase-2S in Saos2-TAp73β inducible cell line (B) and in SH-SY5Y cells stably expressing DNp73β (C). TAp73β was induced by doxocycline (Dox) addition for 14–24 h prior to RNA extraction. RT–PCR was performed to assess expression of caspase-2S, caspase-2L, p73 and mdm2 in both the cell systems. (D and E) ChIP analysis was performed with anti-p73 and anti-HA antibodies using the two cellular systems described earlier. Cells were collected 15 h after TAp73 induction (D). The promoter sequence encompassing the 18-bp elements on the caspase-2S promoter was analysed by PCR (Casp2S). A non-specific site on the caspase-2S promoter was used as negative control. All experiments were repeated at least thrice independently.
Over-expression of DNp73β protects cells from cell death through caspase-2S activation
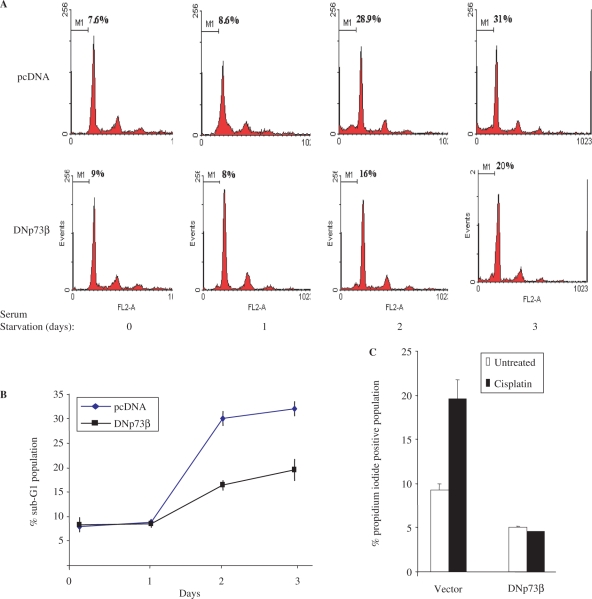
Over-expression of caspase-2S has been shown to protect cells from cell death induced by serum deprivation (27). Hence, we tested if the SH-SY5Y-DNp73β cells, which express higher levels of caspase-2S mRNA, are also more resistant to serum deprivation-induced cell death. Serum deprivation of both SH-SY5Y-pcDNA and SH-SY5Y-DNp73β cells followed by analysis of DNA content to monitor the sub-G1 population—which reflects apoptotic cells—indicated that there was less cell death in the SH-SY5Y-DNp73β cells (% sub-G1 cells → pcDNA versus DNp73β cells: 28.9 versus 16.0 [day2]; 31.0 versus 20.0 [day3]) (Figure 5A and B). Similarly, though treatment with another cellular insult, cisplatin, resulted in an increase in cell death of pcDNA cells, there was no significant death in DNp73β cells (% dead cells → −/+ cisplatin—pcDNA versus DNp73β cells: 8.0/20.0 versus 5.0/4.8) (Figure 5C).
Figure 5.
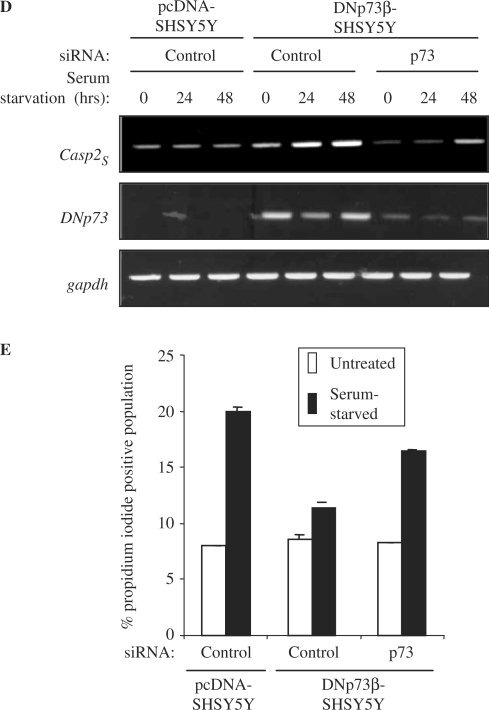
DNp73β over-expressing SH-SY5Y cells are resistant to cell death. (A and B) SH-SY5Y cells stably over-expressing pcDNA or DNp73β were seeded in 6-well plates in triplicates and serum starved in serum free DMEM for up to 72 h. Cultures were harvested at days 0, 1, 2 and 3 during serum starvation for cell cycle analysis. Representative flow cytometric graphics (A). Average of the sub-G1 population, representing apoptotic cells, for each cell line and time point is plotted ± SED (B). (C) These cells were treated with 20 μM cisplatin for 24 h prior to analysis of total cell death by propidium iodide exclusion assay. (D and E) Knockdown of p73 results in reduced caspase-2S expression and increased cell death. The above cells were transfected with control or p73-siRNA and serum-starved for the indicated time periods. The mRNA analysis was performed to determine expression of caspase-2S, DNp73 and gapdh (D), and cells were harvested after 48 h for analysis of cell death by propidium iodide exclusion assay (E). All experiments were repeated at least thrice independently.
In order to verify if the resistance to cell death is due to DNp73β-mediated caspase-2S activation, we silenced the expression of the over-expressed DNp73β using p73-specific siRNA, followed by serum-starvation for up to 48 h. Silencing of p73 resulted in a decrease in the DNp73β levels, as expected (Figure 5D). Importantly, the levels of caspase-2S were also reduced concomitantly (Figure 5D), indicating that DNp73β is indeed responsible for caspase-2S activation. Analysis of cell death revealed that silencing DNp73β expression consistently led to an increase in cell death, compared to control siRNA treated cells (% dead cells upon serum-starvation: SH-SY5Y-pCDNA cells–20.0; SH-SY5Y-DNp73β cells—control versus p73 siRNA: 11.0 versus 16.5) (Figure 5E). These data together indicate that expression of DNp73β, which leads to up-regulation of caspase-2S, contributes to protection of cells against cell death induced by multiple means.
DISCUSSION
The findings presented here highlight two salient points: that the tumour-suppressor TAp73β is able to induce the expression of the anti-apoptotic caspase-2S, and that DNp73β, without the NH2-terminal TA domain, is also capable of activating caspase-2S expression. The former finding, though at first instance perplexing, is entirely compatible with the emerging view that TAp73β may have other roles in supporting cellular growth. The latter finding highlight the fact that some targets can be common to both TAp73β and DNp73β, and hence, may explain why many human tumours co-over-express both TAp73 and DNp73 to provide a strong survival pressure.
It is striking that expression of the tumour-suppressor TAp73β, which induces cell death when over-expressed in many cellular systems, is able to transactivate the anti-apoptotic capase-2S gene. Though loss of all p73 forms result in increased resistance to cell death (9,32), TAp73 is over-expressed in many human cancers (6,11–17). Intriguingly, it is to be noted that physiologically, TAp73β transcripts are not readily detectable (33). These mitigating reasons raise the interesting possibility that TAp73β may have evolved to also support cellular survival under certain conditions, such as seen in human cancers. In support of this, we have recently shown that expression of TAp73 can cooperate with c-Jun to activate AP-1 target genes such as cyclinD1, and hence, promote cellular survival (18). Moreover, absence of p73 led to reduced expression of cyclinD1 and decreased cellular proliferation. In addition, other groups have also raised the possibility that TAp73 may be involved in the activation of genes that are involved in tumourigenesis, such as β-catenin and gastrin (19,20). Thus, these findings together favour the argument for a contributory role of TAp73 in supporting cellular survival, in one way by activating the expression of capase-2S as demonstrated here.
In addition, DNp73β is the variant of p73 that is predominantly expressed in the brain and sympathetic neurons (23). The p73−/− mice display severe defects in their nervous system including massive cell death, suggesting that DNp73β plays an anti-apoptotic role in the brain (33). Over-expression of DNp73β in sympathetic neurons was also able to rescue cell death resulting from nerve growth factor withdrawal (34). Similarly, caspase-2S is highly expressed in the brain and caspase-2S has been shown to protect cells from cell death induced by several means (27,35). Therefore, our results that the SH-SY5Y neuroblastoma cells stably over-expressing DNp73β—which express higher level of caspase-2S compared with pcDNA expressing control cells—are resistant to cell death upon serum starvation and cisplatin treatment are entirely consistent with a protective role for DNp73β in some cellular contexts. Importantly, reducing the exogenously expressed DNp73β levels by gene silencing led to a decrease in caspase-2S expression and a concomitant reversal of resistance to cell death, suggesting that the activation of caspase-2S by DNp73β indeed leads to protection against cell death. However, we have not been able to specifically silence the expression of caspase-2S in DNp73β-over-expressing cells to demonstrate its relevance due to the sequence similarity of caspase-2S and caspase-2L (data not shown). Nonetheless, our data suggest that physiologically, DNp73β may act through caspase-2S to protect neuronal cells from cell death.
Activation of caspase-2S was found to be specific to the p73 family, but not to p53, TAp63α and TAp63β though all three p53 family members share similar DNA-binding domains and are thought to be able to activate a large subset of p53 target genes (6,36). However, the specificity of the p73 family could arise due to subtle differences elsewhere, which may be more critical for binding to the unique site on the caspase-2S promoter, besides the common DNA-binding domain. However, this cannot explain why TAp73α, which also share the same DNA-binding and other domains as TAp73β, and is over-expressed in human cancers as TAp73β, cannot induce caspase-2S expression. The sterile α-domain present in TAp73α but not in TAp73β has been shown to play an inhibitory role in TA, making TAp73α a weaker transactivator of target genes compared to TAp73β (37,38). This could be a reason for the inability of TAp73α to activate caspase-2S in experimental conditions, though it may be able to activate caspase-2S expression in vivo, in conditions where the effect of the sterile alpha motif (SAM) domain is negated by other means. This possibility remains to be explored. Nevertheless, the 18-bp site to which the p73 members bind to on the caspase-2S promoter does not resemble the p53 consensus binding site. Rather, it contains a GC box, a putative binding site for Sp-1 transcription factor. Mutation of the GC box and knockdown of Sp-1 expression did not significantly affect TAp73β's ability to activate the caspase-2S promoter, suggesting that TAp73β does not depend on Sp-1 to activate the promoter. Moreover, comparison of Sp-1 and TAp73β's ability to activate the caspase-2S promoter indicated that TAp73β was a better activator than Sp-1, which only had a marginal effect, thereby excluding any critical role for the latter in caspase-2S activation. Furthermore, DNA-binding mutants of p73 were unable to activate the caspase-2S promoter and conversely, both in vivo ChIP experiments and in vitro DNA-binding assays revealed that p73 was able to bind to this DNA element, indicating that p73 can indeed directly bind to and activate the caspase-2S promoter. Whether this 18-bp element is a unique site that is of general utility for activation of p73-specific (and not p53- or p63-dependent) targets is yet to be explored. Overall, the findings highlight the specificity of TAp73β and DNp73β in the induction of caspase-2S expression.
The ability of both full-length TAp73β and the TA-deficient DNp73β to induce caspase-2S promoter activation is also surprising, but highlights that the DNp73 form may have the ability to activate target genes. There are not many reports that have highlighted that both of these forms can activate classical p53/p73 target genes, as traditionally, it has been thought that the TA domain is essential for transcription factors to recruit co-factors for transactivation of target genes. Therefore, the findings presented here suggest that DNp73β has unique transactivation ability, besides its role as a dominant-negative protein to inhibit p73 and p53 function. Two similar examples were reported, whereby DNp73α was shown to activate the expression of EGR1 and CDC6, and DNp73β was shown to activate classical p53 target genes (5,39). Mechanistically, how this occurs is unclear. It was speculated that the presence of a secondary TA domain in DNp73, which encompass the 13 unique residues at the NH2-terminus together with the PXXP motifs, may form a TA domain responsible for the activity of DNp73 (5). Alternatively, DNp73β may recruit other transcription factors to activate the capase-2S promoter activity. Whatever the mechanism may be, it is evident that DNp73 has the ability to activate target genes, which are unique and does not fall into to the classical ‘p53-target’ gene group.
Another noteworthy point is that both TAp73β and DNp73β were not able to activate the expression from the full-length caspase-2 promoter. Only after truncating the promoter to retain the caspase-2S promoter region specifically did we see an increase in promoter activity, suggesting the existence of repressor elements that may interfere with the induction of caspase-2S by p73β. Moreover, we cannot exclude the possibility that usage of one promoter may be at the expense of the other, though this needs further investigation. This indicates that specific conditions may be required for the induction of caspase-2S by p73β in vivo in the physiological setting.
In conclusion, the data presented here provide evidence for the activation of the anti-apoptotic caspase-2S by both the tumour-suppressive TAp73β and the anti-apoptotic DNp73β, which could be yet another mechanism to promote cellular survival in tumor settings where both p73 forms are over-expressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
T.W.H. was partially supported by a Singapore Millennium Foundation fellowship. We thank the National Medical Research Council of Singapore and the Biomedical Research Council of Singapore for their generous funding and support to K.S. Funding to pay the Open Access publication charges for this article was provided by Biomedical Research Council of Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jost CA, Marin MC, Kaelin W.G., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 2.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 3.Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002;62:3598–3602. [PubMed] [Google Scholar]
- 4.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J. Biol. Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 5.Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol. Cell Biol. 2004;24:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melino G, De Laurenzi V, Vousden KH. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 7.Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18:4993–4998. doi: 10.1038/sj.onc.1202817. [DOI] [PubMed] [Google Scholar]
- 8.Lee CW, La Thangue NB. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 9.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 10.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Becker K, Pancoska P, Concin N, Vanden HK, Slade N, Fischer M, Chalas E, Moll UM. Patterns of p73 N-terminal isoform expression and p53 status have prognostic value in gynecological cancers. Int. J. Oncol. 2006;29:889–902. [PubMed] [Google Scholar]
- 12.Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- 13.Hong SM, Cho H, Moskaluk CA, Yu E, Zaika AI. p63 and p73 expression in extrahepatic bile duct carcinoma and their clinical significance. J. Mol. Histol. 2007;38:167–175. doi: 10.1007/s10735-007-9084-7. [DOI] [PubMed] [Google Scholar]
- 14.Kovalev S, Marchenko N, Swendeman S, LaQuaglia M, Moll UM. Expression level, allelic origin, and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ. 1998;9:897–903. [PubMed] [Google Scholar]
- 15.Zaika A, Irwin M, Sansome C, Moll UM. Oncogenes induce and activate endogenous p73 protein. J. Biol. Chem. 2001;276:11310–11316. doi: 10.1074/jbc.M005737200. [DOI] [PubMed] [Google Scholar]
- 16.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59:3257–3263. [PubMed] [Google Scholar]
- 17.Zaika AI, El-Rifai W. The role of p53 protein family in gastrointestinal malignancies. Cell Death Differ. 2006;13:935–940. doi: 10.1038/sj.cdd.4401897. [DOI] [PubMed] [Google Scholar]
- 18.Vikhanskaya F, Toh WH, Dulloo I, Wu Q, Boominathan L, Ng HH, Vousden KH, Sabapathy K. p73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nat. Cell Biol. 2007;9:698–706. doi: 10.1038/ncb1598. [DOI] [PubMed] [Google Scholar]
- 19.Tomkova K, El-Rifai W, Vilgelm A, Kelly MC, Wang TC, Zaika AI. The gastrin gene promoter is regulated by p73 isoforms in tumor cells. Oncogene. 2006;25:6032–6036. doi: 10.1038/sj.onc.1209610. [DOI] [PubMed] [Google Scholar]
- 20.Tomkova K, Belkhiri A, El-Rifai W, Zaika AI. p73 isoforms can induce T-cell factor-dependent transcription in gastrointestinal cells. Cancer Res. 2004;64:6390–6393. doi: 10.1158/0008-5472.CAN-04-2176. [DOI] [PubMed] [Google Scholar]
- 21.Toh WH, Kyo S, Sabapathy K. Relief of p53-mediated telomerase suppression by p73. J. Biol. Chem. 2005;280:17329–17338. doi: 10.1074/jbc.M500044200. [DOI] [PubMed] [Google Scholar]
- 22.Ramadan S, Terrinoni A, Catani MV, Sayan AE, Knight RA, Mueller M, Krammer PH, Melino G, Candi E. p73 induces apoptosis by different mechanisms. Biochem. Biophys. Res. Commun. 2005;331:713–717. doi: 10.1016/j.bbrc.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 23.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 25.Ramadan S, Terrinoni A, Catani MV, Sayan AE, Knight RA, Mueller M, Krammer PH, Melino G, Candi E. p73 induces apoptosis by different mechanisms. Biochem. Biophys. Res. Commun. 2005;331:713–717. doi: 10.1016/j.bbrc.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 28.Logette E, Wotawa A, Solier S, Desoche L, Solary E, Corcos L. The human caspase-2 gene: alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene. 2003;22:935–946. doi: 10.1038/sj.onc.1206172. [DOI] [PubMed] [Google Scholar]
- 29.Ishii S, Kadonaga JT, Tjian R, Brady JN, Merlino GT, Pastan I. inding of the Sp1 transcription factor by the human Harvey ras1 proto-oncogene promoter. Science. 1986;232:1410–1413. doi: 10.1126/science.3012774. [DOI] [PubMed] [Google Scholar]
- 30.Solier S, Logette E, Desoche L, Solary E, Corcos L. Nonsense-mediated mRNA decay among human caspases: the caspase-2S putative protein is encoded by an extremely short-lived mRNA. Cell Death Differ. 2005;12:687–689. doi: 10.1038/sj.cdd.4401594. [DOI] [PubMed] [Google Scholar]
- 31.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 32.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin W.G., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 33.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 34.Lee AF, Ho DK, Zanassi P, Walsh GS, Kaplan DR, Miller FD. Evidence that DeltaNp73 promotes neuronal survival by p53-dependent and p53-independent mechanisms. J. Neurosci. 2004;24:9174–9184. doi: 10.1523/JNEUROSCI.1588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito A, Uehara T, Nomura Y. Isolation of Ich-1S (caspase-2S)-binding protein that partially inhibits caspase activity. FEBS Lett. 2000;470:360–364. doi: 10.1016/s0014-5793(00)01351-x. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs WB, Kaplan DR, Miller FD. The p53 family in nervous system development and disease. J. Neurochem. 2006;97:1571–1584. doi: 10.1111/j.1471-4159.2006.03980.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu G, Chen X. The C-terminal sterile alpha motif and the extreme C terminus regulate the transcriptional activity of the alpha isoform of p73. J. Biol. Chem. 2005;280:20111–20119. doi: 10.1074/jbc.M413889200. [DOI] [PubMed] [Google Scholar]
- 38.Wang WK, Bycroft M, Foster NW, Buckle AM, Fersht AR, Chen YW. Structure of the C-terminal sterile alpha-motif (SAM) domain of human p73 alpha. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:545–551. doi: 10.1107/s0907444901002529. [DOI] [PubMed] [Google Scholar]
- 39.Kartasheva NN, Lenz-Bauer C, Hartmann O, Schafer H, Eilers M, Dobbelstein M. DeltaNp73 can modulate the expression of various genes in a p53-independent fashion. Oncogene. 2003;22:8246–8254. doi: 10.1038/sj.onc.1207138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.