Abstract
The TAR hairpin of the human immunodeficiency virus type 1 (HIV-1) RNA genome is essential for virus replication. TAR forms the binding site for the transcriptional trans-activator protein Tat and multiple additional TAR functions have been proposed. We previously constructed an HIV-1 variant in which the TAR-Tat transcription control mechanism is replaced by the components of the Tet-ON regulatory system. In this context, the surprising finding was that TAR can be truncated or even deleted, but partial TAR deletions that destabilize the stem structure cause a severe replication defect. In this study, we demonstrate that the HIV-1 RNA genome requires a stable hairpin at its 5′-end because unpaired TAR sequences affect the proper folding of the untranslated leader RNA. Consequently, multiple leader-encoded functions are affected by partial TAR deletions. Upon evolution of such mutant viruses, the replication capacity was repaired through the acquisition of additional TAR mutations that restore the local RNA folding, thus preventing the detrimental effect on the leader conformation.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is a retrovirus with an RNA genome of approximately 9 kb that contains nine open-reading frames and untranslated regions at the 5′- and 3′-end (Figure 1A). The untranslated leader region at the 5′-end encodes several regulatory RNA motifs involved in both early and late replication steps (reverse transcription, transcription, splicing, polyadenylation, translation, RNA dimerization and packaging) (1). This leader RNA can adopt two mutually exclusive structures in vitro (Figure 1B). The energetically favoured structure is formed by a long-distance base-pairing interaction (LDI) between the polyA and dimerization initiation site (DIS) (2). Both the polyA and DIS motifs fold a hairpin in the alternative structure that is termed the branched multiple hairpin (BMH) conformation. Exposure of the DIS hairpin is important for the formation of RNA dimers by kissing-loop base pairing (Figure 1B) (3,4). The kissing-loop complex can rearrange to form an extended dimer that is more stable (5,6).
Figure 1.
The HIV-1 TAR hairpin as part of the untranslated leader RNA. (A) The HIV-rtTA provirus genome is shown, with the Tat-TAR axis of transcription regulation inactivated by mutation of both Tat and TAR (tatm and TARm; crossed boxes). The LTR region is subdivided into the U3, R and U5 region. Transcription and replication of the virus was made dox-dependent by the introduction of tetO elements in the U3 promoter region and replacing the Nef gene by the rtTA gene. The domain structure of the untranslated leader RNA is shown. The untranslated leader RNA of HIV-1 (+1/+368) consists of several regulatory domains (TAR; polyA: polyadenylation hairpin; PAS: primer activation signal; PBS: primer-binding site; SD: splice donor; Ψ: RNA packaging signal; AUG: translation start codon of gag). (B) The leader RNA can adopt two conformations. The polyA and DIS sequences are base paired in the LDI structure. The same sequences form the polyA and DIS hairpin in the BMH structure. The DIS hairpin allows the formation of RNA dimers by kissing-loop base pairing. The riboswitch model for regulated dimerization argues that the ground-state LDI structure must first rearrange into the BMH conformation to expose the DIS hairpin, which mediates subsequent RNA dimerization.
In both the LDI and BMH conformation, the TAR hairpin is located at the extreme 5′-end of the HIV-1 transcripts. This hairpin has an essential role in the activation of transcription from the promoter in the 5′-long terminal repeat (LTR) of the proviral DNA genome (7,8). The TAR hairpin has a highly conserved 3-nucleotide pyrimidine bulge that binds the viral Tat transactivator protein (9), and an apical 6-nucleotide loop that binds the cellular Cyclin T1 protein in a Tat-dependent manner (10,11). The Tat-TAR-Cyclin T1 axis results in enhanced transcription through phosphorylation of the RNA polymerase. In addition to its role in transcription, the TAR hairpin has also been suggested to be involved in dimerization of the viral genome (12), packaging of the viral RNA into virions (13–16) and in the strand transfer step of reverse transcription (17). Mutation of TAR causes severe replication defects due to a lack of LTR transcription (18–20). This dominant effect on transcription precluded the careful analysis of the putative non-transcriptional functions of TAR in HIV-1 replication.
We previously constructed an HIV-1 variant that does not depend on the Tat-TAR interaction for activation of transcription (21–23). In this HIV-rtTA variant, both the Tat protein and its TAR-binding site were inactivated by mutation (Figure 1A) and functionally replaced by the components of the Tet-ON gene regulation system (24). The gene encoding the rtTA transcriptional activator protein was inserted in place of the 3′-terminal nef gene, and tet operator (tetO)-binding sites were introduced in the LTR promoter. Administration of the effector doxycycline (dox) induces a conformational switch in the rtTA protein, which subsequently can bind to the tetO-LTR promoter and activate transcription of the proviral genome. Thus, transcription and replication of HIV-rtTA are critically dependent on the presence of dox.
HIV-rtTA has a mutated TAR hairpin (TARm) with five nucleotide substitutions in the bulge and loop domains to prevent binding of Tat and Cyclin T1 (Figure 2). This virus does not require TAR for activation of gene expression and thus allows us to test for additional TAR functions in viral replication by deleting parts of TAR. We introduced deletions on either the left or the right side of the TARm hairpin (Figure 2: mutants A and C, and mutants B, D and F, respectively). We also made the double mutants (AB and CD) and a nearly complete TAR deletion (mutant E). As previously shown, the mutations did not affect dox-dependent transcription. However, whereas the double mutants did not affect viral replication, the single mutants showed a severe replication defect (25).
Figure 2.
Mutant TAR hairpins. Shown is the wild-type TAR hairpin (TARwt) and the TARm version with bulge and loop mutations that is present in the HIV-rtTA virus. The TARm sequence is partially or near-completely deleted in mutants A–F. The deleted nucleotides are indicated by a grey box. The transcription and replication properties of these mutant viruses are indicated as previously presented (25). In this study, we focused on the structure and function of the TAR-mutated leader transcripts.
It was previously observed that mutations in the leader RNA can affect the equilibrium between the dimerization-incompetent LDI structure and the dimerization-prone BMH structure (2,15,26,27). Although the TAR hairpin itself is not directly involved in the switch from LDI to BMH, as it is present in both structures (Figure 1B), destabilization of TAR may affect the LDI-BMH equilibrium and thus indirectly influence the function of other RNA motifs. To understand why the single side TAR deletions abolish viral replication, we analyzed the effect of these mutations on the leader RNA conformation.
MATERIALS AND METHODS
Mutant and evolved HIV-1 sequences
All TAR mutations were introduced in the infectious HIV-rtTA (25) molecular clone, which is based on the HIV-1 LAI plasmid (28). To start virus cultures, the SupT1 cell line was transfected by electroporation as described previously (29). In brief, 5 × 106 SupT1 cells were transfected with 10 μg of the proviral constructs and cultured in 5 ml medium with 1 μg/ml dox (Sigma D-9891). For the selection of virus variants with improved replication capacity, virus-cell cultures were continued for up to 175 days (25). Cell samples were stored at −80°C for PCR amplification of HIV-1 genome segments and subsequent sequence analysis. Proviral DNA sequences were PCR amplified from total cellular DNA with primers U3-Xba-Not (5′-ACG TCT AGA GCG GCC GCA CTG GAA GGG CTA ATT CAC TC-3′, position −331 to −313) and AD-GAG (5′-ATG GAT CCG TTC TAG CTC CCT GCT TGC CC-3′, position +463 to +442), ligated into pCR2.1-TOPO TA-cloning vector (Invitrogen, Breda, The Netherlands) and sequenced with primer BB3 (5′-GAG TCC TGC GTC GAG AGA GCT CCT CTG GTT-3′, position +245 to +216). We used the 5′ RACE (rapid amplification of cDNA ends) system Version 2.0 (Invitrogen) to analyze the 5′-end of the RNA transcripts, as described previously (25).
In vitro transcription of leader RNA
RNAs were in vitro transcribed from DNA templates in which a T7 promoter was positioned directly upstream of the transcription start site as previously described (27). For the production of these DNA templates, we PCR amplified the HIV-1 LAI (TARwt) and HIV-rtTA plasmids (TARm, mutant and evolved variants) with a forward primer encoding the T7 promoter sequence and the 5′-end of the transcript, and a reverse primer R368 (5′-TCC CCC GCT TAA TAC TGA CGC T-3′) that is complementary to nucleotides +347 to +368 in the wild-type leader sequence. We used forward primer T7-2 (5′-CTA ATA CGA CTC AGT ATA GGG TCT CTC TGG AGC ATT GGA-3′) for the production of TARwt, TARm, B, D and F transcripts, T7-A (5′-CTA ATA CGA CTC AGT ATA GGG TCT CTC TGG AGC ATT GGA-3′) for transcripts A, AB and AR2, T7-C (5′-CTA ATA CGA CTC AGT ATA GGG CCA GAA AGG AGC ATT GGA-3′) for the transcripts C and CD, T7-E (5′-CTA ATA CGA CTC AGT ATA GGG CCC ACT GCT TAA GCC TCA-3′) for transcript E, T7-AR1 (5′-CTA ATA CGA CTC AGT ATA GGG TCT CTC TAG AGC ATT GGA-3′) for transcript AR1, T7-FG3 (5′-CTA ATA CGA CTC AGT ATA GGG GCT CTC TGA CTA GAC CAG-3′) for transcript FG3, T7-FG4 (5′-CTA ATA CGA CTC AGT ATA GGG GGC TCT CTG ACT AGA CCA-3′) for transcript FG4, T7-FG5 (5′-CTA ATA CGA CTC AGT ATA GGG GGG CTC TCT GAC TAG ACC A-3′) for transcript FG5, T7-FG6 (5′-CTA ATA CGA CTC AGT ATA GGG GGG GCT CTC TGA CTA GAC CA-3′) for transcript FG6, T7-ER1 (5′-CTA ATA CGA CTC AGT ATA GGC CAC TGC TTA AGC CTC AAT-3′) for transcript ER1, T7-ER2 (5′-CTA ATA CGA CTC AGT ATA GAC TGC TTA AGC CTC AAT AAA-3′) for transcript ER2 and T7-ER3 (5′-CTA ATA CGA CTC AGT ATA GGG CCA CTG CTT AAG CCT CAA CGC-3′) for transcript ER3. In vitro transcription was performed with the Megashortscript T7 transcription kit (Ambion, Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands). For the production of 32P-labeled transcripts, 1 μl [α32P] UTP (0.33 Mbq/μl, Amersham, GE Healthcare, Diegem, Belgium) was added to the reaction. Transcripts were excised from a 6% denaturing polyacrylamide gel (visualized by UV-shadowing) and eluted from the gel fragment by overnight incubation in 1 × TBE-buffer at room temperature. The RNA was ethanol precipitated and redissolved in water. Quantification of the RNA was done by UV-absorbance measurements or scintillation counting in case of 32P-labeled transcripts. The size and integrity of the transcripts were analyzed on a denaturing sequence gel.
In vitro RNA dimerization and conformation analysis
100 ng 32P-labeled RNA was incubated with or without oligonucleotide CN1 at 85°C for 2 min in 10 μl MO-buffer (125 mM KAc, 2.5 mM Mg Acetate, 25 mM HEPES pH 7.0) and slowly cooled from 65°C to room temperature for renaturation and dimerization (30). CN1 is complementary to nucleotides 123–151 of the HIV-1 leader RNA and CN1 annealing effectively prevents folding of the LDI conformation. After incubation, the samples were chilled on ice and 4 μl non-denaturing loading buffer (MO-buffer with 30% glycerol and BFB dye) was added to the samples. Samples were analyzed on a 4% polyacrylamide gel in 0.25 × TBE (22.5 mM Tris, 22.5 mM boric acid, 0.625 mM EDTA) for LDI/BMH conformation analysis and 0.25 × TBM (22.5 mM Tris, 22.5 mM boric acid, 0.1 mM MgCl2) for dimerization analysis (27,31). Gels were run at 150 V at room temperature, dried and subjected to autoradiography. Quantification of the percentage RNA dimerization was performed on a Phosphor Imager (Molecular Dynamics). The dimerization yield was determined by dividing the amount of dimer by the total amount of RNA (dimer and monomer).
RNA secondary structure prediction
Computer-assisted RNA secondary structure prediction was performed using the Mfold v3.2 algorithm (32,33). Settings were the same for all folding jobs, using standard conditions (37°C and 1.0 M NaCl), a 5% suboptimal range and a maximum distance between paired bases of 200 nucleotides. To analyze the BMH structure of these transcripts, we forced the folding of the DIS hairpin by using the constraint option in the Mfold program. Folding was performed with wild-type, mutant and evolved sequences that started at position +1 and ended at the nucleotide corresponding to position +368 in wild-type HIV-1.
RNA absorbance measurements
UV melting of the T7 transcripts was measured by monitoring the absorbance of UV light at 260 nm on a Varian Cary 300 Bio UV-visible spectrophotometer in a quartz cuvet with a standard 1 cm path length. Prior to the measurement, the 40 ng/μl RNA in 50 mM Na-cacodylate buffer (pH 7.2) was incubated at 85°C for 5 min and then slowly cooled to room temperature for renaturation.
RNA structure probing
The T7 transcribed leader RNAs (nt 1–368) corresponding to the TARm, A, B and AB variants were used for RNA structure probing as described (34). These RNAs (50 pmol) were diluted in 37.5 μl water and mixed with 12.5 μl 4 × MO-buffer. The samples were incubated at 85°C for 3 min and slowly cooled to room temperature. The transcripts were incubated with 10 mM lead (Pb2+) nitrate at room temperature. Samples (10 μl) were taken at 0 and 10 min after the addition of lead (Pb2+) nitrate, and the reaction was stopped by adding 2 μl 0.5 M EDTA. RNA products were purified over a NucAway™ spin column (Ambion), ethanol precipitated and dissolved in 10 μl H2O. Oligonucleotides (30 pmol) CN1, R368 and lys21 (5′-CAA GTC CCT GTT CGG GCG CCA-3′; complementary to nucleotides +182 to +202 in the wild-type leader sequence) were 5′ end labeled with the kinaseMax kit (Ambion) in the presence of 3 μl of [γ-32P] ATP (0.37 MBq/μl, Amersham). Two picomoles of 32P-labeled oligonucleotide was annealed to 1 pmol of Pb2+-treated RNA by incubation at 85°C for 1 min, followed by slow cooling. These primers were extended at 60°C for 2 h using the Thermoscript reverse transcriptase (Invitrogen). After adding 20 μl gel-loading buffer II (Ambion), the samples were heated to 95°C and 3 μl was analyzed on a denaturing 6% polyacrylamide gel. A sequence ladder was produced with the thermo sequenase cycle sequencing kit (USB), 32P-labeled oligonucleotide primer and the DNA fragment corresponding to the TARm leader region as template (T7-2/R386 PCR product).
RESULTS
Single-side TAR deletions abolish HIV-rtTA replication
We previously presented variants of the dox-dependent HIV-rtTA with deletions in the TAR hairpin (Figure 2). Whereas these mutations did not significantly affect dox -dependent transcription, the single mutants (A, B, C, D, F) abolished viral replication (25). In contrast, the double mutants (AB, CD) replicated efficiently and the truncated mutant E showed delayed replication (summarized in Figure 2). The single deletions are possibly toxic because unpaired TAR sequences are generated, which may interfere with the proper folding of viral RNA. In particular, these mutations may alter the structure of the leader RNA that encodes many important replication signals. To test this hypothesis, we made a set of leader transcripts and performed several analyses to determine the impact of the TAR mutations on the structure and function of the leader RNA.
Replication defect of TAR mutants corresponds with an altered LDI–BMH equilibrium
We probed the conformational state of the wild-type and mutant leader RNAs by inspection of the migration on a native gel. The TARwt transcript (corresponding to nucleotides +1 to +368) exhibits the characteristic fast electrophoretic mobility of the LDI conformation, which is converted to a slower migrating BMH structure after annealing of the antisense oligonucleotide CN1 (Figure 3). This antisense DNA oligonucleotide binds the U5 region of the leader RNA and effectively prevents the LDI interaction between the polyA and DIS motifs, thus forcing the RNA into a BMH-like structure that exposes the DIS hairpin (30). The TARm transcript, which contains the TAR bulge and loop modifications as present in HIV-rtTA, displays the same gel migration pattern as TARwt and thus also adopts the LDI conformation. The analysis of the other transcripts is obviously complicated by the fact that the transcripts differ in size due to the TAR deletions. We therefore focus on the migration differences of the transcripts with and without CN1. Some transcripts display a partial shift from LDI to BMH (mutants A and B) or a complete shift (mutants C and E). We marked the transcripts that adopt the slow-migrating BMH conformation with an asterisk (*) in Figure 3. These BMH-structured RNA molecules migrate somewhat faster than the CN1-induced BMH-folded RNAs because they lack the oligonucleotide. While transcripts A and B show a diffuse pattern, which indicates a partial shift from LDI to BMH, only the LDI is formed by the double mutant AB. Likewise, the C mutant adopts the BMH structure, while the double mutant CD folds the LDI conformation. No BMH structure is observed for the mutants D and F. When most of the TAR hairpin is deleted as in mutant E, the BMH conformation is formed. Thus, whereas the majority of the single mutants display the BMH structure, the double mutants fold only the LDI conformation. Apparently, a deletion in one side of the TAR stem can shift the LDI-BMH equilibrium from LDI to BMH.
Figure 3.
TAR destabilization affects the LDI/BMH equilibrium. Non-denaturing gel analysis of leader transcripts (corresponding to nucleotides +1 to +368 in wild-type HIV-1) with different TAR deletions. Presence and absence of the CN1 oligonucleotide is indicated by + or −, respectively. The asterisk (*) marks the position of transcripts in the BMH conformation.
We next compared the conformation of the TARm, A, B and AB transcripts by measuring its UV absorbance at increasing temperature. In this assay, an increase in absorbance corresponds to the melting of RNA structure, and we previously reported that this method can accurately distinguish the LDI from the BMH conformation (2,35). The melting curves in the 30–80°C trajectory are shown as −δA/δT plot, which allows the determination of the melting transition and the Tm. The TARm leader transcript (+1/+368) shows two discrete transitions in the melting profile at Tm = 52.7°C and Tm = 71.1°C (Figure 4). Previous studies showed that the transition at Tm = 71.1°C is due to melting of the TAR hairpin (2). The transition at Tm = 52.7°C corresponds with denaturation of the extended long distance interaction of the LDI structure. These two transitions were also observed for the TARwt transcripts [results not shown, (2,30,31)]. Mutant A and in particular mutant B exhibited a grossly different UV-melting pattern (Figure 4). First, both transcripts lack the prominent TAR signal at Tm = 71.1°C, which is consistent with the partial TAR deletion. The remnant of TAR may still fold a TAR-like structure that explains minor transitions at Tm = 72.7°C and Tm = 66.9°C for mutant A and B, respectively. Second, the typical resonance of the LDI structure at Tm = 52.7°C is partially lost in mutant A and completely lost in mutant B. Other resonances at lower temperature (Tm = 39.0°C and Tm = 37.3°C for mutants A and B, respectively) and higher temperature (Tm = 60.0°C and Tm = 58.4°C, respectively) indicate the BMH-folding pattern (2). Strikingly, the UV-melting pattern reverses to a characteristic LDI signature with two peaks for the AB double mutant. The transition at Tm = 78.6°C is likely to represent the TAR hairpin of this mutant. Compared to the TARm control, the resonance is much reduced because the hairpin is severely truncated. Interestingly, the Tm of this truncated TAR hairpin of mutant AB is greater than that of TARm or TARwt, which is likely due to removal of destabilizing elements such as the 3-nt and 1-nt bulges.
Figure 4.
Thermal melting profiles of the TAR-mutated leader RNAs. Melting was monitored by UV absorption at 260 nm. The curves show the first order derivative (−δA/δT) to highlight the melting transitions. We analyzed transcripts TARm, A, B and AB. The melting temperature Tm is indicated at the top of the peaks.
To better understand the effects of the TAR mutations on leader folding, the structure of the transcripts (+1/+368) was also studied with the Mfold algorithm (32,33). This RNA-folding program predicts alternative structures and their thermodynamic stability (ΔG in kilocalories per mole). These values were used to calculate the ΔΔG (ΔGLDI − ΔGBMH), which describes the status of the LDI/BMH equilibrium (Table 1). Both conformations are predicted to be present in equimolar ratio when the ΔΔG is zero. The ΔΔG value for TARwt and TARm is −3.2 kcal/mole (Table 1), indicating that these transcripts adopt mainly the LDI conformation. The single mutants A, B, C, D and F show a significant shift to ΔΔG = −0.1 kcal/mole, a state in which both LDI and BMH structures are present in approximately equimolar amounts. Most importantly, Mfold predicted a complete reversal to the TARwt/TARm state for the double mutant AB and CD with a ΔΔG of −3.2 kcal/mole, which indicates that these transcripts fold predominantly the LDI conformation. These results are consistent with the gel migration and UV-melting experiments. There is one notable exception, Mfold predicts the LDI conformation for the E mutant, but the in vitro experiments indicated a BMH-like conformation. Taken together, the RNA structure and replication results demonstrate that folding of the LDI conformation correlates with efficient virus replication, whereas an altered LDI-BMH equilibrium correlates with a replication defect.
Table 1.
Thermodynamic stability (ΔG in kcal/mole) of leader RNAs in the LDI and BMH conformation
| ΔGLDI | ΔGBMH | ΔGLDI–BMH | |
|---|---|---|---|
| TARwt | −129.4 | −126.2 | −3.2 |
| TARm | −129.9 | −126.7 | −3.2 |
| Mutants | |||
| A | −111.7 | −111.6 | −0.1 |
| B | −118.3 | −118.2 | −0.1 |
| AB | −117.6 | −114.4 | −3.2 |
| C | −118.5 | −118.4 | −0.1 |
| D | −115.8 | −115.7 | −0.1 |
| CD | −116.7 | −113.5 | −3.2 |
| E | −102.0 | −98.1 | −3.9 |
| F | −114.2 | −114.1 | −0.1 |
| Evolved variants | |||
| AR1 | −114.0 | −110.8 | −3.2 |
| AR2 | −116.1 | −112.9 | −3.2 |
| FG3 | −114.7 | −114.6 | −0.1 |
| FG4 | −115.2 | −115.1 | −0.1 |
| FG5 | −118.8 | −118.7 | −0.1 |
| FG6 | −119.0 | −118.9 | −0.1 |
| ER1 | −94.6 | −92.3 | −2.3 |
| ER2 | −95.0 | −92.7 | −2.3 |
| ER3 | −112.6 | −110.8 | −1.8 |
Distorted leader RNA conformation affects dimerization
The single-side mutations in TAR affect the LDI-BMH equilibrium. Since the DIS motif, which is required for RNA dimerization, is exclusively exposed in the BMH conformation, one may expect an impact of these mutations on the dimerization capacity. We therefore performed RNA dimerization assays with 32P-labeled transcripts. These transcripts showed the expected differences in size on a denaturing polyacrylamide gel (Figure 5A). To determine the RNA dimerization efficiency, the transcripts were incubated in dimerization buffer and analyzed on a native polyacrylamide gel in the presence of Mg2+ (Figure 5B). The monomer and dimer bands were quantified to determine the dimerization efficiency (Figure 5C). The TARwt and the TARm transcripts are predominantly present as monomer under these conditions, which is consistent with LDI folding of these transcripts. The single mutants A and B show increased RNA dimerization, but the AB double mutant returned to the wild-type level. Similarly, the single mutants C and D show increased dimerization efficiency, which is restored to the wild-type level for the CD double mutant. The extended single mutant F also displays increased dimerization. Thus, HIV-1 leader transcripts dimerize more efficiently when only one side of the TAR hairpin is partially deleted. Also when almost the complete hairpin is removed as in mutant E, dimerization is increased significantly. These results are in agreement with the RNA folding experiments, as mutants that adopt the BMH conformation are dimerization-prone (3,4,30,36).
Figure 5.
Destabilization of TAR results in increased in vitro dimerization of leader RNA. (A) In vitro transcribed leader RNA molecules (corresponding to nucleotides +1 to +368 in wild-type HIV-1) with either a wild-type (TARwt), mutant (TARm) or deleted TAR sequence (mutants A–F) were analyzed on a denaturing polyacrylamide gel. (B) The transcripts were allowed to dimerize in the presence of Mg2+. Monomeric (m) and dimeric RNAs (d) were analyzed on a native polyacrylamide gel containing Mg2+. (C) The monomeric and dimeric bands were quantified and dimerization efficiency was determined by calculating the fraction of RNA present as dimer. The average of 2–5 experiments is shown with arrow bars indicating the SD.
Evolution of TAR-deleted HIV-rtTA variants restores the LDI/BMH equilibrium
Our results thus far demonstrate that a modified leader configuration correlates with reduced replication capacity. To obtain further evidence for this correlation, we tried to select fast-replicating variants for the replication-impaired HIV-rtTA mutants carrying the single TAR deletions. For each variant, we started four long-term cultures by transfecting the SupT1 T cell line with 10 μg of the molecular clones. Only in one of the A cultures and all four F cultures we observed virus replication after prolonged culturing, which was apparent from an increased CA-p24 level in the culture supernatant and formation of virus induced multi-nuclear syncytia. The virus was passaged on fresh cells when massive syncytia were observed. Cell samples were stored at several points as a source for integrated HIV-rtTA DNA, of which the U3-R-U5 region was PCR-amplified and sequenced (Figure 6A).
Figure 6.
Evolutionary repair of the hairpin structure at the 5′-end of the viral RNA of mutants A and F. (A) Upon long-term culturing of the A and F mutant viruses, the sequence of the TAR region was analyzed. The viruses in culture A and F II were cultured for up to 169 days. The cultures F I, III and IV were maintained for up to 175, 97 and 91 days, respectively. The LTR region was PCR amplified and cloned into the TA-cloning vector. The nucleotide sequence of the TAR region was determined for 2–5 clones in each culture. The frequency at which each sequence is observed is indicated (#). The −10 to +66 region is shown for the original TARm, the A and F mutants, and the evolved variants of A and F (with +1 indicating the transcription start site). Nucleotide substitutions and insertions are in bold and underlined. (B) Secondary structure of the TAR sequence in the A and F mutant and evolved viruses as predicted by the Mfold program. (C, upper panel) In vitro transcribed leader RNA molecules with either the original (TARm), mutated (A, F) or evolved TAR sequences (AR1-2, FG3-6) were analyzed on a denaturing polyacrylamide gel. (C, lower panel) The transcripts were allowed to dimerize in the presence of Mg2+ and monomeric (m) and dimeric (d) RNAs were resolved on a native polyacrylamide gel containing Mg2+. (D) The monomeric and dimeric bands were quantified and the dimerization efficiency was determined by calculating the fraction of RNA present as dimer. The average of 3–5 experiments is shown with error bars indicating the SD.
Mutant A acquired several point mutations in the R region (Figure 6A). At day 52, the viral quasispecies consisted of a mixture of two sequences that we named AR1 and AR2, each with two mutations. At later times, only the AR2 sequence was observed. We performed 5′-RACE analysis to verify that the wild-type transcription start site (+1 in Figure 6) is used by the A mutant (data not shown). The Mfold program was used to predict the structure and stability of the mutant and evolved TAR structures (Figure 6B). AR1 acquired two mutations that convert weak G–U into more stable A–U base pairs, concomitant with a change in ΔG of the TAR hairpin from −13.0 to −14.2 kcal/mole. AR2 shares one of these mutations, but acquired a unique mutation that changes a G–U into a G–C base pair. The latter mutation reorganizes the top of the stem to form a more stable hairpin of −16.3 kcal/mole. This AR2 variant seems superior over AR1, as it dominates the viral quasispecies at later culture times (days 87 and 169).
The F mutant acquired multiple nucleotide substitutions and insertions of G residues near the transcription start site in all four independent cultures (numbered I–IV in Figure 6A). Culture II acquired four substitutions and a G-insertion over time, which resulted in a gradual increase in the number of G nucleotides at the transcription start site. Because of these changes around the U3/R border, we used 5′ RACE analysis to determine the actual transcription start site of the virus in this culture at day 169. Transcription of the original F mutant, which was included as a control, started at the same G nucleotide as observed for the TARm and A mutant. The evolved F variant started transcription predominantly at the original +1 nucleotide and the immediate upstream G nucleotide, which resulted in the presence of five or six G nucleotides at the 5′-end of the transcript (FG5 and FG6 in Figure 6B, respectively). The structure and stability of the mutant and evolved TAR hairpins were analyzed with the Mfold program. Since we observed an increase in the number of G nucleotides at the transcription start site in this culture, we also analyzed the folding of transcripts with three and four G nucleotides at the 5′-end as putative evolution intermediates (FG3 and FG4, respectively). The TAR sequence of mutant F is predicted to fold two mini-hairpins (Figure 6B). The three-point mutations in variant FG3 destabilize the upstream mini-hairpin and reduce the overall stability from −14.9 to −10.9 kcal/mole (Figure 6B). The additional G residues resulting from base-substitution or G-insertion cause a structure switch to an alternative single hairpin of which the stability is progressively increased from −14.4 (FG4) to −16.7 (FG5) and −20.8 (FG6) kcal/mole.
The stability of complete leader transcripts of the evolved variants was also analyzed with Mfold and the ΔΔG of the LDI–BMH equilibrium was calculated (Table 1). Whereas the ΔΔG of mutants A and F is −0.1 kcal/mole, the AR1 and AR2 variants show a ΔΔG of −3.2 kcal/mole, which is in fact an identical value as determined for TARwt, TARm, AB and CD. These ΔΔG calculations thus indicate that LDI folding is restored for the AR1 and AR2 transcripts. However, the evolved F variants show a ΔΔG of −0.1 kcal/mole, which does not indicate such a shift in the LDI–BMH equilibrium.
To test whether the additional mutations observed in the evolved A and F variants restore leader RNA folding, we determined the dimerization capacity of leader transcripts. On a denaturing gel the transcripts show migration differences according to their size (Figure 6C, top panel). Upon incubation in dimerization buffer, the transcripts were analyzed on a non-denaturing gel (Figure 6C, lower panel) and RNA dimer and monomer bands were quantitated (Figure 6D). While mutant A has an increased dimerization capacity due to the LDI-to-BMH shift, the evolved AR1 and AR2 variants return to the low level of dimerization as seen for TARm and TARwt. The F mutant also showed increased RNA dimerization. Whereas RNA dimerization efficiency was further increased for the putative FG3 and FG4 intermediates, the wild-type level was restored for the FG5 and FG6 variants. Notably, the 5′ RACE analysis revealed that only the FG5 and FG6 transcripts were produced in the virus-infected cells. In fact, the increased dimerization efficiency of the FG3 and FG4 transcripts may explain why these variants were not observed as intermediates during virus evolution. Thus, both the evolved A and F variants return to the wild-type phenotype of restricted RNA dimerization. These results indicate that evolution of both the A and the F mutants resulted in a stabilization of the TAR-like hairpin, which restored the leader RNA folding and thereby dimerization control.
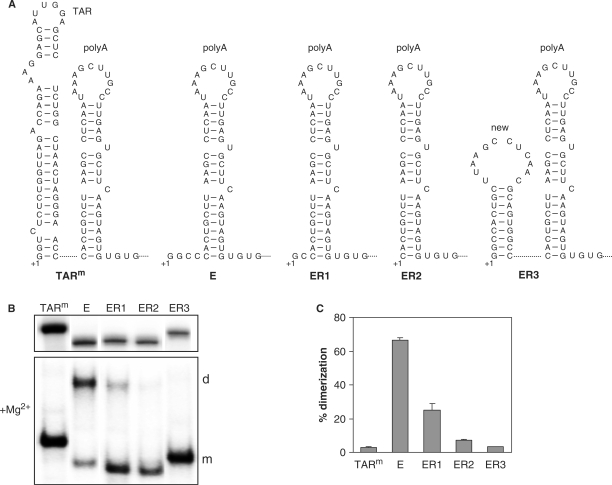
We also tested the dimerization capacity of leader transcripts corresponding to evolved variants of the TAR-deleted mutant E that were described recently (25). We observed distinct evolution paths in two independent cultures. In one culture, the virus acquired additional mutations at the transcription initiation site, which resulted in the modified ER1 and ER2 transcripts (Figure 7A). At an early stage in evolution, both the ER1 and ER2 transcripts were produced, whereas the ER2 variant dominated at a later stage. The mutant E transcript has four unpaired nucleotides upstream of the polyA hairpin and the ER1 variant has only two unpaired nucleotides at this position. In the ER2 transcript these latter nucleotides are deleted as well, and as a result the transcript starts precisely at the first nucleotide of the polyA hairpin. In the other culture, a sequence insertion at the transcription start site resulted in the presence of a new hairpin upstream of the polyA hairpin (ER3 in Figure 7A), which also effectively removes the unpaired nucleotides at the 5′-end of the transcript. We already mentioned that mutant E has a non-wild-type phenotype with a high level of spontaneous RNA dimerization, which is indicative for a BMH-like leader RNA structure. We observed a strong downmodulation of the RNA dimerization capacity of the three evolved variants (Figure 7B and C), which suggests that a wild-type-like LDI structure was restored.
Figure 7.
Re-establishment of a stable hairpin structure at the 5′-end of mutant E restores the wild-type in vitro dimerization level. (A) Secondary structure of the remaining TAR sequences and the polyA hairpin of mutant E and evolved variants. (B, upper panel) In vitro transcribed RNA molecules corresponding to the leader region of HIV-1 rtTA (TARm), the E mutant and the evolved variants (ER1, ER2, ER3) were analyzed on a denaturing polyacrylamide gel. (B, lower panel) The transcripts were allowed to dimerize and analyzed as described in Figure 6C. (C) The dimerization efficiency was determined as described in Figure 6D. The average of 2–4 experiments is shown with error bars indicating the SD.
Extension of the polyA hairpin triggers the BMH leader conformation
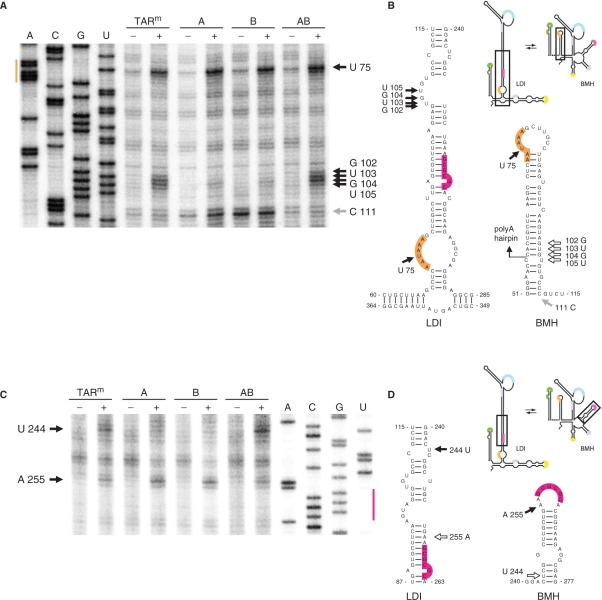
Although the TAR hairpin is not directly involved in the LDI–BMH conformational switch, as it is present in both conformations, our results demonstrate that the single side TAR deletions result in increased BMH folding and dimerization of the leader RNA. To understand how the TAR deletions affected the LDI–BMH equilibrium, we performed RNA structure probing of the TARm, A, B and AB leader transcripts. The RNAs were incubated with lead (Pb2+) nitrate that mainly cleaves at single-stranded nucleotides. The RNA fragments were subsequently analyzed by primer extension analysis. Probing of the polyA region in the TARm transcript (Figure 8A) revealed high Pb2+-induced cleavage at positions G102 to U105, indicating that these nucleotides are predominantly single-stranded, which is characteristic for the LDI conformation (Figure 8B). In contrast, these nucleotides are protected in the A and B mutants, which is in agreement with formation of the polyA hairpin and consequently the BMH conformation. Combined with the results of Mfold analyses, an extended version of the polyA hairpin is proposed for the A and B mutants (Figure 8B). In fact, 3′-terminal TAR nucleotides interact with nucleotides 3′ of the polyA hairpin. In particular the G triplet (positions +51 to +53) interacts with the C triplet (positions +109 to +111). The Pb2+-reactivity of the G102 to U105 nucleotides was restored in the AB double mutant, demonstrating that the LDI conformation was re-established. Nucleotide U75 in the polyA signal was highly reactive in all transcripts, which makes sense as this nucleotide is single stranded in both conformations. With the A and B mutants, a strong stop product is observed both with and without Pb2+ treatment, which probably results from pausing of reverse transcription at C111 at the base of the extended polyA hairpin in the BMH conformation (Figure 8B). This pause product is not observed with the TARm and AB transcripts, which is in agreement with the absence of this hairpin in the LDI conformation. Analysis of the upstream TAR region was more complex because of the introduced deletions. The TARm and AB transcripts fold a stable hairpin with a highly reactive loop sequence, whereas the A and B mutants fold a destabilized structure in which many more nucleotides are reactive (results not shown). Probing of the DIS region revealed reduced reactivity of U244 and increased reactivity of A255 in the A and B mutants compared to the TARm and AB transcripts (Figure 8C). This reactivity pattern is in agreement with the formation of the DIS hairpin and the BMH conformation by the A and B transcripts, and the absence of this hairpin in the LDI conformation of the TARm and AB transcripts (Figure 8D).
Figure 8.
RNA structure probing of TAR mutated HIV-1 leader RNA. Transcripts corresponding to the TARm, A, B and AB variants were incubated with Pb2+ (+) or mock treated (−). The RNA fragments were analyzed by primer extension with oligonucleotide lys21 to analyze the polyA region (A) and with R368 to analyze the DIS region (C). Reactive (black arrows) and protected nucleotides (open arrows) that are discussed in the text are indicated in the LDI and BMH conformation of the leader RNA (Figure 8B and D). These structures were described previously, with minor adaptations (39). The polyA hairpin (nt 58–103 in wild-type HIV) is extended in the A and B mutants, resulting is a strong reverse transcription stop at nucleotide C111 (grey arrow in Figure 8B).
DISCUSSION
The untranslated leader region, of which TAR is a component, is an evolutionary conserved region of the HIV-1 RNA genome. In previous studies, most of the mutations in the TAR hairpin resulted in a replication incompetent virus (20,37,38). Since the TAR hairpin plays a crucial role in Tat-mediated activation of transcription, it was not possible to study other functions of TAR in HIV-1 replication by mutation or (partial) deletion of the TAR sequence. By replacing the TAR-Tat transcription control mechanism with the rtTA-tetO (Tet-ON) system it became possible to study non-transcriptional functions of TAR (23). Our recent study indicates that TAR can be deleted completely when not required for Tat-mediated activation of transcription (25). However, partial TAR deletions resulted in a non-replicating virus, suggesting that left-over TAR sequences may interfere with another viral mechanism. We here demonstrate that the HIV-1 RNA genome requires a stable stem-loop structure at its 5′-end because unpaired nucleotides interact with downstream leader sequences. This interaction results in an unnatural folding of the leader RNA and causes a severe replication defect.
We performed in vitro assays with leader transcripts encoding the full-length or truncated TAR elements to screen for an effect on the LDI/BMH equilibrium. In these assays the wild-type leader adopts predominantly the LDI conformation. Strikingly, transcripts with a deletion in one or the other side of the TAR hairpin demonstrate a shift towards the alternative BMH conformation, in which the DIS hairpin motif is exposed. As a consequence, we measured enhanced RNA dimerization for these mutants. In the double mutants, the wild-type LDI/BMH equilibrium is restored and these mutants show a low level of dimerization, similar to that of the wild-type leader. RNA structure probing revealed that, upon single-side deletion of TAR sequences, some TAR nucleotides base pair with nucleotides directly downstream of the polyA hairpin, resulting in a stabilization of this structure. As a consequence, the leader RNA shifts towards the BMH structure in which the DIS hairpin is exposed, which explains the increased dimerization of the A and B mutants. In the double mutant AB, the TAR nucleotides fold a truncated, but relatively stable TAR-like hairpin and do not disturb the typical LDI folding. Mfold analysis indicate that a similar interaction between TAR sequences and downstream leader sequences can explain the shift toward the BMH conformation for the C, D, E and F mutants. This scenario is strengthened by the evolution of mutant A, E and F viruses. Using alternative evolutionary strategies, the remnant TAR sequences in these mutants are either removed or modified by mutations such that a stable hairpin structure can be formed at the 5′-end of the viral genome, which prevents an interaction with downstream nucleotides. These changes restore LDI folding and restrict RNA dimerization, as in the wild-type RNA.
For the large set of HIV-1 RNA mutants and evolved variants, we observed a strong correlation between RNA properties (LDI/BMH folding, RNA dimerization) and the Mfold analysis. The notable exception is the deletion mutant E, which is predicted to adopt the LDI conformation in Mfold, yet clearly displays the BMH conformation and increased RNA dimerization in the experiments. The unpaired GGCCC nucleotides at the 5′-end of the E transcript may also interact with downstream sequences and stabilize the polyA hairpin. Alternatively, this dangling 5′-end may induce a tertiary base-pairing interaction that stabilizes the BMH conformation, but such interactions are ignored by Mfold. Mutant E displayed delayed replication and the evolved variants remove this ‘toxic’ GGCCC sequence by different means (Figure 7A). These evolved variants restore the wild-type pattern of restricted RNA dimerization. There are also some minor discrepancies between the MFold analysis, in particular the ΔΔG calculation, and the observed dimerization efficiency of these evolved variants, although one has to realize that quite different structural solutions were selected in cultures ER1/2 and ER3.
Also the phenotype of the FG5 and FG6 evolution products of the F mutant is not supported by the MFold prediction. Whereas MFold analysis predicts a similar LDI-BMH equilibrium for the evolved variants and the F mutant (Table 1), we observed reduced RNA dimerization for the FG5 and FG6 variants (Figure 6D), indicating that LDI folding is restored. MFold analysis of the TAR region (Figure 6B) demonstrates that the additional G nucleotides in FG5 and FG6 do restore the stability of this hairpin. This TAR stabilization may prevent the interaction between TAR nucleotides and downstream sequences that probably caused the increased BMH formation of the F mutant. As a result, the evolved leader transcripts can fold the LDI conformation more efficiently and show a wild-type like dimerization efficiency.
We previously discussed that premature RNA dimerization may not be beneficial for the virus (30). If an increased dimerization level in our in vitro assays correlates with increased dimerization in vivo, the unnatural dimerization capacity of the single-side mutants may explain their replication defect. However, it cannot be excluded that an aberrant folding of the viral leader RNA in these mutants does not only affect dimerization but also other processes like RNA stability, transport, packaging, processing and translation. Notably, extension of the polyA hairpin results in partial disruption of the U5-AUG duplex, the interaction of nucleotides +105/+115 (downstream of the polyA hairpin) with nucleotides +334/+344, including the gag-AUG start codon (27,39). The U5-AUG duplex is formed exclusively in the genomic HIV-1 RNA that contains the gag sequences and not in the subgenomic spliced transcripts (39). As said, these adverse effects are neutralized in the TAR double mutants and the evolved variants of mutants A, E and F. In these variants, the remnant TAR sequences can either fold a TAR-like structure that is stable enough to avoid an interaction with downstream sequences or the remnant TAR sequences are completely removed.
In conclusion, we demonstrate that extreme caution is warranted when mutating the highly structured HIV-1 leader RNA. When structured elements are modified, single-stranded regions may be created that cause unwanted side-effects by affecting the proper folding of the other leader RNA replication signals. An effect of leader mutations on the BMH/LDI equilibrium of the HIV-1 leader RNA was previously suggested in a theoretical study that used experimental data obtained for 38 HIV-1 leader mutants (15). We here demonstrate that partial deletions in TAR do indeed affect overall leader structure and thus have detrimental effects on viral replication.
ACKNOWLEDGEMENTS
We thank Wim van Est for art work. This research was sponsored by NWO-CW (Top grant) and the Dutch AIDS Foundation (Aids Fonds grant 2005022). Funding to pay the Open Access publication charges for this article was provided by the Academic Medical Center of the University of Amsterdam.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 2.Huthoff H, Berkhout B. Two alternating structures for the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 4.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 6.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1 LAI RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 7.Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 2005;3:61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 8.Brady J, Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan R, Brodsky A, Williamson JR, Frankel AD. RNA recognition by HIV-1 Tat and Rev. Semin. Virol. 1997;8:186–193. [Google Scholar]
- 10.Richter S, Ping YH, Rana TM. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl Acad. Sci. USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei P, Garber ME, Fang S.-M, Fisher WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 12.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 13.Clever JL, Mirandar D., Jr, Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helga-Maria C, Hammarskjold ML, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell RS, Hu J, Beriault V, Mouland AJ, Laughrea M, Kleiman L, Wainberg MA, Liang C. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type 1 RNA. J. Virol. 2003;77:84–96. doi: 10.1128/JVI.77.1.84-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7:1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berkhout B. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 2000;48:29–73. doi: 10.1016/s1054-3589(00)48003-8. [DOI] [PubMed] [Google Scholar]
- 19.Das AT, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of the human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrich D, Hooker CW, Parry E. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 2000;74:5639–5646. doi: 10.1128/jvi.74.12.5639-5646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das AT, Verhoef K, Berkhout B. A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol. 2004;388:359–379. doi: 10.1016/S0076-6879(04)88028-5. [DOI] [PubMed] [Google Scholar]
- 22.Smith SM, Markham RB, Jeang K.-T. Conditional reduction of human immunodeficiency virus type 1 replication by a gain-of-herpes simplex virus 1 thymidine kinase function. Proc. Natl Acad. Sci. USA. 1996;93:7955–7960. doi: 10.1073/pnas.93.15.7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 2001;75:979–987. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus type-1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 2007;81:7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 2002;277:19967–19975. doi: 10.1074/jbc.M200950200. [DOI] [PubMed] [Google Scholar]
- 27.Abbink TEM, Ooms M, Haasnoot PCJ, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 28.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virol. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 29.Das AT, Zhou X, Vink M, Klaver B, Verhoef K, Marzio G, Berkhout B. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 30.Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooms M, Cupac D, Abbink TEM, Huthoff H, Berkhout B. The availability of the primer activation signal (PAS) affects the efficiency of HIV-1 reverse transcription initiation. Nucleic Acids Res. 2007;35:1649–1659. doi: 10.1093/nar/gkm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooms M, Cupac D, Abbink TE, Huthoff H, Berkhout B. The availability of the primer activation signal (PAS) affects the efficiency of HIV-1 reverse transcription initiation. Nucleic Acids Res. 2007;35:1649–1659. doi: 10.1093/nar/gkm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 37.Berkhout B, Klaver B. Revertants and pseudo-revertants of human immunodeficiency virus type 1 viruses mutated in the long terminal repeat promoter region. J. Gen. Virol. 1995;76:845–853. doi: 10.1099/0022-1317-76-4-845. [DOI] [PubMed] [Google Scholar]
- 38.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]