Abstract
Trinucleotide exchange (TriNEx) is a method for generating novel molecular diversity during directed evolution by random substitution of one contiguous trinucleotide sequence for another. Single trinucleotide sequences were deleted at random positions in a target gene using the engineered transposon MuDel that were subsequently replaced with a randomized trinucleotide sequence donated by the DNA cassette termed SubSeqNNN. The bla gene encoding TEM-1 β-lactamase was used as a model to demonstrate the effectiveness of TriNEx. Sequence analysis revealed that the mutations were distributed throughout bla, with variants containing single, double and triple nucleotide changes. Many of the resulting amino acid substitutions had significant effects on the in vivo activity of TEM-1, including up to a 64-fold increased activity toward ceftazidime and up to an 8-fold increased resistance to the inhibitor clavulanate. Many of the observed amino acid substitutions were only accessible by exchanging at least two nucleotides per codon, including charge-switch (R164D) and aromatic substitution (W165Y) mutations. TriNEx can therefore generate a diverse range of protein variants with altered properties by combining the power of site-directed saturation mutagenesis with the capacity of whole-gene mutagenesis to randomly introduce mutations throughout a gene.
INTRODUCTION
Protein engineering has provided crucial insights into protein structure, function and folding, and has been pivotal in adapting proteins for various biotechnological applications (1). Initial protein engineering strategies focused on site-directed mutagenesis but due to our limited understanding of the complex and cooperative network of weak interactions that comprise a functional protein, it is still difficult to predict the consequence of a mutation. To overcome these difficulties, a new approach was taken that mimicked the natural process of Darwinian evolution and has been termed directed evolution (2–4). Directed evolution involves generating molecular diversity by the introduction of random mutations within a gene followed by selection of desirable protein variants. Unlike site-specific mutagenesis, directed evolution does not require a detailed knowledge of protein structure or have any preconceptions of the importance of certain residues to stability and function. This evolutionary protein engineering strategy has been very successful in generating proteins with new properties, with beneficial mutations observed at positions distant from those predicted to be crucial for generating novel activity (5,6).
Although a variety of methods exist to introduce predominantly single point mutations randomly throughout a whole gene, their limitations include: error, codon and amplification biases, sampling of a restricted set of amino acids, the masking of beneficial mutations by deleterious ones and the inability to sample many of the potential variants due to large and undefined library sizes (7,8). Some of these problems can be overcome by site-saturation mutagenesis, in which diversity is incorporated into a synthetic oligonucleotide. However, this approach inherits the problems of site-directed mutagenesis and mutations are restricted to just a small portion of the protein.
A new method called trinucleotide exchange (TriNEx) is proposed that will overcome the limitations of current methods for generating molecular diversity. TriNEx is a non-PCR-based method that combines the power of whole-gene random mutagenesis with the ability of oligonucleotide-directed mutagenesis to sample an expanded range of amino acid substitutions. Based on a recently developed transposon method for generating single amino acid deletion variants (9), TriNEx involves first the removal of a single contiguous trinucleotide sequence then its replacement with a randomized sequence (Figure 1). This potentially allows the sampling of every codon permutation throughout a gene and defines library diversity and sampling requirements. This method will overcome the error and codon bias inherent in methods such as error-prone PCR and will not be restricted to mutating predefined amino acid residues as is the case with oligonucleotide-directed mutagenesis. There are no PCR steps involved in diversity generation so avoiding any amplification bias. Also, as one transposon is inserted per gene, each variant will contain either a single amino acid or two adjacent substitutions per protein variant, preventing the masking of beneficial mutations by other, distant deleterious mutations. Furthermore, it allows directed evolution to be applied in a similar manner to the powerful site-directed single substitution scanning mutagenesis methods (e.g. alanine scanning mutagenesis) to assess the contribution of individual amino acid residues to the structure, function and folding of proteins.
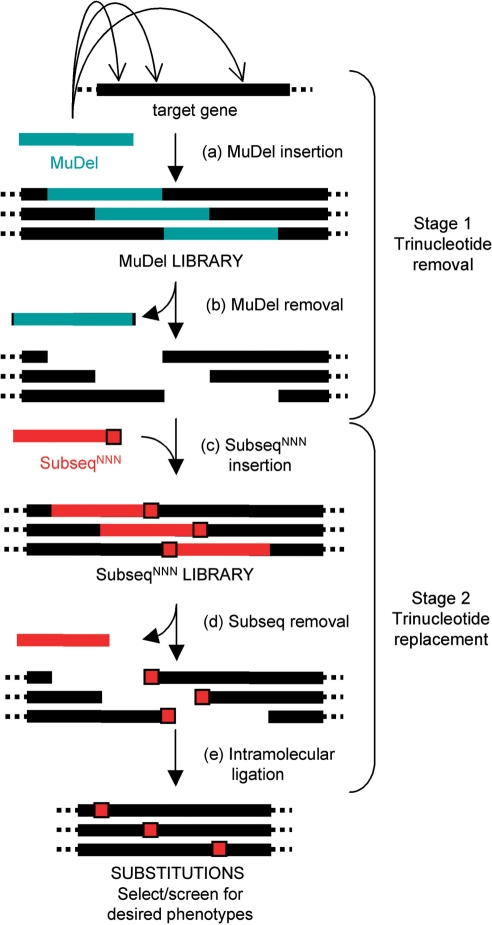
Figure 1.
Schematic outline of the TriNEx procedure. Stage 1. Trinucleotide deletion. The MuDel library is generated by in vitro transposition of the engineered transposon MuDel (teal) into the target DNA. Restriction endonuclease digestion removes MuDel from this library together with 3 bp of the original target DNA to generate a single break per molecule. Stage 2. Trinucleotide replacement. The DNA cassette termed SubSeqNNN (red) is then inserted between the break in the target DNA to generate the SubSeqNNN library. The last three nucleotides at one end of SubSeqNNN (red square) are randomized (NNN). Restriction endonuclease digestion removes all SubSeq, except for the NNN sequence that is now incorporated into the target DNA, which replaces the 3 bp deleted in stage 1. Intramolecular ligation regenerates the complete gene containing new and randomly placed trinucleotide segments. This is followed by selection and/or screening to identify new variants with desired properties.
Here, we demonstrate the TriNEx method using the bla gene encoding TEM-1 β-lactamase. TEM-1 is one of the main enzymes responsible for conferring bacterial resistance to β-lactam antibiotics such as ampicillin but has poor activity towards third generation cephalosporins such as ceftazidime and can be inhibited by clavulanate (10). We successfully generated libraries with triplet nucleotide exchanges present throughout the bla gene. The resulting amino acid substitutions had a range of effects on the in vivo activity of TEM-1, including increased activity towards ceftazidime (CAZ) and improved resistance to clavulanate (CLV). Many of the amino acid substitutions observed were only accessible by two or more adjacent nucleotide changes.
MATERIALS AND METHODS
Construction of libraries with MuDel inserted randomly within the bla gene
The initial small-scale library (termed BLAΔ175) that contained 175 variants with MuDel inserted randomly within the bla gene was a subset of 391 variants generated previously (11). Construction of the larger, extended library (termed BLAΔ1644) comprised of 1644 variants with MuDel inserted randomly within bla was performed essentially as described previously for the single amino acid deletion procedure (9,11). The details of this procedure are described in Supplementary Methods.
Construction of library containing SubSeqNNN randomly placed within bla
The SubSeqNNN cassette was constructed using PCR as described in Supplementary Methods. The library of variants with SubSeqNNN placed within bla was generated by the excision of MuDel from the plasmid followed by the insertion of the SubSeqNNN DNA cassette. The pooled DNA (1 μg) from either the BLAΔ175 or BLAΔ1644 libraries was digested with MlyI to remove MuDel. The cleaved DNA was then treated with APex™ heat-labile alkaline phosphatase (Epicentre Biotechnologies, Madison, WI, USA) for 15 min at 37°C. After inactivation of the phosphatase by heating to 70°C for 10 min, the linearized pNOM DNA was separated by agarose gel electrophoresis and purified. Linear pNOM was ligated with SubSeqNNN in 1:3 molar ratio (pNOM:SubSeqNNN) using the Fast-Link™ DNA ligation kit (Epicentre Biotechnologies). A total of 200 ng of DNA was used in the ligation.
With regards to the BLAΔ175 library, ca 15 ng of the DNA from the ligation reaction was used to transform chemically competent Escherichia coli DH5α cells. The transformed cells were grown on LB agar containing 15 μg/ml kanamycin (Kan) and incubated at 37°C for 16 h. Colonies were picked at random and used to inoculate 5 ml of LB broth containing 15 μg/ml Kan. Plasmid DNA was isolated from each culture using the QIAprep Spin Miniprep kit (Qiagen Ltd, Crawley, UK). SubSeq was removed from the plasmid (ca 2 μg per 50 μl reaction volume) by digestion with MlyI. The plasmid (ca 20 ng) was recircularized directly after digestion using T4 DNA ligase (Promega UK, Southampton, UK). An equivalent of ca 0.4 ng of DNA from the ligation reaction was used to transform chemically competent E. coli DH5α. Half of the cells were then spread on LB agar containing 32 μg/ml Amp and incubated at 37°C for a minimum of 16 h. The bla gene sequence and the Amp MIC for clones producing active TEM-1 variants were determined as described subsequently. The bla gene prior to SubSeq removal was also sequenced for several inactive variants to predict the mutation that resulted in loss of β-lactamase activity.
With regards to the BLAΔ1644 library, the ligation reaction to insert SubSeqNNN within linearized pNOM was then used to transform (ca 15 ng of DNA/transformation) high efficiency chemically competent NovaBlue E. coli cells (Merck KGaA). To calculate the number of transformed cells containing the pNOM-SubSeqNNN plasmid, the equivalent of 1/30 of the transformation mix post-recovery was grown on LB agar plates containing 25 μg/ml Kan and the number of colony forming units (c.f.u.) observed after incubation at 37°C overnight was determined (routinely generated ca 4 × 105 c.f.u./μg DNA). The remainder of the transformation culture was added to 50 ml of LB broth containing 200 μg/ml Kan and incubated at 37°C for 16 h. Plasmid DNA was isolated from the culture using the QIAprep Spin Miniprep kit (Qiagen), and 2 μg of this DNA was digested with MlyI to remove the SubSeq section. The DNA was desalted and 100 ng of the DNA ligated using the Fast-Link™ DNA ligation kit (Epicentre Biotechnologies) to recircularize pNOM. The ligation reaction (10 ng/transformation) was used to transform high efficiency chemical competent NovaBlue E. coli (Merck KGaA, Merck Chemicals Ltd, Nottingham, UK).
Selection of TEM-1 variants with enhanced activity towards ceftazidime and clavulanate
Transformed cells were diluted 10-fold in SOC medium then spread on LB agar containing at least 0.3 μg/ml CAZ or 100 μg/ml Amp and 4 μg/ml CLV. The plates were incubated at 37°C for a minimum of 16 h. Cells transformed with pNOM containing wild-type bla did not grow under these CAZ and CLV selection conditions. Selected colonies were transferred to LB broth containing the equivalent concentration of antibiotic used in the selection. The cultures were grown overnight at 37°C and glycerol added to a final concentration of 25% (v/v) for storage at −80°C.
Characterization of the bla trinucleotide exchange variants
The bla gene of selected variants was isolated by PCR using the GoTaq® system (Promega) with primers flanking the gene (5′-TCCGCTCATGAGACAATAACCCTG-3′ and 5′-CTACGGGGTCTGACGCTCAGTG-3′) and the PCR product sequenced (DNA Sequencing Core, Molecular Biology Unit, Cardiff University) to determine the position and nature of the mutation. The in vivo activity of the selected variants was estimated by measuring the minimum inhibitory concentration (MIC) of Amp, CAZ or Amp and CLV that prevented cell growth. The Amp MIC values for cells producing TEM-1 variants derived from the BLAΔ175 library was determined essentially as described previously (9,11) and detailed in the Supplementary Methods. The CAZ or CLV+Amp MIC values of the selected TEM-1 variants derived from BLAΔ1644 were estimated as follows. The glycerol stocks generated above were used to inoculate LB broth cultures that were incubated at 37°C for 4 h. Each culture was then transferred, using a 96 pronged replicating fork, to LB agar containing 0.15, 0.3, 0.6, 1.2, 2.4, 4.8, 9.6, 19.2 or 38.4 μg/ml CAZ, or 2, 3, 4, 6, 8, 12, 16, 32, 64 μg/ml CLV with 100 μg/ml Amp and the plates incubated at 37°C for 16 h. Four colonies containing pNOM were used as controls. The bla gene was isolated by PCR and sequenced, as described earlier.
RESULTS
Creation of trinucleotide substitution variants from BLAΔ175
The replacement of one contiguous trinucleotide sequence with another was achieved in a two-stage process, as outlined in Figure 1. The first stage involved the removal of a trinucleotide sequence from a target gene using a previously described method based on the use of the engineered transposon MuDel (9). The next stage involves the replacement of the deleted trinucleotide by the insertion of the DNA cassette termed SubSeqNNN into the break within the target gene vacated by MuDel. The mechanism by which trinucleotide deletion and replacement is accomplished is described in Figure 2.
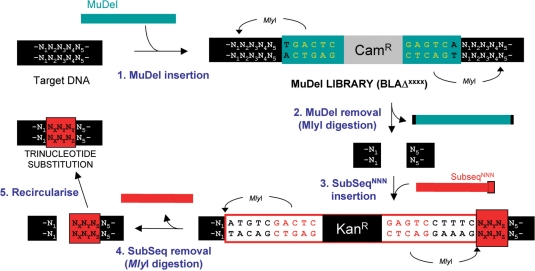
Figure 2.
Mechanism of TriNEx process. Step 1. Two MlyI recognition sites (5′GAGTC(N)5↓) were placed 1 bp from each end of MuDel, 1 bp away from the site of transposon insertion. MlyI cuts 5 bp outside its recognition sequence to generate a blunt end. Insertion of MuDel results in the duplication of 5 bp (N1N2N3N4N5) of the target gene at the insertion point (23,24). Step 2. Digestion with MlyI removes MuDel together with 8 bp of the target DNA (4 bp at each end). This equates to removal of a contiguous 3 bp sequence from the starting target gene (N2N3N4). Step 3. SubSeqNNN is then ligated into the gap vacated by MuDel. SubSeqNNN also contains two MlyI recognition sites strategically placed towards the ends of the cassette. One site is located so that MlyI will cut at the exact point where the target DNA joins SubSeqNNN. The second site will cut 3 bp into SubSeqNNN so donating 3 bp (NXNYNZ) to the target DNA. Step 4. Digestion with MlyI removes SubSeq but with 3 bp of its sequence now replacing the 3 bp deleted from the target gene. Step 5. Intramolecular ligation reforms the target gene but with one contiguous trinucleotide sequence replaced with another.
Initially, TriNEx was demonstrated using a subset of 175 variants, constructed previously (11), with MuDel inserted at random positions within bla. This library was termed BLAΔ175 and MlyI digestion from a DNA pool comprised of the 175 variants removed MuDel. After insertion of SubSeqNNN by ligation and transformation of E. coli, plasmid DNA was isolated from a total of 106 different colonies resistant to Kan and SubSeq removed by digestion with MlyI. Recircularized plasmid was used to transform E. coli, and bacterial resistance to Amp tested by plating on LB agar containing 32 μg/ml Amp. Of these 106, viable cell growth (typically 10–500 colonies per plate) was observed for 73 with no colony growth detected for the remaining 33. Of the 73 bla variants capable of conferring Amp resistance, 34 were sequenced to determine the mutations introduced and to assess their influence on the MIC of Amp that prevented cell growth. To confirm that variants unable to confer resistance were a consequence of potentially deleterious substitution mutations to TEM-1 and not gene frameshifts or trinucleotide deletions, 15 of the 33 bla variants that encoded a TEM-1 variant unable to confer Amp resistance were sequenced prior to SubSeqNNN removal and the TriNEx predicted.
Sequence analysis revealed that the position of the mutations were distributed throughout the bla gene (Figure 3), with 14, 45 and 41% of the variants containing single, double or triple nucleotide changes, respectively. No wild-type or trinucleotide deletion variants of bla were observed, and no secondary mutations were seen. Two variants were predicted to introduce frameshifts. Each of the 49 variants was unique at the genetic level and only two were identical at the protein level (Table 1), which both reconstituted a gene encoding wild-type TEM-1 but with silent mutations within different codons (encoding R120 and L193-L194). The nature of the replacement NNN sequence could be determined exactly for 33 variants, and revealed that 27 were unique with no NNN sequence represented more than twice. Of the 45 variants that encoded amino acid substitutions, mutation of six residues were observed twice and one three times. However, the trinucleotide replaced was not always the same (Table 1). For example, Q278W was generated by exchange of three nucleotides 1 bp upstream of those replaced to produce Q278C. At least 88% of the MuDel insertion positions predicted from the sequence data were judged to be unique, close to that observed previously (11). Just over half (26 from 45) of the TEM-1 variants contained amino acid substitutions only accessible by mutation of at least two of the three nucleotides.
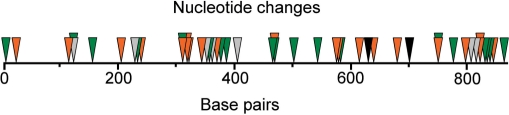
Figure 3.
Position and number of nucleotides changes introduced into the 39 bla TriNEx variants derived from BLAΔ175. The grey, orange and green triangles signify 1, 2 and 3 nucleotide replacements, respectively. The black triangles represent frameshifts due to deletion of two nucleotides from bla.
Table 1.
Sequence variation observed in bla variants derived from the BLAΔ175 library and their influence on TEM-1 activity towards Amp in vivo
| Amino acid substitutionsa | Nucleotide substitutionsb | Base changesc | Amp MIC (μg/ml)d |
|---|---|---|---|
| Wild type | − | − | 4096 |
| M1N-S2G | atgagt→aatggt | 3 | NG |
| R9V | cgt→gtt | 2 | 512 |
| D38E-Q39P | gatcag→gaaccg | 2 | 256 |
| L40ter | ttgggt→tgatgt | 3 | NG |
| G41S | ggt→agt | 1 | 4096 |
| N52K-S53A | aacagc→aaggcc | 3 | 1024 |
| M69R-S70C | atgagc→aggtgc | 2 | NG |
| L76L C77R | ctatgt→ctacgt | 1 | NG |
| C77P | tgt→cca | 3 | NG |
| A79V | gcg→gta | 2 | 2048 |
| V103V E104G | gttgag→gtcggg | 2 | 2048 |
| V103V E104L | gttgag→gtgctg | 3 | 1024 |
| S106T | tca→acc | 2 | 256 |
| S106C | tca→tgt | 2 | 256 |
| P107A | cca→gct | 2 | 256 |
| T114T D115Y | acggat→acctat | 2 | NG |
| M117I | atg→ata | 1 | 1024 |
| T118M-V119I | acagta→atgata | 3 | 512 |
| R120R | aga→agg | 1 | 4096 |
| C123ter | tgcagt→tgatgt | 2 | NG |
| A125G-A126S | gctgcc→ggatcc | 3 | 64 |
| T128T M129G | accatg→actggg | 3 | 512 |
| A135D-N136Y | gccaac→gattac | 3 | NG |
| M155S-G156W | atgggg→agttgg | 3 | 64 |
| M155I-G156Q | atgggg→attcag | 3 | 1024 |
| M155I-G156E | atgggg→atcgag | 2 | 1024 |
| P167P E168L | ccggag→cctttg | 3 | 1024 |
| T181D | acg→gac | 3 | NG |
| R191R K192ter | cgcaaa→cgttaa | 2 | NG |
| L193L-L194L | ctatta→ctccta | 2 | 4096 |
| L194C-T195P | ttaact→tgtcct | 3 | NG |
| R204R Q205R | cggcaa→cgccga | 2 | 4096 |
| W210 fs | gac tgg atg | – | NG |
| →ga––ac atg | |||
| A213G | gcg→ggt | 2 | 4096 |
| P226P A227T | ccggct→ccaact | 2 | 4096 |
| S235 fs | tct gga→tcg ––a | – | NG |
| L250H-G251R | ctgggg→cacagg | 3 | NG |
| L250L G251R | ctgggg→ctacgg | 2 | NG |
| R259I | cgt→atc | 3 | 256 |
| T266K-G267S | acgggc→aagagc | 2 | 512 |
| Q269L | cag→ctg | 1 | 4096 |
| M272R | atg→agg | 1 | 512 |
| D273D E274Q | gatgaa→gaccaa | 2 | 2048 |
| E274P | gaa→cca | 2 | 64 |
| R277R Q278W | agacag→aggtgg | 3 | 4096 |
| Q278C | cag→tgt | 3 | 2048 |
| A280A E281S | gctgag→gcgtcg | 3 | 2048 |
| I282T | ata→acc | 2 | 64 |
| K288N-H289V | aagcat→aacgtt | 3 | 512 |
aSubscripted sequences represent silent nucleotide changes to the codon. fs represent frameshift. Residues numbered according to standard system (25).
bBold and underlined sequences represent the known nucleotides deleted from bla and replaced by SubSeqNNN.
cActual number of nucleotide differences between the variant and wild-type bla.
dNG, no cell growth observed on 32 μg/ml Amp LB agar upon removal of SubSeq and reconstitution of the full length bla.
Exchange of contiguous trinucleotide segments can span two adjacent codons in theoretically 2/3 of cases, potentially leading to a double amino acid substitution. Due to the degeneracy in the genetic code and mutation of either one or two nucleotides, the majority of variants (66%) encoded a single amino acid substitution (Table 1), with just under a half of these (15 from 31) containing a silent mutation.
The in vivo activity of TEM-1 variants also varied depending on the mutation, with Amp MIC conferred on E. coli ranging from 64 μg/ml (e.g. E274P) to 4096 μg/ml (e.g. Q205R and Q278W) observed for wild-type TEM-1 (Table 1). Of the 15 variants that displayed no TEM-1 activity, three were predicted to contain substitutions that generated a stop codon (e.g. K192ter) and another disrupted the initiation codon (M1N-S2G). Many of the other predicted mutations could be classed as potentially disruptive to TEM-1 structure and/or function. The M69R-S70C mutation knocks out the essential catalytic serine. Mutation of C77 in the core helix of TEM-1 will remove the sole disulphide bond in the protein. Others may disrupt organized secondary structure (e.g. L194C-T195P) or introduce a charged residue into the hydrophobic core close to the active site (e.g. A135D-N136Y). TEM-1 has also been shown previously not to tolerate mutations as positions T181 or G251 (12). Two variants were predicted to contain frameshifts due to the removal of two nucleotides from the bla gene (Table 1) but no trinucleotide deletions were predicted. However, it cannot be ruled out that errors introduced post SubSeq removal and the subsequent self-ligation stages were responsible for inactivation of some of the TEM-1 variants.
Construction of an extended TriNEx library and the selection of enhanced TEM-1 variants
The main aim of directed evolution is the creation and sampling of a suitable range of molecular diversity to alter or improve proteins. As a demonstration of the ability of TriNEx to fulfil this aim, it was employed to improve the activity of TEM-1 towards a third generation cephalosporin CAZ, and enhance resistance to the inhibitor CLV. First, a much larger transposon insertion library was required to sample all the potential insertion positions within bla. A new library comprised of 1644 Amp sensitive colonies was constructed with MuDel inserted within bla and was termed BLAΔ1644. The size of this library was deemed sufficient to cover all potential insertion positions within bla (coding region of 858 bp), even when redundancy in transposon insertion positions [10–15%; see above and reference (11)] is factored in. The diversity of transposon insertion was confirmed using a restriction endonuclease procedure described previously (Figure 4a) (11). The digestion products form a smear, as would be expected if MuDel insertion within bla was essentially random (Figure 4b). The BLAΔ1644 members were pooled and MuDel removed by digestion with MlyI and SubSeqNNN ligated into the break. Transformation of E. coli with the ligation mixture routinely generated ca 4 × 105 c.f.u./μg of DNA on agar plates containing Kan. This was deemed sufficiently high to sample the possible 54 912 different genetic variations of bla that can be generated by TriNEx, if every possible contiguous trinucleotide sequence is replaced (gene size of 858 multiplied by 64 different NNN combinations). The transformed cells were subsequently grown under a stringent Kan selection regime (200 μg/ml) to ensure that only cells containing plasmid-borne SubSeqNNN would survive. DNA was then isolated from the culture to generate a plasmid pool of SubSeqNNN inserted at random positions within bla.
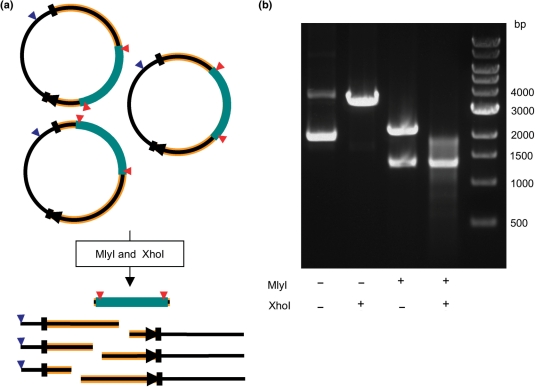
Figure 4.
Restriction endonuclease analysis of the BLAΔ1644 library. (a) Rationale for determining MuDel (teal) insertion diversity within the bla gene (orange with black stripe) of pNOM. Digestion by XhoI (recognition site shown as blue triangle) of the pooled pNOM plasmid with MuDel inserted within the bla gene generates one major product of 3422 bp in size. Digestion with MlyI (recognition site shown as red triangle) generates two major products of 1310 bp (MuDel plus 8 bp from bla) and 2112 bp in size. Digestion with both XhoI and MlyI generates the 1310 bp MuDel fragment together with many different size fragments depending on the insertion position of the transposon. (b) Restriction analysis of the BLAΔ1644 library with XhoI and/or MlyI, as indicated in the figure.
The SubSeq portion was removed by MlyI digestion and the plasmid pool recircularized. This formed the library used for the selection of TEM-1 variants with improved properties. Minimum estimates of 41 000 variants were tested under each condition to select those capable of promoting cell growth on 0.3 μg/ml CAZ or a combination of 100 μg/ml Amp and 4 μg/ml CLV. However, this was probably an underestimate because in the absence of a second selectable marker, the calculation was based on the number of colonies observed on LB agar plates containing 100 μg/ml Amp. While the majority of TEM-1 substitution variants were likely to retain even a low level of activity towards Amp, the previous results of individual TEM-1 variants derived from BLAΔ175 indicated that 31% would be inactive. Using this as a guide, the number of variants screened under each selection condition was likely to be closer to 52 000. Cells producing wild-type TEM-1 (via the parent pNOM plasmid) did not grow at these CAZ or Amp+CLV concentrations. A total of 408 or 93 colonies were observed to grow on LB agar under either the CAZ or Amp+CLV selection conditions, respectively. Of these, 41 colonies selected on CAZ and 27 selected on Amp/CLV were arbitrarily chosen for sequencing and in vivo activity analysis (Table 2 and full list in Supplementary Table 1).
Table 2.
Examples of TEM-1 variants with enhanced properties in vivoa
| Amino acid substitutionb | Nucleotide changes | MIC fold changec |
|
|---|---|---|---|
| CAZ | CLV | ||
| M69I | atg→atc | − | 3 |
| P145R | ccg→cga | 4 | − |
| D163E-R164N | gatcgt→gagaat | 16 | − |
| D163D R164S | gatcgt→gactct | 32 | − |
| D163D R164H | gatcgt→gaccat | 64 | − |
| D163D R164D | gatcgt→gacgat | 32 | − |
| R164P-W165R | cgttgg→cctagg | 32 | − |
| cgttgg→ccgcgg | 4 | − | |
| R164P-W165G | cgttgg→ccaggg | 8 | − |
| W165G | tgg→ggg | 2 | − |
| W165P | tgg→cct | − | 2 |
| W165Y | tgg→tac | − | 3 |
| tgg→tat | − | 3 | |
| E166G | gaa→gga | 8 | − |
| P167H | ccg→cac | 8 | − |
| E171P | gaa→cca | 8 | − |
| R178A | cgt→gca | 4 | − |
| E197P | gaa→cca | 2 | 1.5 |
| G238G E240R | ggtgag→ggcagg | 4 | − |
| S243S R244T | tctcgc→tccacc | − | 3 |
| R275V | cga→gta | − | 8 |
| cga→gtg | − | 8 | |
| cga→gtt | − | 8 | |
| R275T | cga→acc | − | 2 |
aThe full list of sequenced TEM-1 variants are shown in Supplementary Table 1.
bNumbering according to standard system (25).
cFold change compared to MIC observed for cells producing wt TEM-1 encoded in the pNOM plasmid (CAZ MIC 0.3 μg/ml, CLV MIC 4 μg/ml).
Characterization of TEM-1 variants with improved properties
Analysis of the TEM-1 variants with enhanced activity towards CAZ and increased resistance to CLV revealed a range of substitution mutations at different positions instilling various degrees of improvement over wild-type TEM-1 (Table 2 and Supplementary Table 1). Many of the best performing TEM-1 variants had amino acid substitutions only accessible by at least two nucleotide changes per codon. These included the charge-switch mutant R164D with a 32-fold improvement in activity towards CAZ and the semi-conservative aromatic substitution of tryptophan to tyrosine (W165Y), which increased CLV resistance 3-fold (Table 2). The R275V mutation common to variants with the most improved resistance to CLV also required at least two adjacent nucleotide changes to access the amino acid substitution (Table 2). The importance of residue 164 to improving resistance to CAZ (10) was also highlighted here as 30 of the analysed variants were mutated at this residue, with six different substitutions observed (Supplementary Table 1).
Beneficial effects of two adjacent amino acid mutations were also observed. For example, there was up to a 4-fold improvement in the CAZ MIC values for variants containing the W165G mutation when accompanied by R164P (Table 2 and Supplementary Table 1). The R164P mutation was only observed in combination with a substitution at W165. This highlights the advantages of sampling such adjacent mutations. In other cases, the adjacent mutation had little effect. For example, the CAZ MIC value for variants containing the P167H mutation was similar to those that contained the accompanying E168K substitution (Table 2). The use of different codons to specify mutations also appeared to influence the in vivo activities of TEM-1 in some cases (Table 2 and Supplementary Table 1). For example, the R164P-W165R variants were encoded by five different gene sequences but their CAZ MIC values varied 4-fold. It has been noted previously that silent mutations to TEM-1 can also result in such differences (13,14).
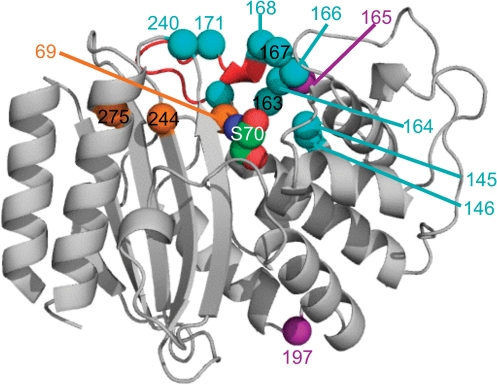
The majority of beneficial mutations occurred close to the active site (Figure 5), of which several were to residues that had previously been identified as important in enhancing activity to CAZ (e.g. R164S and R164H) and resistance to CLV (e.g. M69I) [see (10) and www.lahey.org/Studies/ and references therein]. Beneficial mutations included a run of eight substitutions (six consecutively between 163 and 168) in the Ω-loop (Figure 5). This unexpectedly included three independent variants with enhanced activity to CAZ that had the catalytically important E166 mutated. One mutation, E197P, was relatively distance from the active site (Figure 5) but slightly enhanced activity towards CAZ and resistance to CLV (Table 2). This highlights the power of directed evolution to sample mutations not predicted to be beneficial by structural analysis, and the ability of TriNEx to expand mutational sampling (E→P requires substitution of at least two adjacent nucleotides).
Figure 5.
Position of mutations with respect to the tertiary structure of TEM-1 [PDB code 1BTL (18)]. Mutations that enhance activity to CAZ or CLV or both are shown as cyan, orange or purple balls, respectively. The active site serine (S70) is shown as space-fill and labelled. The Ω-loop is coloured red. The residues are numbered using the recommended numbering system (25).
DISCUSSION
TriNEx as a method for generating molecular diversity
Directed evolution has been proven as an effective strategy for engineering proteins (2–6). Critical to directed evolution is the ability to generate and sample molecular diversity. While whole-gene mutagenesis methods such as error-prone PCR (15) are relatively simple to perform and can introduce mutations throughout a given target DNA sequence, they can only sample a restricted mutational range (with inherent biases) and generate large libraries of which only a fraction can ever be sampled (8). Site-saturated mutagenesis using oligonucleotide-encoded molecular diversity overcomes the limited mutational range of whole-gene mutagenesis but is restricted to sampling just a few residue positions in a protein so requires a detailed knowledge of the structure–function relationship to predict which residues to target.
TriNEx attempts to overcome these problems by providing a useful alternative that combines the benefits of both approaches. The random insertion and deletion (RID) method (16) was also developed to overcome similar problems but this method is difficult to implement (including the requirement of single strand DNA) and uses PCR. Furthermore, many variants contained secondary mutations and frameshifts that reduce the quality of the library. TriNEx does not use PCR at any stage during library construction and utilizes simple DNA manipulation and microbiological methods familiar to most molecular biologists. Only two frameshifts were predicted and no secondary point mutations or trinucleotide deletions were observed in the sequenced clones produced by TriNEx (Tables 1 and 2, Supplementary Table 1).
TriNEx has been successfully demonstrated using the bla gene encoding TEM-1 β-lactamase as our test system. The observed mutations were distributed throughout bla, with the majority of the variants containing 2- or 3-bp changes with respect to wild-type (Figure 3 and Tables 1 and 2). Many of the resulting amino acid substitutions introduced into TEM-1 could only be accessed by more than 1 bp change to a codon. Therefore, TriNEx can expand the molecular diversity sampled by whole-gene mutagenesis procedures to introduce novel amino acid substitutions that may enhance the properties of a protein.
The first transposon step ultimately determines TriNEx library diversity. MuDel is based on the mini-Mu transposon, which has a reported 5-bp target site for insertion based on the consensus sequence 5′ N-Py-G/C-Pu-N 3′ (17). Such a sequence bias may result in hotspots for transposon insertion and therefore introduce a bias in the library. While some duplication of the trinucleotide sequences deleted in separate variants derived from the BLAΔ175 library was observed (Table 1), the general redundancy of transposon insertion positions was estimated to be 10–15%. This agrees with previously observed redundancy but in a different mutational context (11). Restriction analysis of the BLAΔ libraries reported both here (Figure 4) and previously (11) confirm the random nature of MuDel insertion position. Therefore, the diversity of insertion positions sampled by MuDel allows mutations to be distributed throughout bla. A diverse range of replacement NNN encoded as part of SubSeqNNN was also observed (Table 1).
Insertion of MuDel outside of the target gene within the plasmid backbone may also be an issue. This was overcome here by selection for clones resistant to Cam but sensitive to Amp due to the knockout of bla by MuDel insertion. Using linear DNA comprised solely of the target gene can overcome this problem. It has been demonstrated previously that linear DNA acts as an efficient substrate for transposon insertion (17). While recloning the target gene back into pNOM after transposition may reduce the efficiency of the first step in the procedure, the subsequent selection for Cam resistance will result in theoretically 100% of clones containing MuDel inserted within the target gene. This is particularly important when applying TriNEx to proteins in which there is no simple selection or screen available. We have recently demonstrated this strategy, along with other approaches, in our laboratory to implement TriNEx for non-antibiotic resistance proteins (data not shown).
The number of variants that can be generated at each stage is also critical to the ability of TriNEx to sample a suitable range of diversity. The generation of 1644 variants with MuDel inserted within the 858 bp bla gene was deemed more than sufficient even when taking into account redundancy in transposon insertion. Blunt-end ligations akin to that necessary for the insertion of SubSeqNNN can be less efficient than those performed with compatible overhangs, which may potentially reduce the number of subsequent variants sampled. The blunt-end ligation of SubSeqNNN into bla routinely generated 4 × 105 Kan resistant colonies per microgram of DNA after transformation, which was considered high enough to sample the potentially 54 912 different potential genetic variations of bla. The final recircularization ligation to reform the trinucleotide replacement bla gene variants was extremely efficient, routinely producing ca 107 c.f.u./μg DNA. Therefore, TriNEx can generate a suitable number of variants at each stage to sample an extensive range of molecular diversity for a single and, if required, multiple iterative rounds where the target gene may be comprised of several different variants selected from the previous round of TriNEx.
As TriNEx can span two codons in theoretically 2/3 of cases, this can and does lead to substitution mutations in two adjacent amino acid residues (Table 1). Of the 49 sequenced variants derived from BLAΔ175, 30 were known to contain trinucleotide exchanges that spanned two codons (Table 1), yet only 14 encoded a double amino acid substitution. Due to the degeneracy in the genetic code many of the mutations were silent at the protein level. The result is that 66% of the variants contained a single amino acid substitution. Furthermore, sampling such double substitutions may not be consider detrimental but may enhance diversity. For example, a 4-fold increase in TEM-1 activity towards CAZ was observed when W165G was combined with R164P (Table 2).
Mutations introduced by TriNEx alter the activity of TEM-1
TriNEx introduced a wide variety of amino acid substitutions into TEM-1 with varying effects on the enzyme's in vivo activity (Tables 1 and 2). Many of the TEM-1 variants derived from BLAΔ175 retained a high degree of activity, with eight (including the two with wild-type amino acid sequence) of the 34 active variants conferring an Amp MIC value similar to that of wild-type. Interestingly, these included mutations to residues generally conserved in class A β-lactamases (10) or not consider tolerant to a substitution (12) (e.g. P107, A125, G156, M272 and I282). As 69% of the tested variants derived from BLAΔ175 library retained even a small degree of β-lactamase activity towards Amp, this confirms previous work (12) that suggests TEM-1 is largely tolerant to amino acid substitutions.
TriNEx has also been successfully employed to improve the activity of TEM-1 towards the third generation cephalosporin CAZ and increase resistance to the inhibitor CLV. Most of these mutations lay within or close to regions involved in the catalytic process and substrate binding (Figure 5). Many of the beneficial mutations were observed in the Ω-loop (residues 163–178 (18)). This included substitution of the catalytically important E166 (to glycine or lysine) observed in three independent clones that improved TEM-1 activity towards CAZ by up to 8-fold (Table 2 and Supplementary Table 1). A variety of substitutions to R164 were observed in many of the variants with improved activity towards CAZ, including those with highest observed activity (Table 2 and Supplementary Table 1). Mutations to R164 have been demonstrated previously to improve activity towards CAZ by removing a salt bridge that constrains conformation of the Ω-loop (10,19). To our knowledge, some of the specific mutations to R164 identified in this study (to asparagine, aspartate and proline) have not been observed previously. Mutations outside the Ω-loop also contribute towards enhanced activity towards CAZ. These include some novel mutations such as P145R and P145R-K146E that lie close to both the catalytic centre and the Ω-loop (Figure 5).
TriNEx also introduced mutations to residues known to contribute to improved resistance to CLV (M69, W165, R244 and R275) (10). Mutations at these positions are thought to alter substrate binding, including for suicide substrates such as CLV. Mutations to R275 produced the largest improvements, with the R275V substitution increasing resistance by 8-fold (Table 2). Natural variants of TEM-1 resistant to CLV contain either a leucine or glutamine at residue 275 and are found in combination with other beneficial mutations [(10) and www.lahey.org/Studies/]. The ability of TriNEx to expand the mutational range to incorporate R275V may explain why this mutation observed in our study had such a drastic effect alone.
One TEM-1 variant containing the E197P mutation displayed a slightly enhanced activity towards CAZ and resistance to CLV. Its potential mode of action is unclear as it is distant from the active site and to our knowledge has not been observed before. The fact this particular substitution required the exchange of 2 bp of the E197 codon may explain why it has not been observed in natural or engineered variants of TEM-1. However, the relatively small enhancements may not have been sufficient to overcome previous selection pressures.
SUMMARY
We have successfully demonstrated that TriNEx can generate a diverse range of proteins with altered properties. Although the exchange of contiguous trinucleotide sequences has been demonstrated here, the method has the potential to allow the exact replacement of a codon with a defined sequence. This will be useful for implementing scanning mutagenesis or extending directed evolution to allow incorporation of artificial amino acids (e.g. encoded by the amber stop codon) in vivo at random positions within a protein (20). The basic method also provides a novel approach for creating integral fusion proteins that have the potential to act as novel molecular switches by replacing SubSeqNNN with a DNA cassette encoding a protein domain (21,22). These applications are currently being explored.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC). (BB/E007384 and BB/E001084) and the Welsh Assembly Government (HE07POC3007). The Open Access publication charges were waived.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brannigan JA, Wilkinson AJ. Protein engineering 20 years on. Nat. Rev. Mol. Cell. Biol. 2002;3:964–970. doi: 10.1038/nrm975. [DOI] [PubMed] [Google Scholar]
- 2.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 3.Dalby PA. Optimising enzyme function by directed evolution. Curr. Opin. Struct. Biol. 2003;13:500–505. doi: 10.1016/s0959-440x(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 4.Yuen CM, Liu DR. Dissecting protein structure and function using directed evolution. Nat. Methods. 2007;4:995–997. doi: 10.1038/nmeth1207-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005;15:447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Tao H, Cornish VW. Milestones in directed enzyme evolution. Curr. Opin. Chem. Biol. 2002;6:858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- 7.Lutz S, Patrick WM. Novel methods for directed evolution of enzymes: quality, not quantity. Curr. Opin. Biotechnol. 2004;15:291–297. doi: 10.1016/j.copbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Neylon C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res. 2004;32:1448–1459. doi: 10.1093/nar/gkh315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DD. Triplet nucleotide removal at random positions in a target gene: the tolerance of TEM-1 β-lactamase to an amino acid deletion. Nucleic Acids Res. 2005;33:e80. doi: 10.1093/nar/gni077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matagne A, Lamotte-Brasseur J, Frere JM. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem. J. 1998;330(Pt 2):581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simm AM, Baldwin AJ, Busse K, Jones DD. Investigating protein structural plasticity by surveying the consequence of an amino acid deletion from TEM-1 β-lactamase. FEBS Lett. 2007;581:3904–3908. doi: 10.1016/j.febslet.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T. Amino acid sequence determinants of β-lactamase structure and activity. J. Mol. Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 13.Palzkill T, Thomson KS, Sanders CC, Moland ES, Huang W, Milligan TW. New variant of TEM-10 β-lactamase gene produced by a clinical isolate of proteus mirabilis. Antimicrob. Agents Chemother. 1995;39:1199–1200. doi: 10.1128/aac.39.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen BA, Bradford PA, Quinn JP, Wiener J, Weinstein RA, Bush K. Genetically diverse ceftazidime-resistant isolates from a single center: biochemical and genetic characterization of TEM-10 β-lactamases encoded by different nucleotide sequences. Antimicrob. Agents Chemother. 1993;37:1989–1992. doi: 10.1128/aac.37.9.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadwell RC, Joyce GF. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 16.Murakami H, Hohsaka T, Sisido M. Random insertion and deletion of arbitrary number of bases for codon-based random mutation of DNAs. Nat. Biotechnol. 2002;20:76–81. doi: 10.1038/nbt0102-76. [DOI] [PubMed] [Google Scholar]
- 17.Haapa S, Taira S, Heikkinen E, Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jelsch C, Mourey L, Masson JM, Samama JP. Crystal structure of Escherichia coli TEM1 β-lactamase at 1.8 A resolution. Proteins. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 19.Petrosino J, Cantu C., III, Palzkill T. β-Lactamases: protein evolution in real time. Trends Microbiol. 1998;6:323–327. doi: 10.1016/s0966-842x(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Xie J, Schultz PG. Expanding the genetic code. Ann. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 21.Ferraz RM, Vera A, Aris A, Villaverde A. Insertional protein engineering for analytical molecular sensing. Microb. Cell Fact. 2006;5:15. doi: 10.1186/1475-2859-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng. Des. Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 23.Craig NL. Target site selection in transposition. Annu. Rev. Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 24.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu. Rev. Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 25.Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A β-lactamases. Biochem. J. 1991;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.