Abstract
Loss of the maintenance of genetic material is a critical step leading to tumorigenesis. It was reported that overexpression of Aurora-A and the constitutive activation of the epidermal growth factor (EGF) receptor (EGFR) are implicated in chromosome instability. In this study, we examined that when cells treated with EGF result in centrosome amplification and microtubule disorder, which are critical for genetic instability. Interestingly, the expression of Aurora-A was also increased by EGF stimulus. An immunofluorescence assay indicated that EGF can induce the nuclear translocation of EGFR. Chromatin immunoprecipitation (ChIP) and re-ChIP assays showed significant EGF-induced recruitment of nuclear EGFR and signal transducer and activator of transcription 5 (STAT5) to the Aurora-A promoter. A co-immunoprecipitation assay further demonstrated that EGF induces nuclear interaction between EGFR and STAT5. A small interfering (si)RNA knockdown assay also showed that EGFR and STAT5 are indeed involved in EGF-increased Aurora-A gene expression. Altogether, this study proposes that the nuclear EGFR associates with STAT5 to bind and increase Aurora-A gene expression, which ultimately may lead to chromosome instability and tumorigenesis. The results also provide a novel linkage between the EGFR signaling pathway and overexpression of Aurora-A in tumorigenesis and chromosome instability.
INTRODUCTION
Genetic instability is a major event in tumorigenesis. Proteins involved in the cell-cycle checkpoint mechanism or controlling chromosome replication and separation during cell division are believed to be important for maintaining genome integrity and fidelity. Among them, the Aurora kinase family is critical for various events in mitosis and/or meiosis. They play important roles in cell division, including the control of centrosome and spindle function, involvement of kinetochore–microtubule interactions and cytokinesis. Three family members of Aurora kinases, Aurora-A, -B and -C were discovered in mammals. Human Aurora-A is a centrosomal-associated serine/threonine kinase, which is involved in cell-cycle progression, cell survival and malignant transformation. Aurora-A is located on chromosome 20q13.2, a region commonly amplified in malignancies, such as melanomas and cancers of the breast, colon, pancreas, ovary, bladder, liver and stomach. It was reported that Aurora-A is overexpressed in many cancer cells (1–3), suggesting that Aurora-A is involved in tumorigenesis. In proliferating cancer cell lines, the expression of Aurora-A, including messenger RNA (mRNA), protein levels and kinase activities, is under cell-cycle control. Interest in Aurora has intensified since the discovery that transfection of Aurora-A into rodent Rat1 and NIH3T3 fibroblast cell lines is sufficient to induce colony formation in culture and tumors in nude mice (1,4), thus establishing Aurora-A as a bona fide oncogene (4–6). Dysregulation of Aurora kinases has been linked to tumorigenesis. Therefore, the control of Aurora-A expression and activation is an important event for normal cell-cycle progression (7). Previous studies indicated that the increased expression of Aurora-A in cancers occurs through gene amplification, RNA transcriptional upregulation, or protein stabilization (4). Among them, many studies focused on the regulation of protein stability. In addition, several lines of evidence have shown that the E4TF1/hGABP (GA-binding protein) transcription factor plays an important role in the transcriptional regulation of Aurora-A in a cell-cycle-dependent manner (8,9), and the TRAP220/MED1 (thyroid hormone receptor-associated protein complex component/methyl-CpG binding endonuclease) directly interacts with GABP to regulate Aurora-A gene expression in HeLa cells (10). Furthermore, the DUSP6/MKP-3 (dual specificity phosphatase 6/MAPK phosphatase-3, a candidate tumor suppressor gene and a specific phosphatase for MAPK1), can downregulate Aurora-A gene expression in pancreatic cancer (11). But until now, the detailed transcriptional regulatory mechanism of Aurora-A in cancer cells remains largely uncertain.
The epidermal growth factor receptor (EGFR) is a transmembrane glycoprotein containing tyrosine kinase activity. Upon growth factor stimulation, EGFR activates and transfers extracellular signals into cytoplasmic molecules such as mitogen-activated protein kinase (MAPK), phospholipase C-γ (PLCγ) and phosphatidylinositol-3-OH (PI-3) kinase and regulates target gene expressions (12–16). Recently, many studies have shown that the nuclear localization of the EGFR is strongly correlated with highly proliferating tissues (15,17–24). The nuclear EGFR can recognize AT-rich sequence sites (ATRSs) of target gene promoters and activate gene expression. Therefore the function of the nuclear EGFR is that of a transcriptional activator which regulates gene expression required for cell proliferation (21,23), for example cyclin D1 (21), inducible nitric oxide synthase (23) and cyclooxygenase-2 (25). Interestingly, the EGFR lacks a DNA-binding domain (23), and the nuclear EGFR physically interacts with other transcriptional molecules, such as the signal transducer and activator of transcription 3 (STAT3) (23) and E2F1 (26), to activate gene expression. Most importantly, it was reported that the overexpression and constitutive activation of EGFR in cancer cells may be associated with chromosomal instability (27,28), a phenotype just like the overexpressed Aurora-A in cancers (29).
In this study, we demonstrate that EGF stimulation results in centrosome amplification and microtubule disorder as well as the induction of Aurora-A overexpression. The ligand-activated EGFR is translocated into the nucleus and then targets the ATRSs of the Aurora-A promoter through interacting with STAT5. This is the first report to correlate the overexpression of Aurora-A and the nuclear EGFR in malignant cancer cells, and hence may provide a target for cancer therapy.
MATERIALS AND METHODS
Cell culture and drug treatment
A431, 293T, MCF7, MDA-MB-231 and MDA-MB-468 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, GIBCO, Invitrogen, Carlsbad, CA), LS174T cells were cultured in Minimum Essential Medium Alpha medium (MEM-α medium, GIBCO). CHO cells were cultured in F12 medium (GIBCO). Culture media were supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin and 100 U/ml penicillin. In this series of experiments, cells were treated with 60 ng/ml (10 nM) EGF in optimal serum-free conditions.
siRNA transfection and STAT5 overexpression
siRNA oligos for knockdown of endogenous STAT5 proteins were prepared by using the siRNA SMARTpool from Dharmacon (M-005169-02-0005 for STAT5A and M-010539-01-0005 for STAT5B, Dharmacon RNA Technologies, Lafayette, CO), and Ambion (catalog number: 138787 for STAT5A, Ambion, The RNA Company, Austin, TX). EGFR siRNA oligos were purchased from QIAGEN (Hs_EGFR_12_HP Validated siRNA). STAT5A and STAT5B cDNA were cloned from HeLa complementary (c)DNA by a PCR (STAT5A forward primer: 5′-CATAAGCTTATGGCGGGCTGGATCCAGGCC-3′ and reverse primer: 5′-CAACTCGAGTGAGAGGGAGCCTCTGGCAGA-3′; and STAT5B forward primer: 5′-CGGAAGCTTATGGCTGTGTGGATACAAGCT-3′ and reverse primer: 5′-GTTCTCGAGCGATTGTGCTGTCGGGAT-3′), and constructed into a pcDNA3.1™/myc-His expression vector (Invitrogen, Carlsbad, CA) using the HindIII and XhoI restriction enzyme sites. The siRNA and STAT5 expression vectors were transfected into cells using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions.
Quantitative reverse-transcription polymerase chain reaction (RT-PCR) and RT-PCR
Total RNA from cultured cells was prepared from TriZole reagent (Invitrogen) as described by the manufacturer's instructions. For the RT-PCR, 3 μg of total RNA was used to generate cDNA by reverse transcriptase III (Invitrogen) followed by the PCR (Aurora-A forward: 5′-ATGGACCGATCTAAAGAAAAC-3′ and reverse: 5′-CGATTCCTAAGACTGTTTGC-3′; EGFR forward: 5′-CATAGTCAGCAGTGACTTTCTC-3′ and reverse: 5′-GTTACACACTTTGCGGCAAGG-3′; and GAPDH forward: 5′-CCATCACCATCTTCCAGGAG-3′ and reverse: 5′-CCTGCTTCACCACCTTCTTG-3′). For the quantitative real-time RT-PCR, cDNA synthesis was performed using the TITANIUM One-Step RT-PCR kit (Clontech, Palo Alto, CA) containing SYBR Green I (BioWhittaker Molecular Applications; BMA, Rockland, ME). In brief, first-strand cDNA was synthesized at 50°C for 60 min, followed by a 10-min denaturation at 95°C. PCRs were then perfomed in the same tubes using the following conditions for 35 cycles: 95°C for 15 s, 60°C for 15 s and 68°C for 20 s. The sequences of primers used for RT-PCR were as follows: human Aurora-A forward: 5′-AATGCCCTGTCTTACTGTCATTC-3′ and reverse: 5′-TCCAGAGATCCACCTTCTCATC-3′; and human RPL13A forward: 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′ and reverse: 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′. Real-time fluorescence monitoring and the melting curve analysis were performed with LightCycler according to the manufacturer's recommendations (Roche Molecular Diagnostics, Lewes, East Sussex, UK). Negative controls containing no RNA template were included in each experiment. A melting curve was created at the end of the PCR cycle to confirm that a single product had been amplified. Data were analyzed by LightCycler software version 3.5 (Roche Molecular Diagnostics) to determine the threshold cycle (Cp) above the background for each reaction. The relative transcript amount of the target gene, calculated using standard curves of serial RNA dilutions, was normalized to that of RPL13A of the same RNA.
Reporter assay
Cells were transfected with wild type or mutated Aurora-A promoter (–968 ∼ +124) luciferase construct (pGL2-Aurora-A promoter) by LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions with a slight modification. Cells are replated 24 h before transfection at an optimal cell density in 2 ml of fresh culture medium in a 6-well plate. As a transfection efficiency control, the Renilla luciferase reporter plasmid, phRG-TK, was co-transfected into cells. Twenty-four hours following transfection, cells were serum-starved for 24 h, stimulated with 60 ng/ml (10 nM) EGF under serum-free condition for various times, harvested and subjected to the luciferase assay using the dual-luciferase reporter assay kit (Promega, Madison, WI). Normalization with the Renilla luciferase activity, mean luciferase activities, and standard deviations were derived from three independent experiments. For investigating the effect of STAT5A and EGFR, the Myc-STAT5A or/and the EGFR expression vector—pCO11-EGFR was/were co-transfected with pGL2-Aurora-A promoter.
Preparation of cell lysates and cell fractions
For total cell lysates, cells from 10-cm plastic dishes were washed twice with PBS and lysed with RIPA buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA and 2 mM EGTA] containing 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin and 10 μg/ml aprotinin. The lysates were centrifuged at 12 000 × g for 5 min. The supernatants were collected and stored at –70°C until used.
For nuclear extracts, cells were washed twice with PBS and scraped in 500 μl of PBS. Cells were collected by centrifuging at 7500 × g for 30 s, resuspended in 400 μl of buffer A [10 mM Hepes (pH 7.9), 1.5 mM MgCl2 and 10 mM KCl] and placed on ice for 10 min. Nuclei were pelleted by centrifugation at 7500 × g for 30 s, and the resulting supernatant formed the cytosolic fraction. The nuclear pellets were resuspended in 100 μl of buffer C [20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 0.2 mM EDTA, 420 mM NaCl and 25% (v/v) glycerol] and placed on ice for 20 min. The suspension was centrifuged at 7500 × g for 2 min, and the resulting supernatant was termed the nuclear fraction. The cytosolic and nuclear fractions were stored at –70°C until used. Buffers A and C contained 0.5 mM dithiothreitol, 2 μg/ml leupeptin, 1 mM orthovanadate, 2 μg/ml pepstatin A and 0.5 mM phenylmethylsulfonyl fluoride.
Co-immunoprecipitation assay and immunoblotting analysis
For the co-immunoprecipitation assay, nuclear extracts were purified from EGF-treated A431 cells, and then incubated with anti-EGFR antibodies or normal rabbit immunoglobulin G (IgG) in an immunoprecipitation buffer [20 mM Hepes (pH 7.9), 120 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM orthovanadate, 2 μg/ml pepstatin A and 2 μg/ml leupeptin] under gentle shaking at 4°C overnight, and then Protein A agarose beads were added and incubated for a further 2 h. Beads were washed three times with immunoprecipitation buffer and two times with PBS. The immunoprecipitation complexes were resolved by SDS–PAGE and subjected to immunoblotting analysis by anti-EGFR polyclonal antibodies (pAbs) or anti-STAT5 pAbs.
Indirect immunofluorescence assay and confocal microscopy
The immunofluorescence assay was performed as previously described (30). Briefly, cells were grown on coverslips and fixed with 3.7% formaldehyde at room temperature for 10 min. The fixed cells were then probed with anti-Aurora-A pAbs (ab1287, Abcam, Cambridge, MA) and an anti-γ-tubulin mAb (GTU88, Sigma, St Louis, MO), or anti-α-tubulin mAb (mAb) (DM1A-FITC, Sigma). DNA was stained with DAPI (Sigma). After the first antibody incubation, these immunoactive probes were detected with Alexa 568 (Molecular Probes, Cincinnati, OH), a Texas Red-conjugated goat anti-mouse IgG, and Alexa 488 (Molecular Probes), and FITC-conjugated goat anti-rabbit IgG. Coverslips were mounted and observed with a laser scanning confocal system (FV1000, Olympus, PA, USA).
Chromatin immunoprecipitation (ChIP) and sequential ChIP assay
Cells were crosslinked with 1% formaldehyde at 37°C for 15 min, washed twice with PBS, lysed with L1 buffer [50 mM Tris–HCl (pH 8.0), 2 mM EDTA, 0.1% NP-40 and 10% glycerol], and then resuspended with L2 buffer [50 mM Tris–HCl (pH 8.0), 5 mM EDTA and 1% SDS]. The lysates were sonicated to shear the size of the DNA to around 500 bp. Sonicated extracts were diluted 10-fold with a dilution buffer [50 mM Tris–HCl (pH 8.0), 0.5 mM EDTA, 0.5% NP-40 and 0.2 M NaCl]. After pre-cleaning with salmon sperm DNA/BSA-saturated protein A Sepharose, 200 μg of the extracts was used for immunoprecipitation assay. The immunoprecipitated complexes were pelleted and washed with high-salt and low-salt buffers three times each. For ChIP assay, the DNA–protein complex was then eluted in an elution buffer (1X TE buffer containing 1% SDS) with rotation at room temperature for 15 min, and the immune complex crosslinking was reversed by heating at 65°C overnight, followed by treatment with 100 μg/ml proteinase K at 50°C for 1 h. DNA was extracted twice with phenol/chloroform and precipitated with ethanol. The pellet was redissolved in H2O and subjected to PCR amplification using specific primers to the Aurora-A promoter, which were 5′-CTGTTGCTTCACCGATAAATGGC-3′ and 5′-CTCTAGCTAGAAAGCCGATTGGC-3′. For sequential ChIP assay, the DNA–protein complex was eluted from the protein A Sepharose beads by incubating with 10 mM DTT in 37°C for 30 min twice. After diluted 10-fold with dilution buffer, the eluted mixture was used to perform the second immunoprecipitation assay. L1, L2 and dilution buffer all contained 0.5 mM phenylmethylsulfonyl fluoride, 1 mM orthovanadate, 2 μg/ml pepstatin A, 2 μg/ml leupeptin, 5 mM sodium fluoride and 1 μg/ml aprotinin.
DNA affinity precipitation assay (DAPA)
The DNA affinity-binding assay was performed by mixing 200 μg of nuclear extract proteins, 2 μg of biotinylated Aurora-A nucleotides and 20 μl of streptavidin–agarose beads (4%) with a 50% slurry. The mixture was incubated at 4°C for 1 h with rotation. Beads were pelleted and washed with cold PBS containing 0.1% NP-40 three times. The binding proteins were eluted by SDS loading buffer and analyzed by Western blot analysis with specific antibodies.
RESULTS
The EGF induces centrosomes amplification and microtubule disorder in A431 cells
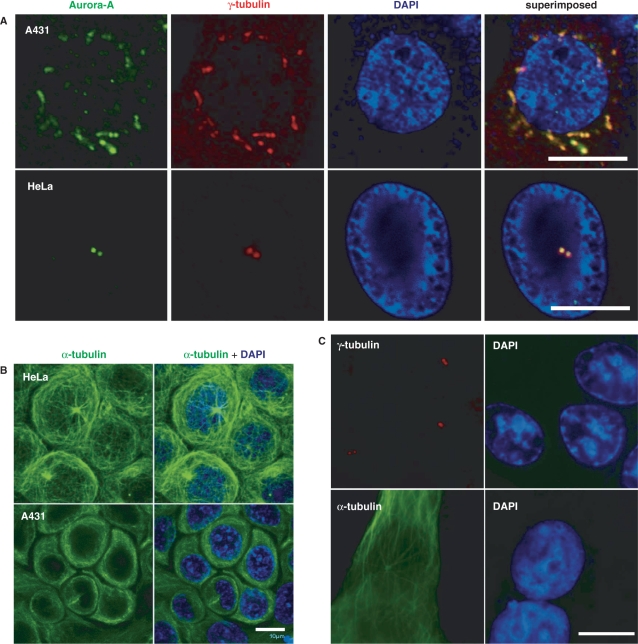
It is well recognized that centrosome abnormalities are one of the main reasons for chromosomal instability through the development of multipolar mitotic spindles (31). It was also reported that activation of the EGFR may result in genetic instability (27,28). These descriptions prompted us to investigate whether activation of the EGFR results in chromosomal instability through abnormal centrosomes. In order to examine this possibility, the biological effects of the EGF in EGFR-overexpressed A431 cells were evaluated by an immunofluorescence assay. Serum-deprived A431 cells were treated with the EGF for 3 or 24 h, and then re-cultured in growth medium for another 24 h to allow the cell cycle to progress. These EGF-treated A431 cells were doubly stained with γ-tubulin (Figure 1A, red) and Aurora-A (Figure 1A, green) antibodies, both of which are well-known centrosomal proteins. The data revealed that the EGF-induced centrosomes amplification in A431 cells (Figure 1A). The situation with the microtubule cytoskeleton was also examined using anti-α-tubulin antibody, and the results indicated that the microtubule network was disarranged (Figure 1B). Both the centrosome number and microtubule organization in A431 cells without EGF treatment were normal (Figure 1C). The quantitative results of EGF-induced centrosomes amplification are shown in Table 1. These results indicated that the EGF can induce centrosomes amplification and microtubule disorder in A431 cells. Besides, the result in Figure 1A also showed that Aurora-A protein is increased.
Figure 1.
Effect of EGF on genetic instability. (A, B) A431 cells were treated with the EGF (10 nM) for 24 h, and then the EGF was washed out, and cells were further cultured in normal culture medium for another 24 h to allow them to undergo one cell cycle. The immunofluorescence assay of cells was then performed using anti-Aurora-A antibodies (green), anti-γ-tubulin antibody (red) and DAPI (blue, a DNA-specific dye) (A), or an α-tubulin antibody (green) (B). HeLa cells were used as a staining control. Scale bars, 10 μm. (C) The centrosome number and microtubule organization are normal in EGF-untreated A431 cells. Serum-starvated A431 cells were further cultured in growth medium for 24 h, and then the immunoflourescence assay was performed by using anti-γ-tubulin antibody (red) and anti-α-tubulin antibody (green) for monitoring the centrosome number and microtubule organization status.
Table 1.
The degree of centrosomes amplification in counted cells
| A431 | Centrosomes amplification |
|---|---|
| +EGF 3h | 47.59% (35/74) moderate |
| +EGF 24h | 60.56% (43/71) excessive |
| −EGF 24h | 6.60% (7/106) |
Each static results were calculated in different field of a coverslip. Three independent experiments were performed to conform the results.
The EGF increases Aurora-A gene expression in EGFR-expressing cell lines
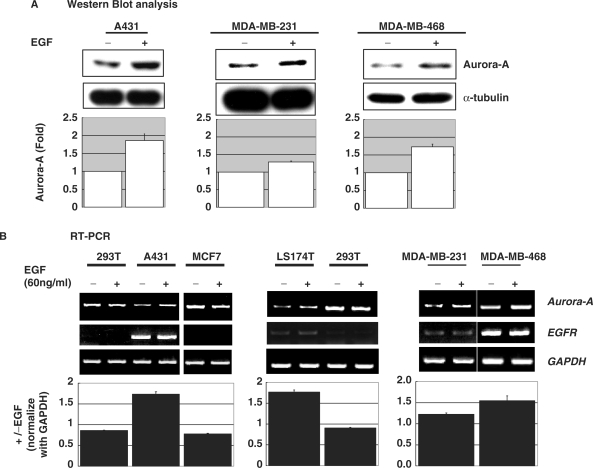
We then analyzed the effect of the EGF on Aurora-A protein expression in three cultured cell lines: A431, MDA-MB-231 and MDA-MB-468. The data revealed that after EGF stimulation, the expression of Aurora-A kinase increased (Figure 2A). Because it is well recognized that the EGF transduces its signaling through binding with the EGFR, expressions of the EGFR and Aurora-A were examined in six cultured cancer cell lines by a RT-PCR assay. The results indicated that the expression of Aurora-A mRNA is increased in four of these cell lines, which express the EGFR, under EGF treatment (Figure 2B). Data of the quantitative real-time RT-PCR further showed that Aurora-A mRNA was indeed elevated under EGF stimulation in A431 cells (Figure 2C). The promoter activity of the Aurora-A gene was also enhanced in A431 cells but not in 293T or MCF7 cells without EGFR expression under EGF stimulation (Supplementary Figure 1). Both the promoter activity and protein expression of Aurora-A were increased in a time-dependent (Figure 2D) manner with EGF treatment. These data demonstrate that the Aurora-A promoter is activated by EGF stimulation in EGFR-expressing cultured cell lines.
Figure 2.
EGF induces Aurora-A gene expression in cultured cells. Serum-deprived cells treated with (+) or without (–) the EGF (10 nM) were analyzed by (A) Western blot analysis, (B) RT-PCR and (C) real-time RT-PCR. The expression of Aurora-A was enhanced by EGF stimulation only in EGF receptor (EGFR)-expressing A431, LS174T, MDA-MB-231 and MDA-MB468 cells but not 293T and MCF7. The quantitative results of A and B are shown below. The time point for EGF treatment in A and B is 24 h. (D) The time-dependent activation of Aurora-A promoter was measured by a dual reporter assay system, and the Aurora-A protein expression levels at the indicated time points were analyzed by Western blot analysis. The quantitative result of the Western blot is shown at the side.
Activation of the EGFR is required for EGF-induced Aurora-A expression
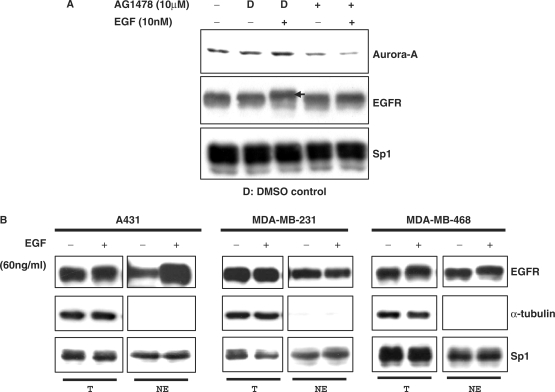
It is known that activation of the EGFR is highly correlated with malignant cancers, and the overexpression of the Aurora-A oncogene plays a pioneering role in the early stage of tumorigenesis (3). Until now, no studies have clarified the possible relationships between activation of the EGFR and overexpression of Aurora-A in cancer cells. Our data demonstrate that EGF treatment can increase Aurora-A expression in EGFR-expressing cell lines (Figure 2). To confirm this phenomenon and verify that activation of the EGFR does increase Aurora-A gene expression, A431 cells were treated with the EGF, and the activation status of the EGFR and the expression of Aurora-A were then analyzed. As shown in Figure 3A, the EGFR was activated (as indicated by the arrow) and expression of Aurora-A was also increased under EGF stimulation. When cells were pretreated with the EGFR inhibitor, AG1478, the expression of Aurora-A was no longer increased by EGF stimulation (Figure 3A). Altogether, these results indicate that EGF treatment can result in EGFR activation and increased expression of Aurora-A.
Figure 3.
Activation of the EGFR is required for EGF-induced Aurora-A expression. (A) A431 cells were pretreated with 10 μM AG1478 (an EGFR inhibitor) for 30 min prior to EGF stimulation. Total cell lysates were subjected to an immunoblot assay with the indicated antibodies. D, DMSO. Immunoblotting of the Sp1 transcription factor was used as a loading control. Arrow indicates the activated/phosphorylated EGFR, which presents a band shift. (B) Subcellular localization of the EGFR. Nuclear fractions from A431, MDA-MB-231 and MDA-MB-468 cells were purified, and then a Western blot analysis was performed using anti-EGFR antibodies, anti-α-tubulin antibody and anti-Sp1 antibodies. T, total cell lysate; NE, nuclear extract. Cells were treated with (+) or without (–) EGF (10 nM) for 30 min.
The EGFR may activate Aurora-A gene expression through a nuclear pathway
It was established that when the EGFR is activated, ligand–EGFR complexes undergo two fates: endocytosis/degradation or recycling to the cell membrane for subsequent activation (32). Endocytosis/degradation of the activated EGFR plays an essential role in its anti-tumor effects (33). Recently, it was reported that the EGFR can be activated and then translocated into the nucleus as a transcriptional activator (21). The nuclear translocation of the EGFR is highly correlated with malignancy (21). The EGFR carries out its biological effects via two signaling pathways: the cytoplasmic/traditional pathway and the nuclear pathway (34). Here, when cells were treated with the EGF, the EGFR still remained at a high level (Figure 3B, ‘T’ part) and existed in nucleus fractions (Figure 3B, ‘NE’ part) of A431, MDA-MB-231 and MDA-MB-468 cells. These results suggest that the ligand-activated EGFR did not undergo the endocytosis/degradation process, but went through the nuclear pathway. This implies that the activated EGFR mostly passes through the nuclear pathway to induce Aurora-A expression. Moreover, anti-phospho-EGFR antibodies (phospho-tyrosine 845) were used to further confirm the nuclear localization of the activated EGFR in these cells (data not shown). These results indicate that the signaling pathway involved in the EGF-induced Aurora-A gene expression in A431 cells (also in MDA-MB-231 and MDA-MB-468) may be the nuclear EGFR pathway.
The nuclear EGFR can target the Aurora-A promoter upon EGF stimulation
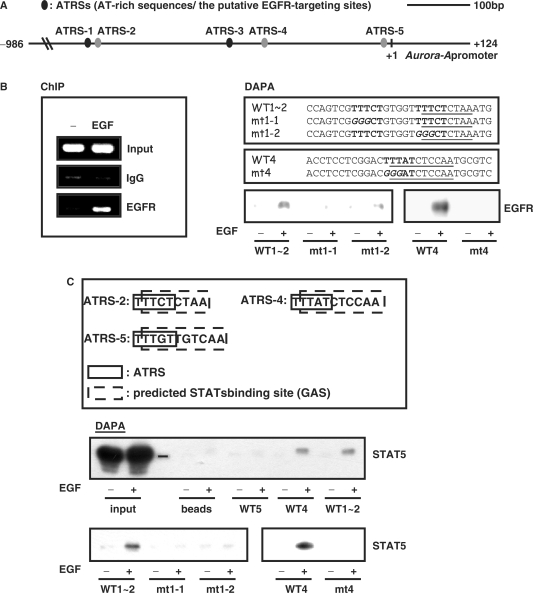
In order to verify the physiological role of the nuclear EGFR in the increased expression of Aurora-A, the promoter region of Aurora-A was analyzed to confirm the potential EGFR-targeting sites (AT-rich sequences, ATRSs): TNTTT or TTTNT (21). Sequence analysis revealed that there are five putative ATRSs on the proximal region of the Aurora-A promoter (between -576 and +124, ATRS-1 ∼ 5, Figure 4A). The distance between ATRS-1 and ATRS-2 is only five nucleotides. To directly verify whether the nuclear EGFR can bind to and activate the Aurora-A promoter, we performed an in vivo ChIP assay. The nuclear EGFR was recruited to the Aurora-A promoter by EGF stimulation (Figure 4B). Next, we identified the regulatory elements responsible for nuclear EGFR targeting in the Aurora-A promoter upon EGF stimulation by a DNA-affinity precipitation assay (DAPA). Four probes containing the ATRSs were designed to assay the binding of the nuclear EGFR, among which probe WT1∼2 contains two putative ATRS sites, ATRS-1 and ATRS-2. The results indicated that only WT1∼2 (ATRS-1 and ATRS-2) and WT4 (ATRS-4) could recruit the nuclear EGFR but not the WT3 (ATRS-3), WT5 (ATRS-5) (data not shown), ATRS-1∼2 mutants (mt1-1 and mt1-2) or ATRS-4 mutants (mt4) (Figure 4B). These data revealed that the nuclear EGFR may activate expression of the Aurora-A gene through binding with ATRS-1∼2 and ATRS-4.
Figure 4.
The nuclear EGFR/STAT5 complex is recruited to the Aurora-A promoter and regulates gene expression. (A) Five predicted EGFR-targeting sites (the ATRS-1 ∼ 5) are located in the proximal region of the Aurora-A promoter. This graph shows the wild-type Aurora-A promoter region from –986 to +124. The black oblongs mean the putative ATRSs, and the gray oblongs mean the ATRSs contain the consensus STAT protein-binding sequences. ATRS-1: –479 ∼ –475; ATRS-2: –469 ∼ –465; ATRS-3: –260 ∼ –256; ATRS-4: –206 ∼ –201; ATRS-5: –15 ∼ –11. (B) A431 cells were treated with (+) or without (–) EGF (10 nM) for 30 min, and then lysed and performed the in vivo ChIP assay by using anti-EGFR antibodies (left panel and Figure 4D, lane 5). The nuclear proteins were mixed with biotin-labeled oligonucleotides to perform the DNA-affinity precipitation assay (DAPA) (right panel). After incubation, the protein-bound probes were precipitated and subjected to immunoblot analysis with anti-EGFR antibodies. The nucleotides sequences of ATRS1-2 (WT1 ∼ 2) and ATRS-4 (WT4) and their mutants (mt1-1, mt1-2 and mt4, the italic letters are the mutated nucleotide sequences) are shown in the upper panel. (C) The consensus signal transducer and activator of transcription (STAT) protein-binding sequences (dashed-line box) within AT-rich sequences sites (ATRSs) (solid-line box) of the Aurora-A promoter are shown in the upper panel. Association of STAT5 with the Aurora-A promoter ATRSs was evaluated by DAPA as described in Figure 4B. (D) An in vivo ChIP assay demonstrated EGF-induced STAT5 recruitment (lanes 2 and 7). Cells were treated with (+) or without (–) EGF (10 nM) for 30 min, and the EGFR/STAT5 complex targeting the Aurora-A promoter was detected by the sequential ChIP assay (lane 8). Following the first immunoprecipitation with anti-EGFR antibodies, the EGFR–DNA complex was eluted and subjected to the second immunoprecipitation with anti-STAT5 antibodies. The quantitative result of IgG, STAT5 ChIP assay and sequential ChIP (lane 6 ∼ 8) were showed below. Lane 3 is an input control. (E) The transcriptional activity of Aurora-A promoter ATRS mutants. A431 cells were transfected with the indicated mutant constructs. After serum starvation, cells were treated with (+) or without (–) EGF (10 nM) for 18 h, and the promoter activity of Aurora-A was then measured by a dual reporter assay system. The upper panel shows the wild-type Aurora-A promoter region from –986 to +124. The black oblongs mean the putative ATRSs, the gray oblongs mean the ATRS contain the consensus STAT protein-binding sequences, and the white oblongs are the mutated ATRSs. IgG, normal rabbit IgG. *P < 0.05.
The nuclear EGFR cooperates with STAT5 and targets the Aurora-A promoter to regulate its expression
Because of a lack of a DNA-binding domain, the nuclear EGFR need to cooperate with specific transcription factors to target the gene promoter region (23). Analysis of the DNA sequences of the five proximal ATRSs of the Aurora-A promoter revealed that three of them contain the consensus of STAT protein-binding sites (ATRS-2, ATRS-4 and ATRS-5, Figure 4C). The DAPA analysis showed that STAT5 is associated with probe WT1∼2 (ATRS-1 and ATRS-2), and WT4 (ATRS-4) but not WT5 (ATRS-5), ATRS-1 and -2 mutants (mt1-1 and mt1-2), or the ATRS-4 mutant (mt4) (Figure 4C). The ChIP assay further demonstrated that STAT5 (Figure 4D) can be recruited to the Aurora-A promoter upon EGF stimulation. Although it was reported that STAT3 (23) or E2F1 (26) can interact with the EGFR in activating gene expression, the ChIP assay and DAPA analysis indicated that the two transcription factors are not involved in EGF-induced Aurora-A gene expression (data not shown). The association of the nuclear EGFR/STAT5 complex with the Aurora-A promoter was directly demonstrated by the sequential ChIP assay (Figure 4D). The reporter assay further revealed that ATRS-2 and ATRS-4 are important for EGF-induced Aurora-A expression (Figure 4E).
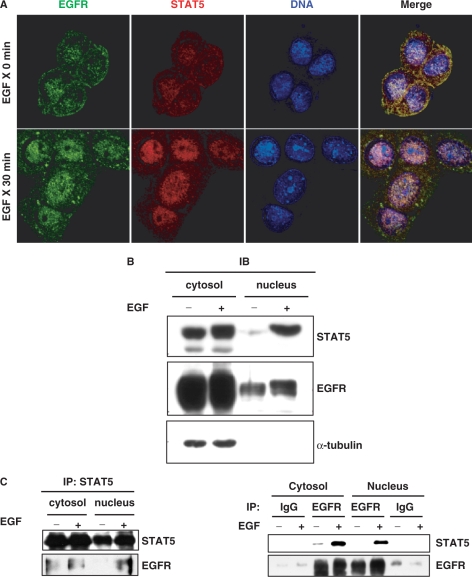
In order to verify the nuclear interaction of EGFR with STAT5, an immunofluorescence assay was performed. As shown in Figure 5A, the EGFR was translocated into the nucleus where it co-localized with STAT5 after EGF stimulation. Western blot analysis indicated the nuclear co-localization of EGFR and STAT5 in purified cell fractions (Figure 5B). The co-immunoprecipitation assay coupled with an immunoblot analysis also showed the nuclear interaction between EGFR and STAT5 (Figure 5C). These data strongly support the notion that the nuclear translocation of EGFR and STAT5 is significantly enhanced by EGF stimulation. It was also established that the EGFR can physically interact with STAT5 in the nucleus.
Figure 5.
Interaction between nuclear EGFR and STAT5. (A) To detect the subcellular localization of EGFR and STAT5, A431 cells were fixed and then doubly stained with anti-EGFR (green) and anti-STAT5 (red) antibodies. DAPI is a DNA-specific dye. Scale bars, 20 μm. (B, C) A431 cells were treated with (+) or without (–) EGF for 30 min, and then fractionated into nuclear and cytosolic fractions for the immunoblot (IB) analysis (B) and immunoprecipitation/Western blot assay (C) using the indicated antibodies.
The EGFR and STAT5 are important for EGF-induced Aurora-A gene expression
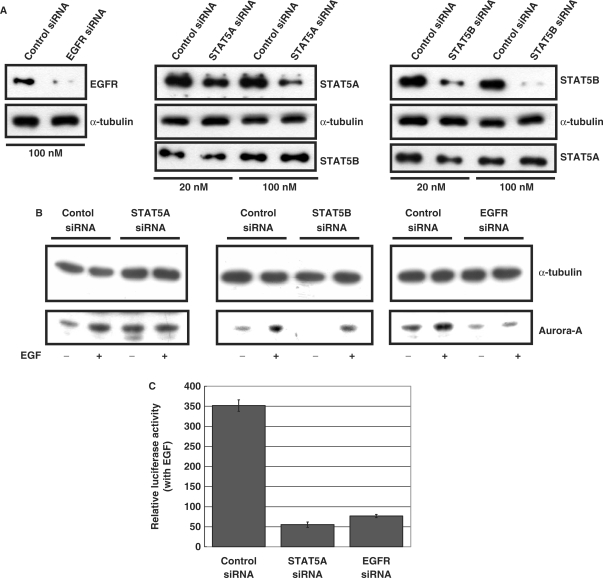
Since the nuclear EGFR can interact with STAT5 in the activation of Aurora-A gene expression, we next tried to verify the importance of EGFR and STAT5 in EGF-induced Aurora-A expression by an siRNA assay. The efficiency of the EGFR and STAT siRNA was first tested. Immunoblot analysis revealed that the EGFR siRNA possesses the ability to substantially attenuate expression of the EGFR, and the ability and specificity of STAT5A and STAT5B siRNA were also examined (Figure 6A). In addition, the control siRNA did not reduce expression of the EGFR, STAT5A or STAT5B (Figure 6A). As shown in Figure 6B, when the EGFR or STAT5A proteins were reduced, the effect of EGF-induced Aurora-A expression dropped, whereas STAT5B siRNA did not influence the effect of EGF-induced Aurora-A expression (Figure 6B). The reporter assay also demonstrated that EGF-induced Aurora-A promoter activation was reduced by EGFR siRNA and STAT5A siRNA (Figure 6C). When Myc-STAT5A was ectopically expressed, the induced expression of Aurora-A by EGF was enhanced, and also Aurora-A promoter activity was increased by EGF stimulation (Figure 6D). To further demonstrate the specific role of STAT5A, but not STAT5B, in the induction expression of Aurora-A under EGF stimulus, a second STAT5A siRNA species was used to rule out the possibility of siRNA off targeted effect, and a rescue experiment was also performed. The result showed that the expression of a siRNA-insensitive Myc-STAT5A can restore the decreased activity of Aurora-A promoter in the STAT5A knockdown cells under EGF treatment (Figure 6E). In order to further confirm the cooperative effect of EGFR and STAT5A in induction the expression of Aurora-A, EGFR and STAT5A were co-transfected into a no EGFR expressed CHO (Chinese hamster ovary) cells (23). The data indicated that ectopic expression of STAT5A or EGFR, but not the STAT5B, can increase the promoter activity of Aurora-A. Moreover, co-expression of STAT5A and EGFR has a synergic effect in increase Aurora-A promoter activity, whereas co-expression of STAT5B and EGFR has not (Figure 6F). Most importantly, the recruitment of nuclear EGFR to Aurora-A promoter was abolished in the STAT5A knockdown cells (Figure 6G, lane 5), but not in the control and STAT5B knockdown cells (Figure 6G, lanes 4 and 6). Therefore, these results suggest that the EGFR and STAT5A play critical roles in EGF-induced Aurora-A gene expression.
Figure 6.
The EGFR and STAT5A play important roles in EGF-induced Aurora-A expression. Protein expression levels (A, B) of cells transfected with the indicated small interfering (si)RNA. The effects of EGF on the protein expression level (B) and promoter activity (C) of Aurora-A in the EGFR, STAT5A or 5B knockdown cells were examined. (D) A431 cells were transfected with a vector only or Myc-STAT5A. Forty-eight hours after transfection, cells were harvested, and the effects of EGF on protein expression (left) and Aurora-A promoter activity (right) were analyzed. **P < 0.01. (E) The protein expression levels of cells transfected with the second STAT5A siRNA species were examined (left panel). A431 cells were transfected with a second species of STAT5A siRNA, a si-RNA-insensitive Myc-STAT5A or both (right panel). The protein expression level (upper) and the effects of EGF on Aurora-A promoter activity (lower) were analyzed. **P < 0.01. (F) EGFR, Myc-STAT5A or Myc-STAT5B was transfected into CHO cells, and the effect of EGF on Aurora-A promoter activity (left panel) was then analyzed as described above. The protein expression levels of cells overexpressing EGFR, Myc-STAT5A or Myc-STAT5B were examined (right panel). (G) The in vivo ChIP assay demonstrated that nuclear EGFR is recruited to the promoter region of Aurora-A only in control and STAT5B knockdown cells (lanes 4 and 6) but not in the STAT5A knockdown cells (lane 5). Cells were treated with EGF (10 nM) for 30 min, and then harvested for performing the ChIP assay as described in Figure 4B. The protein expression levels of cells transfected with the indicated small interfering (si)RNA were examined (lanes 1–3).
DISCUSSION
In this study, we put forth the idea that in some EGFR-expressing cancer cells, the activated EGFR can be translocated to the nucleus to serve as a transcriptional activator and cooperate with STAT5 to enhance the expression of Aurora-A. The overexpressed Aurora-A may result in centrosomes amplification and microtubule disorder, both of which are critical steps for aneploidy and chromosome instability, ultimately leading to tumor formation. It is well recognized that the expression of Aurora-A is cell-cycle-dependent and is involved in centrosome functions including maturation, duplication and separation as well as spindle assembly and stability. Therefore the accurate control of Aurora-A expression may be very important for maintaining genetic stability (35). In addition, it was reported that the EGFR is overexpressed in some malignant tumors, and the overexpressed EGFR can be detected in the nucleus where it serves as a transcriptional activator (17,21). This phenomenon is highly correlated with cell proliferation (21). Furthermore, the constitutively activated EGFR is thought to play a role in chromosome instability (27,28). These descriptions encouraged us to investigate whether any possible correlation between overexpressed EGFR and Aurora-A exists in tumor cells.
It is known that the activated EGFR carries out its biological effects via two signaling pathways: the cytoplasmic/traditional pathway and nuclear pathway (34). Activation of the cytoplasmic/traditional EGFR pathway leads cells to tumorigenesis, metastasis, high proliferation and resistance to chemotherapy and radiation. In some cases, the membrane EGFR is translocated into the nucleus by ligand stimulation, and after nuclear translocation, the nuclear EGFR interacts with other transcriptional factors, which possess DNA-binding ability, thus activating gene expression (34). There are a lot of reports indicating that nuclear localization of the EGFR is highly correlated with tumor malignancy, cell proliferation, cell-cycle progression and patient survival (34,36). The nuclear importation of the membrane receptor was not only observed for the EGFR, but also for many other receptor tyrosine kinases, such as HER-2 (25,37), FGFR (38), HER-3 (39) and cytokine receptors (40). In this study, EGF enhanced Aurora-A gene expression only in EGFR-expressing cell lines. The activated EGFR was located into the nuclei of these tested cells (Figures 3B and 5A), and cooperated with STAT5 to enhance Aurora-A gene expression (Figure 4).
STAT5 is activated by many growth factors and cytokines (41), thereby suggesting the involvement of STAT5 in growth signaling. Activation of STAT5 is correlated with cell proliferation and differentiation. Although the target genes which are activated by STAT5 in pathogenesis are consistent with normal cells, constitutive activation of STAT5 leads to target gene overexpression, resulting in enhanced expression of anti-apoptosis and cell-cycle progression genes (41). There are two STAT5 proteins, STAT5A and STAT5B, which are encoded by two distinct but closely related genes. STAT5A has a higher DNA-binding affinity than STAT5B, and it is mostly involved in prolactin-directed mammary gland maturation (42), whereas STAT5B is believed to be involved in the response to the growth hormone (42). This study is the first report to demonstrate that STAT5 can transactivate the expression of the Aurora-A gene. To our surprise, according the results, only STAT5A not STAT5B was involved in EGF-induced Aurora-A overexpression (Figure 6). This is an additional evidence to prove that STAT5A and STAT5B may participate in different cell functions.
It was previously reported that the EGF can induce rapid centrosomal separation, but the underlying mechanism was not clear (43). Based on this study, we believe that Aurora-A kinase may be the major player causing centrosomal separation which ultimately results in centrosomes amplification under EGF stimulation. It is well recognized that the EGFR and Aurora-A are overexpressed in many cancers (44,45), and this is the first study to examine their correlation. By treating cells with the EGFR-specific inhibitor, AG1478, the EGFR could not further activate or induce Aurora-A gene expression (Figure 3A). When the endogenous EGFR was decreased by the siRNA technique, EGF-induced Aurora-A overexpression was also attenuated (Figure 6B). These results support the notion that the activated EGFR indeed plays a role in activating Aurora-A gene expression.
This study demonstrated that the nuclear EGFR can cooperate with STAT5A to target the promoter region of Aurora-A and enhance its expression in cancer cells. The nuclear EGFR hence may indirectly play a functional role in cell proliferation and chromosome instability. These results further provide a link between the activated EGFR and chromosomal instability. However, we cannot rule out the possibility that the effect of the traditional/cytoplasmic EGFR signaling pathway on EGF-induced Aurora-A overexpression. In fact, LS174T cells, which express the membrane EGFR (Figure 2B), underwent EGFR endocytosis after EGF stimulation (data not shown), but the phenomena of EGF-induced Aurora-A overexpression also existed in these cells (Figure 2B). The traditional membrane EGFR pathway transduces the extracellular signals into the nucleus through activation of multiple signaling cascades, whereas the nuclear EGFR pathway directly transmits extracellular signals from the cytoplasmic membrane to its target genes in the nucleus (34). How cells determine the fate of the EGFR to undergo the cytoplasmic or nuclear pathway remains largely unknown. It was reported that the nuclear localization of receptor tyrosine kinases is growth-factor-dependent and may play a role in transcriptional activation (17). There are many reports indicating that the pathological and clinical importance of a cellular protein is not only dependent on its expression level but also on its subcellular localization (36). Nuclear localization of the EGFR is usually co-expressed with Ki-67 and cyclin D (36), both of which are indicators of cell proliferation. Therefore, detection of the nuclear EGFR may indicate a poor survival rate in cancer patients and activated cell proliferation (36). However, using an immunohistochemistry assay, the expressions of Aurora-A and the EGFR in breast and colorectal cancer tissues were analyzed, and our preliminary results indicated that overexpression of Aurora-A has a correlation trend with EGFR expression, of around 66.67% (10/15) in breast cancers and 75% (9/12) in colorectal cancers (Hung, L.Y. et al., unpublished data). Combined with the siRNA data (Figure 6), we concluded that the EGFR is indeed involved in EGF-induced Aurora-A expression.
In recent years, an EGFR mAb has been used to treat many types of cancer patients (46), including colorectal cancers (47,48). The small molecular inhibitor of the Aurora kinases is also used to treat acute myelogenous leukemia and chronic myelogenous leukemia (49,50). Although it was reported that the EGFR and Aurora-A are overexpressed in many cancers, including colorectal, breast and lung cancers, none of them provided a linkage between them. This report may provide new insights for cancer therapeutics targeting both the EGFR and Aurora-A in cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Ju-Ming Wang for helpful comments and discussion. We also thank Mr Dan Chamberlin for English editing help. This work was supported by grants (NSC94-2314-B-006-121, NSC95-2320-B-006-084 and NSC95-2320-B006-066-MY3) from the National Science Council, and the Ministry of Education Program for Promoting Academic Excellent of University under grant 91-B-FA0190104 of the Republic of China, and National Cheng Kung University Project of Promoting Academic Excellent and Developing World Class Research Centers. Funding to pay the Open Access publication charges for this article was provided by National Science Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 3.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 4.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat. Rev. Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 5.Littlepage LE, Wu H, Andresson T, Deanehan JK, Amundadottir LT, Ruderman JV. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl Acad. Sci. USA. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, Ward IM, Saya H, Fang G, van Deursen J, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 8.Kimura M, Matsuda Y, Yoshioka T, Okano Y. Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 1999;274:7334–7340. doi: 10.1074/jbc.274.11.7334. [DOI] [PubMed] [Google Scholar]
- 9.Kimura M, Uchida C, Takano Y, Kitagawa M, Okano Y. Cell cycle-dependent regulation of the human aurora B promoter. Biochem. Biophys. Res. Commun. 2004;316:930–936. doi: 10.1016/j.bbrc.2004.01.178. [DOI] [PubMed] [Google Scholar]
- 10.Udayakumar TS, Belakavadi M, Choi KH, Pandey PK, Fondell JD. Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J. Biol. Chem. 2006;281:14691–14699. doi: 10.1074/jbc.M600163200. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa T, Kanai N, Shiwaku HO, Soga N, Uehara A, Horii A. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25:4831–4839. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 12.Anderson D, Koch CA, Grey L, Ellis C, Moran MF, Pawson T. Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- 13.Boonstra J, Rijken P, Humbel B, Cremers F, Verkleij A, van Bergen en Henegouwen P. The epidermal growth factor. Cell Biol. Int. 1995;19:413–430. doi: 10.1006/cbir.1995.1086. [DOI] [PubMed] [Google Scholar]
- 14.Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J. Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol. Cell. Biol. 1992;12:981–990. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roskoski R., Jr The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem. Biophys. Res. Commun. 2004;319:1–11. doi: 10.1016/j.bbrc.2004.04.150. [DOI] [PubMed] [Google Scholar]
- 16.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr. Opin. Cell. Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 18.Fischer OM, Hart S, Gschwind A, Ullrich A. EGFR signal transactivation in cancer cells. Biochem. Soc. Trans. 2003;31:1203–1208. doi: 10.1042/bst0311203. [DOI] [PubMed] [Google Scholar]
- 19.Gusterson B, Cowley G, McIlhinney J, Ozanne B, Fisher C, Reeves B. Evidence for increased epidermal growth factor receptors in human sarcomas. Int. J. Cancer. 1985;36:689–693. doi: 10.1002/ijc.2910360612. [DOI] [PubMed] [Google Scholar]
- 20.Kamio T, Shigematsu K, Sou H, Kawai K, Tsuchiyama H. Immunohistochemical expression of epidermal growth factor receptors in human adrenocortical carcinoma. Hum. Pathol. 1990;21:277–282. doi: 10.1016/0046-8177(90)90227-v. [DOI] [PubMed] [Google Scholar]
- 21.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell. Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 22.Lipponen P, Eskelinen M. Expression of epidermal growth factor receptor in bladder cancer as related to established prognostic factors, oncoprotein (c-erbB-2, p53) expression and long-term prognosis. Br. J. Cancer. 1994;69:1120–1125. doi: 10.1038/bjc.1994.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Tervahauta A, Syrjanen S, Syrjanen K. Epidermal growth factor receptor, c-erbB-2 proto-oncogene and estrogen receptor expression in human papillomavirus lesions of the uterine cervix. Int. J. Gynecol. Pathol. 1994;13:234–240. doi: 10.1097/00004347-199407000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Wang SC, Lien HC, Xia W, Chen IF, Lo HW, Wang Z, Ali-Seyed M, Lee DF, Bartholomeusz G, Ou-Yang F, et al. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–261. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 27.Ma PC, Zhang X, Wang ZJ. High-throughput mutational analysis of the human cancer genome. Pharmacogenomics. 2006;7:597–612. doi: 10.2217/14622416.7.4.597. [DOI] [PubMed] [Google Scholar]
- 28.Tomida S, Yatabe Y, Yanagisawa K, Mitsudomi T, Takahashi T. Throwing new light on lung cancer pathogenesis: updates on three recent topics. Cancer Sci. 2005;96:63–68. doi: 10.1111/j.1349-7006.2005.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida N, Nagasaka T, Kashiwagi K, Boland CR, Goel A. High copy amplification of the Aurora-A gene is associated with chromosomal instability phenotype in human colorectal cancers. Cancer Biol. Ther. 2007;6:525–533. doi: 10.4161/cbt.6.4.3817. [DOI] [PubMed] [Google Scholar]
- 30.Hung LY, Chen HL, Chang CW, Li BR, Tang TK. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol. Biol. Cell. 2004;15:2697–2706. doi: 10.1091/mbc.E04-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loffler H, Lukas J, Bartek J, Kramer A. Structure meets function- -centrosomes, genome maintenance and the DNA damage response. Exp. Cell Res. 2006;312:2633–2640. doi: 10.1016/j.yexcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur. J. Cancer. 2001;37(Suppl. 4):S3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 33.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 34.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br. J. Cancer. 2006;94:184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutertre S, Descamps S, Prigent C. On the role of aurora-A in centrosome function. Oncogene, 2002;21:6175–6183. doi: 10.1038/sj.onc.1205775. [DOI] [PubMed] [Google Scholar]
- 36.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 37.Xie Y, Hung MC. Nuclear localization of p185neu tyrosine kinase and its association with transcriptional transactivation. Biochem. Biophy. Res. Commun. 1994;203:1589–1598. doi: 10.1006/bbrc.1994.2368. [DOI] [PubMed] [Google Scholar]
- 38.Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J. Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW. c-erbB-3: a nuclear protein in mammary epithelial cells. J. Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krolewski JJ. Cytokine and growth factor receptors in the nucleus: what's up with that? J. Cell. Biochem. 2005;95:478–487. doi: 10.1002/jcb.20451. [DOI] [PubMed] [Google Scholar]
- 41.Debierre-Grockiego F. Anti-apoptotic role of STAT5 in haematopoietic cells and in the pathogenesis of malignancies. Apoptosis. 2004;9:717–728. doi: 10.1023/B:APPT.0000045785.65546.a2. [DOI] [PubMed] [Google Scholar]
- 42.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 43.Sherline P, Mascardo RN. Epidermal growth factor induces rapid centrosomal separation in HeLa and 3T3 cells. J. Cell Biol. 1982;93:507–512. doi: 10.1083/jcb.93.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Normanno N, Bianco C, De Luca A, Maiello MR, Salomon DS. Target-based agents against ErbB receptors and their ligands: a novel approach to cancer treatment. Endocr. Relat. Cancer. 2003;10:1–21. doi: 10.1677/erc.0.0100001. [DOI] [PubMed] [Google Scholar]
- 45.Warner SL, Bearss DJ, Han H, Von Hoff DD. Targeting Aurora-2 kinase in cancer. Mol. Cancer Ther. 2003;2:589–595. [PubMed] [Google Scholar]
- 46.Karamouzis MV, Grandis JR, Argiris A. Therapies directed against epidermal growth factor receptor in aerodigestive carcinomas. JAMA. 2007;298:70–82. doi: 10.1001/jama.298.1.70. [DOI] [PubMed] [Google Scholar]
- 47.Adams R, Maughan T. Predicting response to epidermal growth factor receptor-targeted therapy in colorectal cancer. Expert. Rev. Anticancer Ther. 2007;7:503–518. doi: 10.1586/14737140.7.4.503. [DOI] [PubMed] [Google Scholar]
- 48.Elfiky AA, Saif MW. The developing trend of monoclonal antibodies in the treatment of colorectal cancer. Expert Opin. Biol. Ther. 2007;7:871–883. doi: 10.1517/14712598.7.6.871. [DOI] [PubMed] [Google Scholar]
- 49.Cheetham GM, Charlton PA, Golec JM, Pollard JR. Structural basis for potent inhibition of the Aurora kinases and a T315I multi-drug resistant mutant form of Abl kinase by VX-680. Cancer Lett. 2007;251:323–329. doi: 10.1016/j.canlet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Ikezoe T, Yang J, Nishioka C, Tasaka T, Taniguchi A, Kuwayama Y, Komatsu N, Bandobashi K, Togitani K, Koeffler HP, et al. A novel treatment strategy targeting Aurora kinases in acute myelogenous leukemia. Mol. Cancer Ther. 2007;6:1851–1857. doi: 10.1158/1535-7163.MCT-07-0067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.