Abstract
Rare AGA or AGG codons close to the initiation codon inhibit protein synthesis by a tRNA-sequestering mechanism as toxic minigenes do. To further understand this mechanism, a parallel analysis of protein synthesis and peptidyl-tRNA accumulation was performed using both a set of lacZ constructs where AGAAGA codons were moved codon by codon from +2, +3 up to +7, +8 positions and a series of 3–8 codon minigenes containing AGAAGA codons before the stop codon. β-Galactosidase synthesis from the AGAAGA lacZ constructs (in a Pth defective in vitro system without exogenous tRNA) diminished as the AGAAGA codons were closer to AUG codon. Likewise, β-galactosidase expression from the reporter +7 AGA lacZ gene (plus tRNA, 0.25 μg/μl) waned as the AGAAGAUAA minigene shortened. Pth counteracted both the length-dependent minigene effect on the expression of β-galactosidase from the +7 AGA lacZ reporter gene and the positional effect from the AGAAGA lacZ constructs. The +2, +3 AGAAGA lacZ construct and the shortest +2, +3 AGAAGAUAA minigene accumulated the highest percentage of peptidyl-tRNAArg4. These observations lead us to propose that hungry codons at early positions, albeit with less strength, inhibit protein synthesis by a minigene-like mechanism involving accumulation of peptidyl-tRNA.
INTRODUCTION
Codon usage is determined by the relative abundance of cognate tRNAs (1,2). A low cognate tRNA concentration corresponds to a rare codon and the frequency of rare codons in a gene determines the rate of protein synthesis of the corresponding protein (3). Usually, synthesis is enhanced by over-expression of tRNA(s) cognate to the rare codon(s) (4,5). A computational search of rare AGA/AGG codons in Escherichia coli genes showed that they are located preferentially within the first 25 codons. More than 100 genes containing a single AGG or AGA codon within the first 25 codons are associated with essential functions (6). Protein synthesis modulation by minor codons close to the initiator has been proposed by a number of authors. For example, Chen and Inouye (7) demonstrated that the closer AGG codons were to the initiation codon, the stronger the effect on protein expression. They used synthetic constructs containing from one to five codons inserted after the 10th codon of the lacZ gene. They also showed that single and particularly tandems of two to five AGG have stronger effects when placed closer to the translation start (7). Similar results were obtained when groups of two to five AGG codons were inserted after codons 13, 223 and 307 in a synthetic codon test system. Insertion of AGG codons right after codon 13 had the most severe effects on translation (8). To explain these effects Chen and Inouye (6) have proposed that the rare codons close to the initiator may stall the ribosome and prevent the entry of new incoming ribosomes. A model in which some mRNA is released from ribosomes during 5′-translational blockage by low usage AGG codons has been suggested (9). In addition to the codon position, the presence of unfavorable codons downstream from the first rare codon is also relevant to the degree of tRNA sequestration (10). Over-expressing genes containing clusters of AGG codons in wild-type cells can significantly exacerbate the depletion of specific tRNAs in the ternary complex pool (11,12). By contrast, no inhibition of translation is found when AGG codons are interspersed or scattered through the reading frame (12). Lambda int, which encodes the phage integrase, is a natural gene containing a high frequency of single and clusters of minor codons including 33 rare arginine codons. Three arginine tandems containing AGA and/or AGG codons are located at positions 3–4, 108–109 and 176–177. Over expression of int in wild-type E. coli cells inhibits cell growth and protein synthesis (13). Changing the rare arginine codons at positions 3 and 4 to common arginine (CGT) codons is enough to enhance Int translation as much as tRNAArg4 supplementation does for the wild-type gene (14). These results suggested that int-mediated inhibition entails reduction of the tRNA pool provoked by AGA AGG triplets located at positions 3 and 4. Indeed, int causes a stronger cell growth and protein synthesis inhibition in Pth defective cells, which are reversed by tRNAArg4 or Pth supplementation. These effects are linked to peptidyl-tRNAArg4 (pep-tRNAArg4) accumulation mainly in the soluble fraction (15). In Escherichia coli NGG codons (CGG, AGG, UGG or GGG) but not GGN or GNG (where N is non-G) have been associated with low expression of a reporter gene, if located at positions +2 to +5. NGG codons (CGG, AGG, GGG and UGG), in the early coding region downstream the initiation codon of a highly expressed reporter gene, inhibit growth of a pth thermo-sensitive (Ts) mutant strain (16). This effect is reverted by supplementing the cells with the specific tRNA and is not seen for other codons or when NGG codons are placed at a later position (+7). Although not demonstrated, the authors suggested that the reduced gene expression associated with NGG in the early coding region of mRNA is the result of pep-tRNA drop-off from the ribosome during translation. Protein synthesis inhibition by AGA hungry codons at positions 2+ or 3+ of lacZ has been observed both in vivo and in vitro (3). The inhibition has been unambiguously linked to pep-tRNA accumulation and low levels of Pth activity and/or the cognate tRNA concentration (3).
The idea of translation regulation by rare codons and/or codon usage around the initiation region has been suggested by several authors (17,18). To explain these negative effects, several possibilities have been advanced including ribosome pausing, tRNA sequestration and premature translation termination (6,8,19). Stalling of the int translating ribosome at the rare arginine AGA codon at position 3 of the int ORF may also block the binding of new ribosomes in the ribosome-binding site. Interestingly, these observations parallel the reported characteristics of mini-genes (20–23). Mini-gene expression in pth defective (rap) cells inhibits cell growth, arrests protein synthesis and accumulates pep-tRNA (21,22,24). Mini-gene-mediated toxicity is reversed by specific tRNAs or Pth supplementation (20,22). Thus, as minigenes do, overexpression of int and other genes containing AGA or AGG hungry codons near the initiation codon causes a strong cell growth and protein synthesis inhibition in pth defective cells, which are reversed by tRNAArg4 or Pth supplementation. These effects are also accompanied with pep-tRNAArg4 accumulation mainly in the soluble cell fraction. This analogy may be explained by the same underlying mechanisms which involve ribosome stalling and pep-tRNA drop off. To further substantiate these observations we analysed the inhibition of protein synthesis and the accumulation of pep-tRNA from a set of constructs with double AGAAGA codons at positions from 2+ to 7+ and a series of 3–8 codon minigenes containing AGAAGA before the stop codon. Here, we found a dependency between the position of the AGAAGA hungry codons and the degree of protein synthesis inhibition and between the length of the minigene and its capacity to inhibit β-galactosidase (β-gal) expression from a +7 AGA lacZ reporter gene. The closer the hungry codons were located to the initiation codon or the shorter the minigene, the stronger the inhibition of protein synthesis. Interestingly, the highest accumulation of pep-tRNA was produced both by hungry codons at positions +2, +3 and by the shortest minigene and decreased at later positions or as the minigene lengthened. Thus, both the AGAAGA hungry codons and the minigenes showed an inverse relationship between the degree of protein synthesis inhibition and the accumulation of pep-tRNA. These observations lead us to propose that hungry codons at early positions of the reading frame inhibit protein synthesis by a mechanism similar to that of minigenes.
MATERIALS AND METHODS
Strains and growth conditions
Escherichia coli strains and plasmids used in this study are listed in Table 1. P90C pth(rap), a Pth-defective strain, contains 10 times less Pth activity than wild-type cells (25). Unless different growth conditions are indicated, bacterial cultures were grown at 32°C in Luria–Bertani medium (26). When required, the medium was supplemented with ampicillin (Amp) 100 µg/ml.
Table 1.
Strain and plasmid characteristics
| Strains or plasmids | Genotype and relevant characteristics | Source |
|---|---|---|
| P90C | araA(lac-pro)thi zch::Tn10 | Lab collection |
| P90C pth(rap) | araA(lac-pro)thi pth(rap)zch::Tn10 | Lab collection |
| C600 | F- (leu-B6 thi-1 supE44 lac Y1 tonA21) zch::Tn10 | (41) |
| C600 pth(rap) | F- (leu-B6 thi-1 supE44 lac Y1 tonA21) pth(rap)zch::Tn10 | (41) |
| Plasmids | Description | |
| pKQV4 | bla lacI ptac | (43) |
| placZwt | pKQV4 containing EcoRI/HinDIII lacZ gene insert from plexlacz | (3) |
| LacZ derivatives | Description | |
| placZ7AGA | Derivative of placZwt with TTA7AGA substitution in lacZ | This work |
| placZ2-3AGAAGA | Derivative of placZwt with ACC2AGA, GAT3AGA substitutions in lacZ | This work |
| placZ3-4AGAAGA | Derivative of placZwt with GAT3AGA, CCC4AGA substitutions in lacZ | This work |
| placZ4-5AGAAGA | Derivative of placZwt with CCC4AGA, GTC5AGA substitutions in lacZ | This work |
| placZ5-6AGAAGA | Derivative of placZwt with GTC5AGA, GTT6AGA substitutions in lacZ | This work |
| placZ6-7AGAAGA | Derivative of placZwt with GTT6AGA, TTA7AGA substitutions in lacZ | This work |
| placZ7-8AGAAGA | Derivative of placZwt with TTA7AGA, CAA8AGA substitutions in lacZ | This work |
| AGAAGA Minigenes | Description | This work |
| placZ2-3AGAAGAUAA | Derivative of placZwt with ACC2AGA, GAT3AGA, CCC4UAA substitutions in lacZ | This work |
| placZ3-4AGAAGAUAA | Derivative of placZwt with GAT3AGA, CCC4AGA, GTC5UAA substitutions in lacZ | This work |
| placZ4-5AGAAGAUAA | Derivative of placZwt with CCC4AGA, GTC5AGA, GTT6UAA substitutions in lacZ | This work |
| placZ5-6AGAAGAUAA | Derivative of placZwt with GTC5AGA, GTT6AGA, TTA7UAA substitutions in lacZ | This work |
| placZ6-7AGAAGAUAA | Derivative of placZwt with GTT6AGA, TTA7AGA, CAA8UAA substitutions in lacZ | This work |
| placZ7-8AGAAGAUAA | Derivative of placZwt with TTA7AGA, CAA8AGA, CGT9UAA substitutions in lacZ | This work |
PLASMID CONSTRUCTS
placZ plasmid containing lacZ gene was constructed by PCR amplification of lacZ gene from pLEX/lacZ vector (Invitrogen) as previously described (27). The lacZ constructs and minigenes containing AGAAGA codons used in this work (Table 1) were constructed from placZ by site-directed mutagenesis (Stratagene) using complementary oligonucleotides. For each construct only the forward 5′ oligonucleotide is shown in Table 2. For example: 5-LacZ 2,3 AGAAGA and 3-LacZ 2,3AGAAGA were used to obtain the lacZ construct containing AGAAGA at +2, +3 positions and the pair of oligonucleotides 5-LacZ 2,3 AGAAGAUAA and the complementary 3-LacZ 2,3 AGAAGAUAA were used to synthesize AUGAGAAGAUAA minigene and so forth. The +7 AGA reporter gene was also constructed by site-directed mutagenesis using 5-LacZ 7 AGA and 3-LacZ 7 AGA oligonucleotides. All the substitutions indicated here, correspond to E. coli chromosomal lacZ gene sequence positions. All plasmid constructs were transformed in the indicated E. coli strains by the RbCl2 method and the expression of proteins was under the control of the IPTG-inducible ptac promoter.
Table 2.
Oligonucleotide characteristics
| Oligonucleotides | Sequence | Characteristics |
|---|---|---|
| 5-LacZ7AGA | 5′-catgaccgatcccgtcgttagacaacgtcgtgactggg-3′ | lacZ +7 AGA substitution |
| 5-LacZ2-3AGAAGA | 5′-ggaaacagaattcatgagaagacccgtcgttttacaacg-3′ | lacZ +2,+3 AGAAGA substitution |
| 5-LacZ3-4AGAAGA | 5′-cagaattcatgaccagaagagtcgttttacaacgtcg-3′ | lacZ +3,+4 AGAAGA substitution |
| 5-LacZ4-5AGAAGA | 5′-cagaattcatgaccgatagaagagttttacaacgtcgtgactggg-3′ | lacZ +4,+5 AGAAGA substitution |
| 5-LacZ5-6AGAAGA | 5′-cagaattcatgaccgatcccagaagattacaacgtcgtgactggg-3′ | lacZ +5,+6 AGAAGA substitution |
| 5-LacZ7-8AGAAGA | 5′-ccgatcccgtcgttagaagacgtcgtgactgggaaaaccc-3′ | lacZ +7,+8 AGAAGA |
| 5-LacZ2-3AGAAGAUAA | 5′-ggaaacagaattcatgagaagauaagtcgttttacaacg-3′ | lacZ +2,+3 AGAAGAUAA substitution |
| 5-LacZ3-4AGAAGAUAA | 5′-cagaattcatgaccagaagauaagttttacaacgtcg-3′ | lacZ +3,+4 AGAAGAUAA substitution |
| 5-LacZ4-5AGAAGAUAA | 5′-cagaattcatgaccgatagaagauaattacaacgtcgtgactggg-3′ | lacZ +4,+5 AGAAGAUAA substitution |
| 5-LacZ5-6AGAAGAUAA | 5′-cagaattcatgaccgatcccagaagauaacaacgtcgtgactggg-3′ | lacZ +5,+6 AGAAGAUAA substitution |
| 5-LacZ6-7AGAAGAUAA | 5′-catgaccgatcccgtcagaagauaacgtcgtgactggg-3′ | lacZ +6,+7 AGAAGAUAA substitution |
| 5-LacZ7-8AGAAGAUAA | 5′-ccgatcccgtcgttagaagauaacgtgactgggaaaaccc-3′ | lacZ +7,+8 AGAAGAUAA substitution |
Cell-free transcription–translation
In vitro reactions (50 µl) were prepared with 2 mg of plasmid DNA in TE (10 mM HCl, pH 8.0, 1 mM EDTA), 20 ml of premix [87.5 mM Tris-Ac pH 8.0, 476 mM potassium glutamate, 75 mM NH4(Ac), 5 mM dithiothreitol, 20 mM Mg(Ac)2, 1.25 mM each of 20 amino acids, 5 mM ATP, 1.25 mM each of CTP, UTP, GTP, 50 mM phosphoenol pyruvate, 250 µg/ml E. coli tRNA, 87.5 mg/ml polyethylene glycol (8000 Mr), 2.5 mM cAMP, 50 µg/ml folinic acid] and 15 μl of S30 prepared as previously described (28,29). Radiolabeled proteins were synthesized using a premix lacking methionine. A 10 μCi of [35S]-methionine (1170 Ci/mmol) were then added to a final concentration of 2 × 10−11 M. Protein was precipitated from 15 μl reactions by addition of 60 μl of acetone followed by centrifugation. The protein pellets were dried and re-suspended in SDS sample buffer prior to gradient polyacrylamide gel electrophoresis (30). Immediately following electrophoresis, wet gels were dried and then visualized by autoradiography. When indicated, dried gels were analysed in a radioactivity scanner (Typhon, Molecular Dynamics) to quantify β-gal radioactivity levels as electronic density values (pixels).
Isolation and detection of pep-tRNAArg4
IPTG-induced cells were collected by centrifugation, resuspended in 3 ml of 0.1 M NaOAc pH 4.5, 0.1% SDS and sonicated (Soniprep 150 MSE Sanyo) to lysis (four 30-s periods at 12 microns with 1 min intervals). After centrifuging at 20 000g for 20 min at 4°C, the supernatant was extracted with an equal volume of acid-equilibrated phenol, vortexed for 1 min, and centrifuged at 20 000g for 20 min at 4°C. The aqueous phase was ethanol precipitated with 0.066 volumes of 3 M NaOAc pH 4.5 and 2.5 volumes of ethanol for 1 h at −20°C and was then spun at 14 000 g for 15 min at 4°C. The pellet was resuspended in 3 µl of 0.3 M NaOAc pH 4.5, with 2.5 volumes of ethanol and pelleted as before. It was finally resuspended in 3 µl of 10 mM NaOAc pH 4.5 and stored at −70°C.
Northern assay
Total RNA from various transformants isolated under acidic conditions as described above was resolved on 6.5% acid urea PAGE gels at 4°C, and electroblotted onto a nylon membrane (31). tRNA-specific oligonucleotide probes were synthesized according to the complementary DNA sequence reported by Dong et al. (2). Hybridization with 5′-32P end-labeled oligonucleotides was performed as described by Varshney et al. (31). Where indicated, samples were treated with 100 mM CuSO4 (Cu++). The radioactive signals were quantified using a Typhoon Scan (Amersham Biosciences). Under no treatment, samples showed two bands. The lower band corresponded to free tRNAArg4 and the upper band to aa-tRNAArg4 and/or pep-tRNAArg4. Treatment with Cu++ chases the aminoacyl-tRNA fraction from the upper band to the lower band. The respective percentages of uncharged, aminoacylated or peptidylated tRNAs were estimated relative to the total tRNA for each electrophoretic lane using the following formula:
% tRNA species (uncharged-‚ aminoacyl- or peptidyl-)
RESULTS
Close dependence of protein synthesis on the position of hungry codons
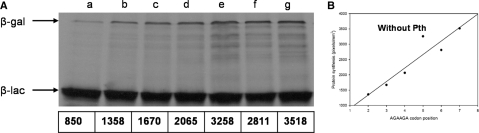
AGG and/or AGA codons at positions +2, +3 of lacZ inhibit protein synthesis and cell growth in Pth defective cells (27). NGG codons, where N is non-G, are associated with a very low gene expression if located at positions +2, +3 and +5 but with higher expression if placed a few codons further downstream (32). We took advantage of these observations to examine the effect of AGA rare codons at early positions on the translation of β-gal. lacZ variants, containing tandems of rare AGAAGA codons at positions 2–3 to 7–8, were cloned in the pKQV4 vector downstream the ptac promoter, which allows expression to be induced by the addition of isopropyl β-d-thiogalactopyranoside (IPTG). The vector carries the laclq gene to maintain repression in the absence of IPTG. The resulting constructs were expressed in a Pth-defective synthesizing S30 cell-free extract prepared as described in ‘Materials and Methods’ section but without exogenous tRNA. The synthesis was carried out in the presence of [35S]-methionine and the synthesized proteins were resolved by SDS–PAGE and visualized by autoradiography. The +2, +3 AGAAGA lacZ construct showed lower synthesis of β-gal, although higher than the wild-type lacZ control (Figure 1A). β-Gal synthesis increased gradually as the AGAAGA tandem changed from +3, +4 to +7, +8 positions. These results show a tendency in which the closer the AGAAGA codons to the initiator the lower the synthesis of β-gal. That the positional effect took place at a translational level is indicated by the fact that the same set of constructs showed an inverted tendency in protein synthesis when tested in a wild-type Pth in vitro reaction (data not shown). Similar results were obtained in the equivalent experiments described below where Pth-defective in vitro reactions were supplemented with Pth. Thus, the differences in protein synthesis were due to a deficient Pth activity or tRNA concentration and not to effects on transcription or mRNA stability. The growing tendency in the synthesis of β-gal, as the AGAAGA codons moved away from the start codon was more apparent in the chart (Figure 1B) where the levels of protein synthesis represented as electronic density values/mm2 (pixels) were plotted versus the position of the AGAAGA tandem in the lacZ constructs.
Figure 1.
Positional effect of AGAAGA codons in the synthesis of β-gal. (A) In vitro transcription–translation reactions deficient in Pth activity, not supplemented with tRNA and carried out as described in ‘Materials and Methods' section, were directed by: placZ (a); placZ2-3AGAAGA (b); placZ3-4AGAAGA (c); placZ4-5AGAAGA (d); placZ5-6AGAAGA (e); placZ6-7AGAAGA (f); placZ7-8AGAAGA (g). After 1 h incubation, samples were processed for autoradiography as described in ‘Materials and Methods’ section. The mobility of β-galactosidase (β-gal) and β-lactamase (β-lac) is indicated. β-gal absolute radioactivity levels indicated as electronic density values (pixels; shown below each lane) were quantified as indicated in ‘Materials and Methods’ section. (B) β-gal synthesis from the AGAAGA constructs was plotted against the AGAAGA codon position.
The inhibition of protein synthesis by minigenes depends inversely on the length of their open reading frame
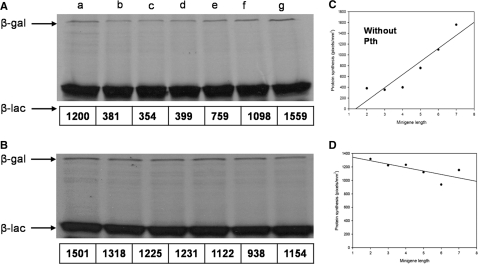
Previous studies have shown that multiple parameters related to the sequence of the mini-gene contribute to the growth inhibitory effect (21). In addition to these parameters, the effect of the length of minigenes on pep-tRNA accumulation and in vivo toxicity has also been studied (33). Thus, the rate of pep-tRNA drop-off and ribosome recycling without dissociating from the ribosome increases as the size of the minigene is shortened from 8 to 2 codons while the rate of translation termination decreases. These antecedents and the observation that the degree of minigene-mediated inhibition of protein synthesis correlates with the number of hungry codons in the reporter gene (3) provided the basis to predict an inverse relationship between the length of the minigene and the degree of protein synthesis of a gene containing a codon starved by the minigene. To test this prediction, we analysed in a two-template based in vitro system, the effect of each of a series of minigenes containing from 3 to 8 sense codons including the tandem AGAAGA before the stop codon (plasmid placx-yAGAAGAUAA, where x–y represents codon positions from 2–3 to 7–8; Table 1) on the expression of a lacZ reporter gene containing AGA at position +7 (placZ7AGA; Table 1). When the analysis was first performed as in the previous section (without exogenous tRNA) a complete inhibition of β-gal was produced by all minigenes (data not shown). It was necessary to supplement the in vitro reaction with tRNA (0.25 µg/µl) to reveal a gradual effect of the minigenes on β-gal synthesis. As expected, the shortest minigene (+2, +3 AGAAGAUAA) produced the strongest inhibition of β-gal and as the length of the minigene increased the synthesis built up to a level comparable to that of wild-type β-gal (Figure 2A). No difference in the expression of β-gal was observed when the +7 AGA lacZ was substituted by wild-type lacZ (Figure 2B and D) or when the experiment was performed in a wild-type in vitro reaction (data not shown) indicating that the effect is mediated by a tRNA-sequestering mechanism. Thus, the results showed a clear dependency on the length of the minigene, in which, the shortest minigene produced the highest inhibition. Unlike the AGAAGA lacZ constructs, minigenes consumed higher levels of tRNAArg4 as indicated by the necessity to supplement the in vitro reaction with tRNA. This tendency and its analogy with that produced by the AGAAGA lacZ constructs are clearer in the charts (compare Figure 1B with 2C).
Figure 2.
The inhibition of protein synthesis from the reporter +7 AGA lacZ construct depends inversely on the length of the AGAAGAUAA minigenes. (A) In vitro transcription–translation reactions were directed by: +7 AGA lacZ (a); +7 placZ plus: placZ2-3AGAGAUAA (b); placZ3-4AGAAGAUAA (c); placZ4-5AGAAGAUAA (d); placZ5-6AGAAGAUAA (e); placZ6-7AGAAGAUAA (f); placZ7-8AGAAGAUAA (g). The mobility of β-galactosidase (β-gal) and β-lactamase (β-lac) is indicated. β-Gal absolute radioactivity levels are indicated as electronic density values (pixels, shown below each lane). (B) In vitro reactions were primed as in (A) but lacZ replaced +7 AGA lacZ. β-Gal synthesis from the reporter +7 AGA lacZ (C) or lacZ (D) constructs was plotted against minigene length.
Positional effect of hungry codons close to the initiation codon on the accumulation of pep-tRNA
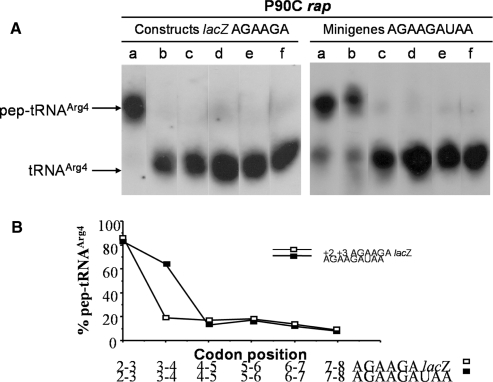
Lambda int gene and lacZ constructs containing AGAAGG and AGAAGA codons at +3, +4 and +2, +3 positions, respectively, were previously associated with a high accumulation of pep-tRNAArg (15,27). Thus, it was interesting to find out if the level of β-gal synthesis showed by the AGAAGA lacZ variants correlated with the production of pep-tRNA and whether this kinetics paralleled the accumulation of pep-tRNAArg4 by the AGAAGAUAA minigenes. For this purpose Pth defective cells (P90C rap) were transformed with the plasmids containing the AGAAGA lacZ variants or AGAAGA minigenes. pep-tRNAArg4 was analysed in total cell extracts from IPTG-induced cells by a northern blot assay and the percentages of pep-tRNA relative to aminoacyl-tRNA and uncharged tRNA were calculated as indicated in ‘Materials and Methods’ section. The results of a representative experiment are presented. The largest amount of pep-tRNAArg4 was produced by both the shortest +2, +3 AGAAGAUAA minigene and the +2, +3 AGAAGA lacZ construct (Figure 3A, lane a). The accumulation of pep-tRNAArg4 was still evident for minigene +3, +4 AGAAGAUAA, however, the level of pep-tRNAArg4 abruptly decreased either as minigene length increased or the AGAAGA tandem moved away from the start codon in lacZ constructs. Albeit not clear in the northern blots, the percentages of pep-tRNA relative to the total tRNA, estimated for each electrophoretic lane using the formula described in ‘Materials and Methods’ section, indicated the presence of decreasing amounts of pep-tRNAArg4 and that the lowest percentages were generated both by the longest minigene (+7, +8 AGAAGAUAA) and the +7, +8 AGAAGAUAA lacZ construct (Figure 3B). No pep-tRNA was observed when the lacZ constructs or minigenes were expressed in wild-type pth cells (data not shown).
Figure 3.
The positional effect of AGAAGA codons in the lacZ constructs is similar to the length-dependent effect of the AGAAGAUAA minigenes in the production of pep-tRNAArg4. (A) The order of the AGAAGA lacZ constructs or the AGAAGAUAA minigenes is the same as that shown for lanes b–g, panel A from Figures 1A and 2A, respectively. Pth defective cells (P90C rap) were processed as described in Materials and Methods to detect pep-tRNAArg4. Samples were treated with Cu++ and pep-tRNAArg4 was revealed by northern blotting using a specific 32P-labeled oligo-probe in samples of total RNA. The relative locations of the various tRNAArg4 derivatives are arrowed. The absolute amount of material applied to each lane varied according to the efficiency of recovery of the tRNA in each manipulation. Band intensities of the different tRNA forms should only be compared within lanes. (B) Percentages of pep-tRNAArg4 calculated as described in ‘Materials and Methods’ section are outlined against AGAAGA codon positions or the length of the minigene.
It was previously reported that different parameters affecting pep-tRNA accumulation like the dissociation rate of pep-tRNA and ribosome recycling without leaving the ribosome are inversely correlated with the length of the coding sequence of the minigene (33). This tendency was also observed with in vivo toxicity, which decreased (growth rate increased) with the mini-gene length. Thus, the growing tendency in the synthesis of β-gal, both as the AGAAGA codons moved away from the start codon or the minigene lengthened correlated inversely with the percentages of pep-tRNAArg, calculated as indicated in ‘Materials and Methods’ section.
The positional effect of the AGAAGA rare codons is reverted by Pth
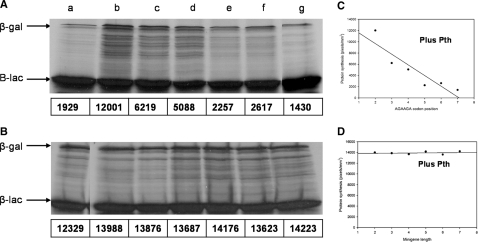
The expression of toxic mini-genes entails specific pep-tRNA accumulation and protein synthesis inhibition in Pth defective cells. The inhibition is averted by over-production of Pth or supplementation with tRNA (20,22). Likewise, lambda int gene which contains AGAAGG codons at positions +3, +4 inhibits protein synthesis and cell growth of Pth defective cells and these effects are suppressed by over-production of Pth or tRNAArg4. These observations made it interesting to analyse whether the positional effect observed on the inhibition of protein synthesis and the accumulation of pep-tRNA, is primarily caused by the lack of Pth. As expected, the length-dependent inhibition of the minigenes was averted by the addition of Pth to the Pth-defective cell-free system (Figure 4B). The reversion is also apparent in the chart where the levels of protein synthesis were plotted versus the length of the minigene (Figure 4D). In other words, β-gal synthesis from the +7 lacZ reporter gene did not show great variations regardless of the minigene expressed. As to the AGAAGA lacZ constructs, not only was the synthesis of β-gal restored by the addition of Pth to the in vitro reaction but the positional effect was also inverted (Figure 4A and C). Thus, the tRNA recycling activity of Pth unveiled a promoting effect of the same codons, which was stronger as the AGA codons got closer to the initiation codon. The same results were obtained when the set of constructs were expressed in a wild-type in vitro reaction, which is equivalent to the Pth defective in vitro reaction supplemented with Pth (data not shown).
Figure 4.
Peptidyl-tRNA hydrolase prevents the inhibition of β-gal caused by the AGAAGA codons. (A) In vitro transcription–translation reactions, supplemented with Pth (3 µg/50 µg reaction), were primed with AGAAGA lacZ constructs as described in Figure 1 legend. (C) Absolute radioactivity levels indicated as electronic density values (pixels) from each construct were quantified as indicated in ‘Materials and Methods’ section and plotted against the AGAAGA codon position. (B) In vitro reactions, supplemented with Pth (3 µg/50 µl reaction), were directed with the +7 AGA lacZ construct and the AGAAGAUAA minigenes as in Figure 2 legend. The mobility of β-galactosidase (β-gal) and β-lactamase (β-lac) is indicated. (D) β-gal absolute radioactivity levels from the reporter gene indicated as electronic density values (pixels) were plotted against minigene length.
DISCUSSION
The results presented in this work indicate that the positional effect on the inhibition of protein synthesis by hungry codons close to initiation codon resembles the length-dependent inhibition caused by minigenes, i.e. pep-tRNA accumulation. While the production of β-gal from AGAAGA lacZ constructs declined as the AGAAGA codons were closer to AUG codon, the inhibition of β-gal from the reporter +7 AGA lacZ gene became stronger as the AGAAGAUAA minigene shortened. In addition, the production of pep-tRNAArg4 followed the same tendency according to which the +2, +3 AGAAGA lacZ construct or the +2, +3 AGAAGAUAA minigene accumulated the highest percentage of pep-tRNAArg4. Unlike the AGAAGA lacZ constructs, AGAAGAUAA minigenes consumed more tRNAArg as indicated by the necessity to supplement the in vitro reaction with tRNA. Without additional tRNA, a complete inhibition of β-gal is produced (not shown). In addition, high levels of pep-tRNAArg4 were still apparent with the +3, +4 AGAAGAUAA minigene. As expected, Pth reverted the length-dependent minigene effect on the expression of β-gal from the +7 AGA lacZ reporter gene. In the case of the AGAAGA lacZ constructs, Pth not only restored the synthesis but the tendency was inverted. Thus, β-gal synthesis was higher the closer the AGAAGA codons to the initiator. We discuss below these results and the fact that even the +2, +3 AGAAGA lacZ construct produced more β-gal than wild-type lacZ (Compare lanes a and b in Figures 1A and 4A).
Protein synthesis modulation by minor codons close to the initiator has been proposed by different authors. However, the parallel analysis of protein synthesis and pep-tRNA production had not been performed on a set of minigenes of various lengths and lacZ constructs where the hungry codons are moved away codon by codon from the initiator. Thus, the series of AGAAGA lacZ constructs analysed in this work clearly showed a direct relationship between the degree of in vitro β-gal synthesis and the position of the hungry codons and this behavior was similar to that presented by the minigenes.
A number of parameters have been shown to affect the rate of pep-tRNA accumulation and the toxicity of minigenes. Among these, the length of the minigene affected directly four important parameters that determine the pool of pep-tRNA: (i) the average number of times an mRNA is recycled and translated before it is degraded, without leaving the 30S subunit; (ii) the rate of pep-tRNA dissociation from the ribosome at each translation round; (iii) normal termination catalysed by release factors, the principal reaction competing with drop-off and (iv) Pth activity, which allows recycling of pep-tRNA thereby avoiding tRNA sequestration. In addition, we have previously shown that the expression of toxic minigenes inhibits primarily the synthesis of proteins encoded by genes that contain the codon recognized by the tRNA which has been sequestered by the minigene (3). Therefore, these antecedents predict a gradual inhibition of a reporter gene according to the length of the minigene. This hypothesis was tested with a set of minigenes derived from the wild-type lacZ where three codon groups were substituted by AGAAGAUAA codons so that a set of minigenes spanning from 3 to 8 codons including the AGAAGA tandem before the stop codon was generated. Thus, the shortest minigene produced the highest inhibition of β-gal paralleling the same tendency produced by the AGAAGA lacZ constructs suggesting that the same mechanism is underlying these phenomena. To further understand this mechanism, pep-tRNAArg4 accumulated by the AGAAGA lacZ constructs or the AGAAGAUAA minigenes was analysed. Interestingly, the accumulation profile of pep-tRNAArg4 was similar for both series. The percentages of pep-tRNAArg4 dropped steeply as the AGAAGA tandem moved from +2, +3 to +3, +4 for the AGAAGA lacZ constructs or from +3, +4 to +4, +5 in the case of the minigenes, thereafter, decreased slowly to a minimum for the +7, +8 AGAAGA lacZ construct or the +7, +8 AGAAGAUAA minigene. These results and the fact that AGAAGAUAA minigenes consumed more tRNAArg as indicated by the necessity to supplement the in vitro reaction with tRNA suggest that minigenes produce more pep-tRNA which makes them more toxic than the AGAAGA lacZ constructs. These results also indicated that the positional inhibition of the AGAAGA codons resembles the length-dependent inhibition of the minigenes as to the depletion of specific tRNA. The abrupt drop in the accumulation of pep-tRNA when the AGAAGA tandem is moved from +2, +3 to +3, +4 positions or when the minigene enlarges from 4 to 5 codons poorly correlated with the more gradual effects observed in protein synthesis experiments (Figures 1 and 2). However, the profile of pep-tRNA accumulation (including the low levels produced by the constructs containing AGAAGA at +3, +4 or later positions) should be produced specifically since no pep-tRNA was observed when the same constructs or minigenes were expressed in a wild-type Pth background (data not shown). That a tRNA-sequestering mechanism is also mediating the effect in the intermediate lacZ constructs or minigenes is supported by the fact that no inhibition in protein synthesis was observed when the same set of minigenes was expressed in a wild-type cell-free reaction or an inverted effect was produced in the case of the lacZ constructs (data not shown). This is equivalent to the Pth-defective background experiments where supplemental Pth also reverted the inhibitory effect of the AGAAGAUAA minigenes on the expression of the lacZ reporter gene (Figure 4B and D). In the case of the AGAAGA lacZ constructs, the expression of β-gal not only was reverted by Pth but the tendency was inverted in such a way that the closer the AGAAGA codons to the initiator, the higher the expression (compare Figures 1A and B with Figures 4A and C). A stimulatory effect of lacZ variants containing AGA and AGG codons at positions +2 and +3 on the expression of the lacZ reporter gene in wild-type bacteria was observed in a previous study (27). The same variants were deficient in gene expression in pth defective cells due to pep-tRNAArg4 accumulation and ribosome stalling at these codons. In addition, the expression in wild-type cells was comparable to that attained by the lacZ variant that carries +2, +3 AAAAAA, a codon configuration frequently found in the highly expressed genes of E. coli (34–36). The correlation of adenine-rich regions with efficient translation in E. coli was first noted by Dreyfus (37), and in later studies workers reported stimulatory effects on translation due to the addition of adenines (38–40). Several studies have analysed the promoting effect of adenine-containing codons downstream the initiation codon during translation initiation or in early elongation (27,34,35). It has even been proposed that the promoting effect is due to the direct binding of the mRNA with the 16S RNA (38,39). We favor the hypothesis that adenines close to the initiation codon promote an efficient binding of the mRNA which is eventually reflected in an efficient protein synthesis provided that tRNA is not exhausted. Thus, AGA and AGG codons containing adenines either promote the efficient translation when tRNA is available (in pth wild-type in vivo or in vitro systems) or lead to the inhibition of protein synthesis under limiting concentrations of tRNA (in pth defective systems or when the construct is over-expressed in a wild-type background). Thus, the inhibition of protein synthesis has been observed when the constructs containing rare AGA and/or AGG (hungry) codons are expressed under conditions where a specific tRNA becomes scarce (7,8) and the inhibition is reversed by tRNA (14,15). The inhibitory effect by AGA codons has also been observed when the constructs are expressed in Pth defective cells (3,15,27). In these cases, the translational down-regulation is mostly caused by a tRNA sequestering mechanism as indicates the reversion with the specific tRNA. On the other hand, the promoting effect by AGA or adenine-containing codons has been reported to occur under conditions where the pool of tRNA is not exhausted and the constructs are over-expressed in wild-type cells (27,38,39). For example, the lacZ construct containing AGAAGA at +2, +3 positions over expresses β-gal in wild-type cells but inhibits protein synthesis and cell growth in Pth defective cells and this effect is reverted by supplementing the cells with tRNAArg4 (27). In this work, a limited expression of β-gal is produced from the AGAAGA lacZ constructs in a Pth defective in vitro transcription–translation system. If the pool of tRNAArg4 were further reduced, β-gal synthesis from the AGAAGA constructs would even be lower than that expressed from wild-type lacZ (Figure 1, lanes a and b). Consequently, supplementation with Pth, a condition that increases the pool of aminoacylable tRNAArg4 inverses the effect in such a way that the closer the AGA codons to the initiation codon the stronger the synthesis of β-gal (Figure 4). Unpublished results (manuscript in preparation, Castillo Mendez et al.) indicate that not only AGA or AGG codons but in general adenine-containing codons (AAA, AUA, etc.) either enhance translation under conditions where the pool of tRNA is not exhausted or lead to the so called tRNA-sequestering mechanism when the cognate tRNA is scarce and/or Pth activity is defective. Thus, we propose that the adenine-containing transcripts compete efficiently with S30 subunits during initiation of translation conferring them advantage over other mRNAs. Under conditions where the pool of tRNA is not exhausted, the ribosomal complexes translate efficiently. However, if the adenines are part of rare codons (AUA, AGA) and/or the construct is expressed in a Pth-defective background, the pool of tRNA becomes exhausted and the tRNA-sequestering mechanisms is manifested. The pool of tRNA eventually becomes exhausted even in a wild-type background when a number of factors (high-copy plasmid, translational enhancer) contribute to optimize the level of expression of the construct. Thus, the rare codon effect resembles the adenine promoting effect because in both cases the effect is stronger (inhibition or stimulation depending on the levels of tRNA) the closer the adenine-containing codons are to the initiation codon.
The results presented here corroborate and extend the parallelism in the observed characteristics of mini-genes and hungry codons near the initiator (20–22). Like minigenes, the lacZ constructs containing AGA codons close to the initiation codon arrest protein synthesis in a Pth-defective cell-free system, accumulate pep-tRNA, and the inhibition is reverted by supplemental Pth. In addition, the positional effect of AGAAGA in the lacZ constructs is similar to the length-dependent effect of AGAAGAUAA minigenes in protein synthesis inhibition and pep-tRNA accumulation. This may be explained because the mechanisms underlying these effects, at least involve ribosome stalling, pep-tRNA accumulation and possibly ribosome recycling.
Finally, messengers containing clusters of AGA or CGA codons can be tagged by the SsrA system (42). An essential requirement for tagging at rare AGA codons is a scarcity for the cognate tRNA. Supplemental tRNAAGA suppresses tagging and depleting the available pool of tRNA enhances tagging and reveals tagging caused by single rare AGA codons. The results presented here indicate that the positional effect of the AGA codons in the synthesis of β-gal is not relevant for tagging, otherwise triggering of the tmRNA rescue system by the AGA rare codons would compete for the ribosomal complexes stalled at the AGA codons preventing the release of pep-tRNA. Whether the tmRNA rescue system is working in our system or not, the results presented here indicate that its participation does not greatly interfere with the tRNA-sequestering mechanism.
ACKNOWLEDGEMENTS
The authors wish to thank Marco A. Magos-Castro, José G. Bueno-Martínez and Guadalupe Aguilar González for skilful technical support. J.H.S. and G.G. were supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico). E.J.L. was recipient of loan-fellowships from CONACyT and Consejo del Sistema Nacional de Educación Technológica (COSNET). Funding to pay the Open Access publication charges for the article was provided by Centro de Investigación y de Estudios Avanzados.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ikemura T. Correlation between the abundance of E. coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 2.Dong H, Nilsson L, Kurland CG. Covariation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 3.Delgado-Olivares L, Zamora-Romo E, Guarneros G, Hernandez-Sanchez J. Codon-specific and general inhibition of protein synthesis by the tRNA-sequestering minigenes. Biochimie. 2006;88:793–800. doi: 10.1016/j.biochi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen K, Peters TC, Walker JR. A minor arginine tRNA mutant limits translation preferentially of a protein dependent on the cognate codon. J. Bacteriol. 1990;172:2504–2510. doi: 10.1128/jb.172.5.2504-2510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zdanovsky AG, Zdanovskaia MV. Simple and efficient method for heterologous expression of clostridial proteins. Appl. Environ. Microbiol. 2000;66:3166–3173. doi: 10.1128/aem.66.8.3166-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen GT, Inouye M. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 1994;8:2641–2652. doi: 10.1101/gad.8.21.2641. [DOI] [PubMed] [Google Scholar]
- 7.Chen GT, Inouye M. Suppression of the negative effects of arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the E. coli genes. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao W, Tyagi S, Kramer FR, Goldman E. Messenger RNA release from ribosomes during 5′-translational blockage by consecutive low-usage arginine but not leucine codons in Escherichia coli. Mol. Microbiol. 1997;25:707–716. doi: 10.1046/j.1365-2958.1997.5081871.x. [DOI] [PubMed] [Google Scholar]
- 10.Varenne S, Lazdunski C. Effect of distribution of unfavorable codons on the maximum rate of gene expression by a heterologous organism. J. Theor. Biol. 1986;120:99–110. doi: 10.1016/s0022-5193(86)80020-0. [DOI] [PubMed] [Google Scholar]
- 11.Robinson M, Lilley R, Little S, Emtage JS, Yarranton G, Stephens P, Millican A, Eaton M, Humphreys G. Codon usage can affect translation rate in Escherichia coli. Nucleic Acids Res. 1984;12:6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varenne S, Baty D, Verheij H, Shire D, Lazdunski C. The maximum rate of gene expression is dependent on the downstream context of unfavorable codons. Biochimie. 1989;71:1221–1229. doi: 10.1016/0300-9084(89)90027-8. [DOI] [PubMed] [Google Scholar]
- 13.Zahn K. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J. Bacteriol. 1996;178:2926–2933. doi: 10.1128/jb.178.10.2926-2933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zahn K, Landy A. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol. Microbiol. 1996;21:69–76. doi: 10.1046/j.1365-2958.1996.6201335.x. [DOI] [PubMed] [Google Scholar]
- 15.Olivares-Trejo JJ, Bueno-Martinez JG, Guarneros G, Hernández-Sanchez J. The pair of arginine codons AGA AGG close to the initiation codon of the lambda int gene inhibits cell growth and protein synthesis by accumulating peptidyl-tRNAArg4. Mol. Microbiol. 2003;49:1043–1049. doi: 10.1046/j.1365-2958.2003.03611.x. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez de Valdivia EI, Isaksson LA. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. FEBS J. 2005;272:5306–5316. doi: 10.1111/j.1742-4658.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- 17.Bergmann JE, Lodish HF. A kinetic model of protein synthesis: application to hemoglobin synthesis and translational control. J. Biol. Chem. 1979;254:11927–11937. [PubMed] [Google Scholar]
- 18.Liljenstrom H, von Heijne G. Translation rate modification by preferential codon usage: intragenic position effects. J. Theor. Biol. 1987;124:43–55. doi: 10.1016/s0022-5193(87)80251-5. [DOI] [PubMed] [Google Scholar]
- 19.Goldman E, Rosenberg AH, Zubay G, Studier FW. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J. Mol. Biol. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 20.Hernández-Sánchez J, Valadez JG, Vega-Herrera J, Ontiveros C, Guarneros G. λ bar minigene-mediated inhibition of protein synthesis involves accumulation of peptidyl-tRNA and starvation for tRNA. EMBO J. 1998;17:3758–3765. doi: 10.1093/emboj/17.13.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinçbas V, Heurgué-Hamard V, Buckingham RH, Karini R, Ehrenberg M. Shutdown in protein synthesis due to the expression of mini-genes in bacteria. J. Mol. Biol. 1999;291:745–759. doi: 10.1006/jmbi.1999.3028. [DOI] [PubMed] [Google Scholar]
- 22.Tenson T, Vega-Herrera J, Kloss P, Guarneros G, Mankin AS. Inhibition of translation and cell growth by minigene expression. J. Bacteriol. 1999;181:1617–1622. doi: 10.1128/jb.181.5.1617-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz-Vera LR, Hernandez-Ramon E, Perez-Zamorano B, Guarneros G. The rate of peptidyl-tRNA dissociation from the ribosome during minigene expression depends on the nature of the last decoding interaction. J. Biol. Chem. 2003;278:26065–26070. doi: 10.1074/jbc.M301129200. [DOI] [PubMed] [Google Scholar]
- 24.Cruz-Vera LR, Magos-Castro MA, Zamora-Romo E, Guarneros G. Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. Nucleic Acids Res. 2004;32:4462–4468. doi: 10.1093/nar/gkh784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Vera LR, Toledo J, Hernández-Sánchez J, Guarneros G. Molecular basis for the temperature sensitivity of Escherichia coli pth (Ts) J. Bacteriol. 2000;182:1523–1528. doi: 10.1128/jb.182.6.1523-1528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: a Laboratory Manual., Vol. 3, 2nd edn. p. A1. [Google Scholar]
- 27.Zamora-Romo E, Cruz-Vera LR, Vivanco-Domínguez S, Magos-Castro MA, Guarneros G. Efficient expression of gene variants that harbour AGA codons next to the initiation codon. Nucleic Acids Res. 2007;35:5966–5974. doi: 10.1093/nar/gkm643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zubay G. In vitro synthesis of protein in microbial systems. Annu. Rev. Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- 29.Mackie GA, Donly BC, Wong PC. Coupled transcription–translation of ribosomal proteins. In: Spedding G, editor. Ribosomes and Protein Synthesis, A Practical Approach. Oxford: IRL Press; 1990. pp. 191–211. [Google Scholar]
- 30.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNA in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 32.Gonzalez de Valdivia EI, Isaksson LA. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heurgue-Hamard V, Dinçbas V, Bukingham RH, Ehrenberg M. Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J. 2000;19:2701–2709. doi: 10.1093/emboj/19.11.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenstrom CM, Holmgren E, Isaksson LA. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 35.Stenstrom CM, Isaksson LA. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 36.Sato T, Terabe M, Watanabe H, Gojobori T, Hori-Takemoto C, Miura K. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. 2001;129:851–860. doi: 10.1093/oxfordjournals.jbchem.a002929. [DOI] [PubMed] [Google Scholar]
- 37.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 38.Brock JE, Paz RL, Cottle P, Janssen GR. Naturally occurring adenines within mRNA coding sequences affect ribosome binding and expression in Escherichia coli. J. Bacteriol. 2007;189:501–510. doi: 10.1128/JB.01356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Pomeroy-Cloney L, Bjerknes M, Tam J, Jay E. The influence of adenine rich motifs in the 3′ portion of the ribosome binding site on human IFN-gene expression in Escherichia coli. J. Mol. Biol. 1994;240:20–27. doi: 10.1006/jmbi.1994.1414. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Farmer JA, Janssen GR. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 1999;31:1025–1038. doi: 10.1046/j.1365-2958.1999.01228.x. [DOI] [PubMed] [Google Scholar]
- 41.Guarneros G, Guzman P, Garay E. Genetic and physical location of the Escherichia coli rap locus, which is essential for growth of bacteriophage lambda. J. Bacteriol. 1987;169:5188–5192. doi: 10.1128/jb.169.11.5188-5192.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauch MA, Spiegelman GB, Perego M, Johnson WC, Burbulys D, Hoch JA. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]