Abstract
p100 protein (SND1, Tudor-SN) is a multifunctional protein that functions as a co-activator for several transcription factors, has a role in mRNA processing and participates in RNAi-induced silencing complex (RISC) with yet unknown function. In this study we identified a novel function for p100 as a regulator of angiotensin II type 1 receptor (AT1R) expression. The binding of p100 to AT1R 3′-untranslated region (3′-UTR) via staphylococcal nuclease-like (SN-like) domains increased receptor expression by decreasing the rate of mRNA decay and enhancing its translation. Overexpression of p100 increased AT1R expression, whereas decrease in p100 binding to 3′-UTR either by p100 silencing or by the deletion of p100 binding site downregulated receptor expression. The effect of p100 through AT1R 3′-UTR was independent of Argonaute2 (Ago2), a known p100 partner, and was thus RISC-independent. Nucleotides 118 to 120 of the AT1R 3′-UTR were found to be critical for the binding of p100 to 3′-UTR. In summary, p100 is a multifunctional regulator of gene expression that regulates transcription, mRNA maturation, and as described in this article, also mRNA stability and translation.
INTRODUCTION
The control of mRNA stability is a potent regulatory mechanism, because small changes in mRNA half-life result in dramatic changes in the mRNA available for translation. Such posttranscriptional regulation can be achieved by different means including mRNA processing, nucleo-cytoplasmic export, mRNA localization, mRNA stabilization and translational regulation (1,2). These processes are mediated by conserved sequence elements located in the untranslated regions (UTRs) of the mRNA molecules together with specific mRNA-binding proteins (RBPs) that can bind to and affect mRNA properties (2,3).
Recently it was demonstrated that microRNAs (miRNAs) provide an additional posttranscriptional mechanism by which protein expression can be regulated. In animals, miRNAs usually control gene expression through partial complementary elements in the 3′-UTRs of their target mRNAs (4–8). miRNAs are loaded onto an Argonaute (Ago)-containing effector ribonucleoprotein complex, referred to as RISC (RNA-induced silencing complex), which is capable of recognizing cognate mRNAs and inhibiting protein expression. All four mammalian Ago proteins associate with miRNAs and are implicated in translational repression (9–11). However, only Ago2 can mediate specific endonucleolytic cleavage of a target mRNA (9,10,12).
Angiotensin II type 1 receptor (AT1R) is a G-protein-coupled receptor that mediates most of the known physiological actions of angiotensin II, including direct pressor response, renal tubular sodium transport and aldosterone secretion (13). Several recent studies suggest that AT1R expression level is principally regulated by posttranscriptional mechanisms (14–19), which are for the most part mediated by the 3′-UTR. When fused to a reporter gene, the AT1R 3′-UTR increases mRNA decay and decreases AT1R mRNA translation. This effect is mediated by both RNA-binding proteins (RBPs) and miR-155 (20–22), respectively.
During the course of the present study, we purified AT1R 3′-UTR-binding proteins from cytoplasmic extracts, using AT1R 3′-UTR as an affinity probe. One of these proteins was identified as p100 (SND1, Tudor-SN). p100 protein consists of four similar domains with homology to the staphylococcal nucleases (SN), followed by a C-terminal Tudor (TD) domain showing homology to the TD of Drosophila melanogaster tudor gene (23). The SN-like domains of p100 have been demonstrated to possess nuclease activity (23,24) and to mediate protein interactions (25). The TD is generally found in proteins with putative functions in RNA binding or protein binding during RNA metabolism and transport (26,27). p100 was originally identified as a co-activator for Epstein-Barr virus nuclear antigen 2 (EBNA-2) (28), but it was now found to be a multifunctional protein with a role in mRNA regulation.
Interestingly, p100 has been found also to be a part of RISC, but p100 function in this complex has remained unknown (24,29). In the present article we show that the p100 interaction with 3′-UTR leads to both stabilization and enhanced translation of AT1R mRNA and this effect is independent of RISC pathway.
EXPERIMENTAL PROCEDURES
Cell culture, luciferase assay and protein extraction
HEK293 and COS-1 cells were grown in DMEM that was supplemented with 10% fetal bovine serum (FBS), ampicillin/streptomycin and glutamine. Coronary artery vascular smooth cells (VSMCs) were purchased from Lonza Bioscience and cultured on smooth muscle growth medium-2 with 5% FBS, ampicillin/streptomycin and glutamine. Cells were used for 6–10 passages before replacement with fresh early passage stocks. Expression constructs were transiently transfected into HEK293 and COS-1 cells using a standard Fugene 6 protocol (Roche). Silencing RNA for p100 (SND1) was from Invitrogen. Other siRNAs and miRNAs including siAgo2 (siEIF2C2), negative control siRNAs, partially double-stranded RNAs that mimic endogenous precursor miRNAs, hsa-miR-155 and miRNA Negative Control #1, and also the anti-miR miRNA inhibitor anti-miR-155, designed to inhibit endogenous miR-155, were all obtained from Ambion. SiRNAs and miRNAs were transfected using Lipofectamine2000 reagent (Invitrogen) according to manufacturer's instructions. Cells were harvested 24–72 h after transfection and firefly luciferase activities were measured using the Dual Luciferase Assay System (Promega). The luciferase activity was normalized to the activity of the cotransfected renilla luciferase plasmid. Cytoplasmic protein lysates were prepared using TX100 lysis buffer (50 mM Tris, pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM EDTA, 1% TX100 and protease and phosphatase inhibitors). VSMC were transfected as described (30). For AT1R immunoprecipitation, proteins were extracted from 107 of VSMCs with 1% TX100, 0.5% SDS, 0.5% sodiumdeoxycholate. Protein concentrations were determined by Bradford assay (Rio-Rad).
Constructs
The coding region of human AT1R was amplified by a proofreading Pfu polymerase from a HeLa cDNA library, and sequenced to certify authenticity. All the luciferase and AT1R constructs were based on pGL3 (Promega) vector. 3′-UTR (nucleotides 1–887, respectively) was cloned into a XbaI–XbaI site downstream of the luciferase or AT1R coding region (in pGL3 vector luciferase was replaced with AT1R). A simple TATA promoter with an enhancer was added to upstream of the luciferase coding region and the SV40 late polyA signal was added downstream of the 3′-UTR sequence. Progressive deletions of AT1R 3′-UTR were generated using a PCR-based technique. Luciferase constructs expressing AT1R 3′-UTR mutants were generated by PCR-based amplification of a fragment followed by subcloning into 3′-UTR. The orientation and sequence of all constructs were confirmed by sequencing. The pSG5 vector expression plasmids containing full-length p100, SN-like domains (aa 1–639) or TD (aa 640–855) tagged with Flag sequence were generated as previously described (31).
Real-time PCR
In RNA half-life experiments, COS-1 cells were immersed in fresh media containing 1 μg/ml actinomycin D (Tocris) to inhibit transcription and incubated at 37°C. At the indicated times, cells were harvested and total cellular RNA was extracted with Nucleospin RNA II kit (Macherey-Nagel). RNA was treated with DNase I (Ambion). The first-strand cDNA was synthesized with Superscript II First Strand Synthesis System for RT-PCR and oligo(dT)12–18 primer (Invitrogen). Primer sequences used for PCR amplification of GAPDH and β-actin genes have been published earlier (32,33). AT1R oligos used were gcacaatgcttgtagccaaa (upstream) and ccctatcggaagggttgaat (downstream). After the first strand cDNA synthesis, serial dilutions were made. The PCR reactions were performed according to manufacturer's instructions. The annealing temperatures were 59°C for GAPDH oligos and 60°C for AT1R and β-actin oligos. The fluorescence emitted by SYBR Green I was measured in every cycle at the end of the elongation step. The second derivative maximum method was used to determine the crossing-point values. The concentrations of unknown samples were then calculated by setting their crossing points to the standard curve. For normalization of the expression levels, the expression of GAPDH or β-actin was measured. The relative expression level was obtained by dividing the values for the gene of interest with the GAPDH or β-actin value. In addition to the melting curve information obtained from the LightCycler, the PCR products were run in agarose gel electrophoresis to ensure that only a right-sized product was amplified.
MicroRNA isolation and detection
MicroRNAs were extracted by using mirVana miRNA Isolation Kit (Ambion). MirVana qRT–PCR miRNA Detection Kit with primer sets for miR-155 and loading control 5S rRNA was used for detection of isolated RNAs according to manufacturer's instructions (Ambion).
RNA probe preparation
cDNA was used as a template for PCR reactions whereby T7 RNA polymerase promoter sequence was added to the 5′-end of all fragments. RNA transcripts were synthesized from PCR-generated templates including the following 3′-UTR regions: 1–100, 1–300, 1–600, 1–857 (full length 3′-UTR) as well as from 100 to 300, 300 to 600, 600 to 857, 100–200, 150–250, 118–200, and 121–200 (MEGAscript, Ambion), Table 1. For producing polyadenylated RNA probes 30 bases long polyA tail was included in the 3′-oligos described above. RNA probes with or without biotin label (bio-11-UTP, Ambion) were generated by MEGAshortscript or MEGAscript kit according to manufacturer's instructions (Ambion). Transcription reaction was run at least for 2 h and then the products were run on denaturing PAGE. The gel was stained with SYBR Green II RNA gel stain (Molecular Probes, Invitrogen) and the band corresponding to the correct size was excised and eluted with 500 μl of 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% SDS followed by phenol-chloroform extraction and ethanol precipitation.
Table 1.
Primers used in the generation of PCR products that served as templates in in vitro transcription. Numbering starts from the beginning of 3′-UTR
| AT1R 3′-UTR sense primers (number denotes the first nucleotide of the 3′-UTR included in the oligo) | |
| 1 | TAATACGACTCACTATAGGG catgttc gaaacctgtc cataaagtaa ttttgtgaaa gaagg. |
| 100 | TAATACGACTCACTATAGGG gaattga aggagaaaat gcattatgtgg |
| 118 | TAATACGACTCACTATAGGG cattatgtg gactgaaccg acttttctaa |
| 121 | TAATACGACTCACTATAGGG tatgtg gactgaaccg acttttctaa |
| 150 | TAATACGACTCACTATAGGG tctgaac aaaagctttt ctttccttttgcaacaagac |
| 300 | TAATACGACTCACTATAGGG agcctgc ttttgtcctg ttatttttta tttccacata aagg |
| 600 | TAATACGACTCACTATAGGG gtataat ggtgttacta aagtcacata taaaagttaa ac |
| AT1R 3′-UTR antisense primers (number denotes the last nucleotide included in the oligo) | |
| 100 | ctgaaaagta gctaatgctc atttggtagt gaag |
| 200 | tgtggctttgctttgtcttgttgc |
| 250 | attcatcgagtttctgacattgttcttcgagcagcc |
| 300 | cagtaaaatt tctcaaatca acacattcat cgagtttc |
| 600 | attgttttgg cagtgtaaac ctataagaca caggttg |
| 857 | aaacaggaag gcatacttta tgatatataa tctttttaccac |
| Control (part of the luciferase coding region) | |
| S | TAATACGACTCACTATAGGG gcgccattctatccgctggaagatggaacc |
| AS | cgcccaacaccggcataaagaattgaagag |
RNA affinity purification
Two micrograms of in vitro transcribed polyadenylated RNA probes were incubated with polystyrene latex beads with dC10T30 oligonucleotides covalently linked to the surface (Oligotex, Qiagen), in a buffer containing 20 mM Tris–HCl, pH 7.5, 1 M NaCl, 2 mM EDTA and 0.2% SDS. After 2 washes in a wash buffer 10 mM Tris–HCl, pH 7.5, 150 mM NaCl and 1 mM EDTA, the RNA-coated beads were incubated with 100–250 μg cytoplasmic extracts in the RNA-binding buffer (5 mM Hepes, pH 7.9, 7.5 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiotreitol (DTT), 0.1 mg/ml yeast tRNA, 0.1 mg/ml bovine serum albumin, BSA). After incubation for 30 min on ice, the beads were extensively washed with RNA-binding buffer without BSA. Proteins bound to the RNA probe were resolved on 4–20% SDS–PAGE and subjected either to silver staining according to the manufacturer's instructions (Silver Stain Plus, Bio-Rad) or to western blotting.
Mass spectrometry
Silver stained protein bands of interest were cut out of the SDS–PAGE gel and ‘in-gel’ digested essentially as described by Shevchenko et al. (34). Proteins were reduced with DTT and alkylated with iodoacetamide before digestion with trypsin (Sequencing Grade Modified Trypsin, V5111, Promega). The recovered peptides were, after desalting using Millipore C18 ZipTipTM, subjected to matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometric analysis. MALDI-TOF mass spectra for mass fingerprinting and MALDI-TOF/TOF mass spectra for identification by fragment ion analysis were acquired using an Ultraflex TOF/TOF instrument (Bruker-Daltonik GmbH, Bremen, Germany). Protein identification with the generated data was performed using Mascot® Peptide Mass Fingerprint and MS/MS Ion Search programs (http://www.matrixscience.com).
Antibodies, immunoprecipitation (IP) and western blotting
Antibodies used in IPs and western blotting were as follows: polyclonal anti-p100 (C-17, Santa Cruz), polyclonal anti-Ago2 (Upstate), monoclonal anti-Flag (Sigma), polyclonal anti-Luciferase (Abcam), monoclonal anti-AT1R (TONI-1, Abcam), and monoclonal anti-β-tubulin (Upstate). IPs were performed with specific primary antibodies for 2 h at 4°C, followed by incubation with protein G Sepharose beads (Amersham Pharmacia Biotech) for 30 min at 4°C, and several washes with the TX100 lysis buffer. Affinity-purified proteins, IPs and cell lysates were electrophoresed in 4–20% SDS–PAGE and transferred into nitrocellulose membrane (Hybond ECL). Immunodetection was performed using specific antibodies, followed by incubation with biotinylated secondary antibodies (Dako Cytomation), streptavidin-biotin HRP-conjugated secondary antibodies (Amersham Biosciences), and enhanced chemiluminescence (ECL, Amersham).
Purification of Flag-tagged proteins
Immunoaffinity purification of Flag-tagged p100, SN-like, and TD proteins was performed with Flag affinity resin according to manufacturer's instructions (Sigma).
RNA electrophoretic mobility-shift assay (REMSA)
REMSA was performed incubating biotinylated probes together with immunoaffinity-purified p100-Flag, SN-Flag and TD-Flag proteins. In brief, 100 ng of different Flag-tagged proteins were incubated with 1 μg of 3′-UTR probe in 5 mM Hepes, pH 7.9, 7.5 mM KCl, 0.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 0.1 mg/ml yeast tRNA, 0.1 mg/ml BSA for 30 min on ice. The unbound probe was digested with RNAse A for 1 h at RT. Glycerol was added to the mixture and the samples were separated by 6% native PAGE with 0.5 × Tris borate-EDTA (TBE) running buffer. The gels were incubated in 5 × SSC/10 mM NaOH for 30 min, in 1 × TBS, pH 7.6 for 2 × 15 min and in 1 × TBE for 30 min. Finally the binding products were transferred from gels into BrightStar-Plus nylon membranes, followed by detection with BrightStar Biodetect (Ambion).
Protein IP for AT1R mRNA detection
The assay was performed essentially as described (35). In this assay, p100-Flag was immunoprecipitated from a cytoplasmic lysate prepared from 107 of COS-1 cells with the use of 1 μg of anti-Flag antibody or with unrelated IgGs as a negative control. The samples were first precleared with protein G Sepharose beads. p100-Flag was then immunoprecipitated after which protein G Sepharose beads were added. The IPs were extensively washed with 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% NP-40, 1 mM DTT and 100 U/ml RNasin RNAse inhibitor as well as 2 mM vanadyl ribonucleoside complexes solution (Sigma) and protease and phosphatase inhibitor cocktails. Proteins were digested with 0.1% SDS and 30 μg of proteinase K followed by RNA extraction with phenol-chloroform-isoamyl alcohol mixture. RNA was precipitated with 10 μg of yeast tRNA as a carrier. RT–PCR was then performed with poly-dT oligos, followed by PCR reactions with primers for coding region of AT1R mRNA or for β-actin for normalization (as described in real-time PCR section). The PCR products were separated on agarose gel.
In vitro translation
In vitro translation reactions were performed using Flexi Rabbit Reticulocyte Lysate System (Promega) as instructed by the manufacturer. Proteins synthesized in vitro were labelled with biotinylated lysine tRNA. A 100 ng of BSA control or immunoaffinity-purified p100 was added to each translation reaction. The results were evaluated by luciferase assay as well as by western blotting. In the western blot, the in vitro translation products were detected using streptavidin-HRP antibodies according to manufacturer's protocols (Promega).
35S labelling
For 35S labelling COS-1 cells were transfected either with p100-Flag, sip100 or with appropriate controls. After 72 h, 100 μCi/ml of 35S-methionine was added and the cells were incubated for additional 7 h. Cells were then lysed and subjected to luciferase IP with anti-luciferase antibody, followed by SDS–PAGE and autoradiography.
Ligand binding assay
AT1R binding was measured using subconfluent cells grown in 24-well plates. The assay was performed as described previously (36).
RESULTS
p100 interacts with AT1R 3′-UTR
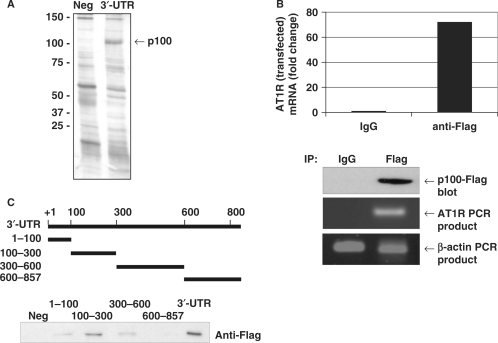
To search for proteins that bind to AT1R 3′-UTR, we performed protein separation from HEK293 cell lysates by RNA affinity purification with full-length AT1R 3′-UTR probe. The 100 kDa band, seen only in the lane corresponding to binding products with the 3′-UTR probe, was identified by mass spectrometry as p100 (Figure 1A). The association of AT1R mRNA with p100 was monitored by extracting RNA from the IP material and subjecting it to reverse transcription (RT) and quantitative real-time PCR analysis. Because the expression of AT1R is very low in COS-1 and HEK293 cells and because the commercial p100 antibodies did not immunoprecipitate p100, we transfected cells with p100-Flag together with a construct where AT1R coding region was fused to 3′-UTR, and made IPs with anti-Flag antibodies under conditions that retained the integrity of ribonucleoprotein (RNP) complexes. As shown in Figure 1B, the AT1R PCR product was dramatically enriched in p100 IP samples compared with control IgG IP samples. Also endogenous AT1R was co-precipitated with p100-Flag with 4-fold difference to control (data not shown). The amplification of β-actin PCR product was found in all samples and this low-level contaminating housekeeping transcript served to monitor the evenness of sample input. These data indicate that p100 forms RNP complex with AT1R mRNA.
Figure 1.
p100 interacts with AT1R 3′-UTR. (A) Search for AT1R 3′-UTR-binding proteins using affinity purification. The details of the purification are given under ‘Experimental Procedures’. A negative control probe (Neg) consisting of the segment of luciferase coding region and a probe corresponding to nucleotides 1–857 of AT1R 3′-UTR (3′-UTR) were incubated with HEK293 cell lysates and run into SDS–PAGE, which was then silver-stained. The differentially expressed 100 kDa protein was analysed by mass spectrometry. (B) Amplification of AT1R mRNA from p100 IPs. p100-Flag was immunoprecipitated with anti-Flag antibody or control IgG. RNA was extracted from the IPs, subjected to RT–PCR, followed by quantitative PCR analysis of AT1R and β-actin (loading control) mRNAs. The AT1R mRNA levels were normalized to β-actin mRNA levels. Results are expressed as a fold change compared to AT1R mRNA precipitated by IgG. Lower panels show control anti-Flag western blot and agarose gels with AT1R and β-actin PCR products. (C) p100 interacts with 100–300 of AT1R 3′-UTR. A schematic representation of different 3′-UTR transcripts used as probes in affinity purification reactions is shown. Probes were incubated with p100-Flag protein containing cell lysates. The results are shown as western blot demonstrating bound Flag-p100.
To identify the AT1R 3′-UTR region recognized by p100, we performed affinity purification assays with RNA probes corresponding to different regions of the 3′-UTR from p100-Flag overexpressing COS-1 cell lysates. We saw that the full-length 3′-UTR had the highest affinity for p100 and 100–300 region was sufficient for p100 binding (Figure 1C).
SN-like domains of p100 mediate the interaction with 3′-UTR and have a broader binding specificity than full-length p100
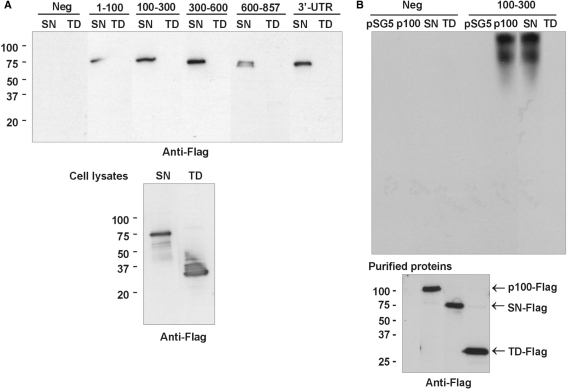
SN-like domains of p100 have been known as protein-interaction domains (25), and recently they were shown to mediate RNA interactions also (37). The TD is found in proteins with putative functions in RNA binding or protein binding (26,27). Next, we evaluated the interaction of SN-like domains and TD with 3′-UTR. To study the binding of different domains of p100 to 3′-UTR, we performed affinity purification reactions with different 3′-UTR transcripts. The anti-Flag western blot (Figure 2A, upper panel) demonstrated that SN-like domains mediate the interaction with 3′-UTR, whereas TD showed no binding. It was also seen that in addition to 100–300 region, being the principal interaction area for full-length p100, SN-like domains bind strongly also to 3′-UTR bases in the 300–600. It might be that although TD does not directly interact with the 3′-UTR, it is important for the binding specificity of p100.
Figure 2.
SN-like domains mediate the p100 interaction with 3′-UTR. (A) Mapping of the binding sites of SN-like domains and TD. The same 3′-UTR transcripts as in Figure 1C were used in the affinity purification reactions with cell lysates prepared from SN-like domains or TD overexpressing COS-1 cells. The results are shown as a western blot of Flag-tagged protein domains bound to transcripts (upper panel). Below is the western blot of Flag-tagged TD and SN-like domains from cell lysates. (B) REMSA. COS-1 cells were transfected with empty pSG5 vector, or with p100-Flag, SN-Flag, or TD-Flag expression constructs. Immunoaffinity-purified Flag-tagged proteins were incubated with biotinylated probes consisting of the segment of luciferase coding region (Neg, four first lanes) or 100–300 probe (four last lanes), as indicated. The unbound probe was digested by RNAase A. The relative levels of purified proteins used in REMSA are shown in anti-Flag western blot (bottom panel).
REMSA confirms the p100 interaction with 100–300 region of AT1R 3′-UTR
To confirm the binding of p100 and SN-like domains to 100–300 region, we performed electrophoretic mobility-shift assay (REMSA). We first purified TD and SN-like domains using GST system, but we did not observe any binding of either domains with AT1R 3′-UTR with this approach. The absence of interaction may be due to the labile nature of the p100-AT1R mRNA complex, improper folding of p100 domains in bacterial cells or lack of other binding proteins needed for the interaction. Due to these problems, we used affinity-purified Flag-tagged proteins. The proteins were incubated with biotinylated probes corresponding to negative control transcript or specific 100–300 transcripts. The unbound probe was digested and the interaction between the transcript and different domains or full-length p100 was assessed by REMSA. The negative control transcript did not show any gel shift in the absence or presence of p100, SN-like domains or TD (Figure 2B, upper panel). In contrast, the 100–300 fragment of AT1R 3′-UTR showed a strong shift with full-length p100 and SN-like domains. This result indicated that SN-like domains mediate the interaction of p100 with 100–300 region of 3′-UTR.
p100 and Ago2 co-immunoprecipitate and both bind to 100–300 region of AT1R 3′-UTR
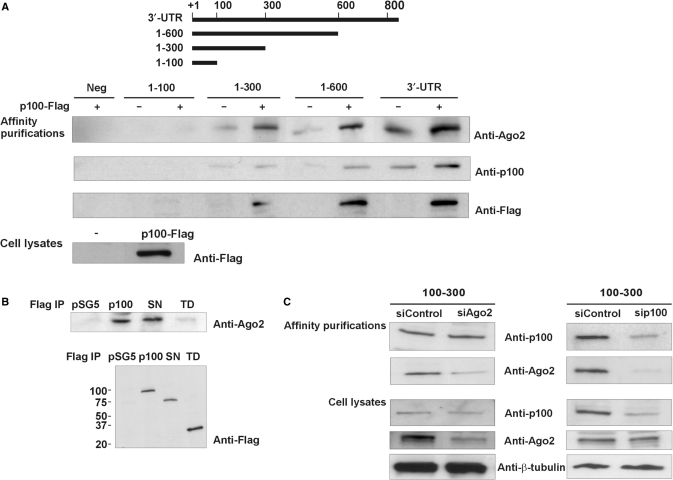
Because of the published interaction of p100 with Ago2, we tested for the presence of Ago2 in 3′-UTR affinity purifications. As shown in Figure 3A, Ago2 was found to affinity purify with the same set of probes as p100. In addition, the binding of Ago2 was further increased when p100 was overexpressed. Ago2 was also found to be present in immunoaffinity-purification reactions with full-length p100 and SN-like domains (Figure 3B, upper panel). Thus, it seems that Ago2 binds to AT1R 3′-UTR in complex with p100 and that SN-like domains are critical for this interaction.
Figure 3.
Ago2 co-purifies with p100. (A) Ago2 is present in same affinity purification reactions as p100. A schematic representation of different 3′-UTR transcripts used in affinity purification reactions is shown. In the first lane there is the control reaction where a region from luciferase coding region was used as a probe (Neg). The affinity purifications were performed with lysates from normal HEK293 cells or from HEK293 cells overexpressing p100-Flag. Western blots of proteins isolated by RNA affinity purification were done with anti-Ago2, anti-p100 and anti-Flag antibodies. The expression of p100-Flag was also monitored from cell lysates (bottom panel). (B) Ago2 and p100 interact. COS-1 cells were transfected with empty pSG5 vector or with p100-Flag, SN-Flag or TD-Flag expression constructs, followed by immunoaffinity purification. Co-precipitated Ago2 was detected by anti-Ago2 western blotting (upper panel). The relative levels of purified Flag-tagged proteins were detected by anti-Flag western blotting (lower panel). (C) Silencing of Ago2 has no effect on p100 binding to 100–300, whereas silencing of p100 inhibits Ago2 binding. COS-1 cells were transfected with negative control siRNA (30 nM) or with siRNA targeting Ago2 or p100 (30 nM). Corresponding cell lysates were affinity purified with a probe spanning 100–300 nucleotides of 3′-UTR. Western blots of the bound proteins with anti-p100 and anti-Ago2 antibodies are shown. Below are the control blots of Ago2, p100 and β-tubulin expression levels in cell lysates.
p100 binding to 100–300 region of AT1R 3′-UTR is Ago2 independent
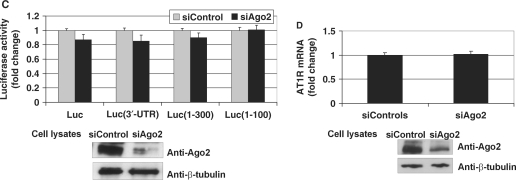
Given that p100 binds 3′-UTR in complex with Ago2, we next tested whether p100 binding to 100–300 region of AT1R 3′-UTR is due to the RNA binding of Ago2 or vice versa. For this purpose we silenced either Ago2 or p100 expression. We found that silencing of Ago2 expression did not affect the binding of p100 to the 100–300 region of 3′-UTR (Figure 3C, left panel). In contrast, silencing of p100 expression reduced Ago2 binding to this same region (Figure 3C, right panels). These results suggest that at least in the absence of RISC activation, Ago2 is recruited to AT1R 3′-UTR by p100.
RISC activation by miR-155 does not require the binding of p100 to AT1R 3′-UTR
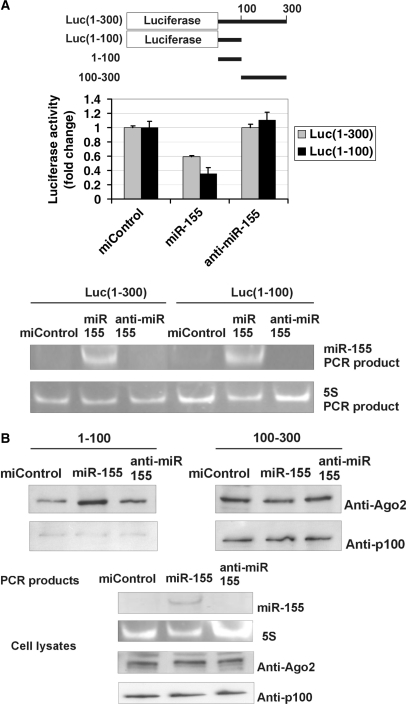
Taken into account the RISC-relatedness of published p100–Ago2 interactions, we next wanted to evaluate whether p100 had a function in miR-155-induced RISC activation. We could not detect expression of endogenous miR-155 in HEK293 (data not shown) or COS-1 cells (Figure 4A), indicating that the binding of p100 with 3′-UTR detected was not dependent on miR-155. MiR-155 binds to nucleotides 70–90 of AT1R 3′-UTR (22). We therefore analyzed the miR-155 effect on the luciferase construct with 100 or 300 first 3′-UTR nucleotides fused after luciferase coding sequence [Luc(1–100) and Luc(1–300), respectively]. While the transfection of miR-155 had no effect on the expression of luciferase 3′-UTR-less construct (data not shown), it attenuated the expression of both Luc(1–100) and Luc(1–300) (Figure 4A). The finding that miR-155 reduced also the expression of Luc(1–100) construct indicates that the area of p100 binding (100–300) is not required for miR-155 function and the formation of active RISC complex. Anti-miR-155 had no effect consistent with the lack of miR-155 in these cells.
Figure 4.
MiR-155 does not require p100 to function. (A) MiR-155 inhibits the activity of Luc(1–300) and Luc(1–100) constructs. A schematic representation of the luciferase constructs used in Figure 4A and 3′-UTR probes used in Figure 4B is shown. COS-1 cells were transfected with a Luc(1–300) construct with 1–300 nucleotides of 3′-UTR, or Luc(1–100) construct with 1–100 nucleotides of AT1R 3′-UTR and co-transfected with 30 nM of negative control miRNA (miControl), miR-155 or anti-miR-155 together with plasmid expressing renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The mean luciferase value from miControl-transfected cells was set to one. Below are the agarose gel figures showing the expression of miR-155 and 5S detected by PCR. (B) MiR-155 specifically increases the binding of Ago2 to 1–100 region of 3′-UTR, but not to 100–300 region. MiR-155 has no effect on p100 binding to either probe. COS-1 cells were transfected with miControl, miR-155 or anti-miR-155. Corresponding cell lysates were affinity purified by probes spanning 1–100 or 100–300 nucleotides of the 3′-UTR, as indicated. The binding results were analysed by western blot analysis with anti-Ago2 or anti-p100 antibodies (upper two panels). Below are the control gels illustrating the expression levels of miR-155 and 5S (PCR products in agarose gels), and Ago2 and p100 (western blots) in cell lysates.
As a further evidence for the formation of active RISC at 1–100 region, we observed a clear increase in the binding of endogenous Ago2 to 1–100 region after miR-155 transfection in subsequent affinity purification experiments (Figure 4B, left panels). In contrast, at the same time, there was no increase in binding of p100, pointing out that p100 binding to this UTR region is not induced by miR-155. These results imply that p100 is not a part of miR-155-induced functional RISC complex. To further control the specificity of p100 and Ago2 recruitment by miR-155, we used 100–300 transcript that lacks the miR-155 binding site. As was expected, neither the binding of p100 nor Ago2 was increased after addition of miR-155 (Figure 4B, right panels). These results indicated that the binding of p100 to 3′-UTR is independent of miR-155 and that p100 is not critical for miR-155 function. Thus p100 interaction with 3′-UTR is most probably not related to miR-155-induced RISC pathway. It appears that there exist two independent ways to recruit Ago2 to AT1R mRNA: the traditional miR-155-mediated way and p100-mediated way. At the light of our results these two ways are not competing but p100 creates an additional way to recruit Ago2 to receptor mRNA.
Overexpression of p100 increases expression of luciferase construct containing AT1R 3′-UTR
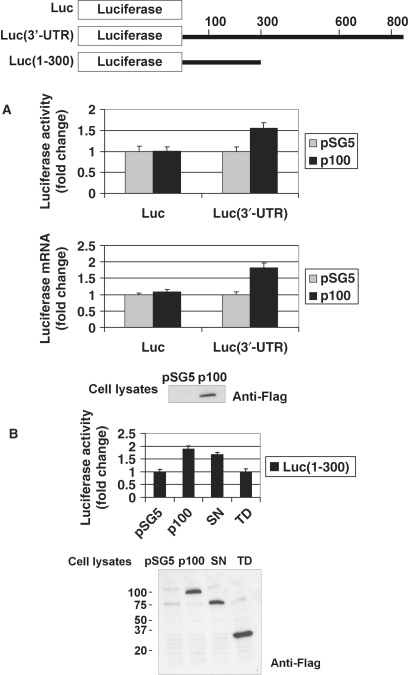
To assess the functional consequences of p100–3′-UTR interaction, cellular p100 levels were increased by overexpression in COS-1 cells and the role of p100 on the expression of the heterologous reporter bearing the AT1R 3′-UTR was examined. In this assay, the levels of both luciferase activity and mRNA expressed from the luciferase-AT1R 3′-UTR constructs were monitored. As shown in Figure 5A (first column), the abundance of luciferase synthesized from Luc(3′-UTR)-transfected cells increased after p100 overexpression. This elevation of protein expression was associated with increased levels of chimeric mRNA (Figure 5A second column). We also tested whether SN-like domains, which mediate the p100 interaction with 3′-UTR, were sufficient for p100 functional effects. When TD or SN-like domains were expressed together with luciferase construct, including the area showing strongest interaction with p100 (1–300 region), we saw that SN-like domains were functional alone, whereas the overexpression of TD had no effect (Figure 5B). Based on these results, p100 increases AT1R expression and this effect is mediated by SN-like domains. These data support the notion that p100 upregulates AT1R expression by increasing AT1R mRNA levels via 3′-UTR.
Figure 5.
Overexpression of p100 or SN-like domains increases the activity of luciferase construct containing AT1R 3′-UTR. A schematic representation of luciferase constructs used is shown. (A) Overexpression of p100 increases the expression of luciferase construct containing 3′-UTR. COS-1 cells were transfected either with a luciferase coding region (Luc) or with Luc(3′-UTR) construct where luciferase gene was fused with full-length AT1R 3′-UTR, and co-transfected with empty pSG5 vector, or pSG5 plasmid expressing p100-Flag, together with renilla luciferase. Cells were either lysed and assayed for firefly and renilla luciferase activities (upper column) or subjected to RNA extraction, followed by RT–PCR and quantitative luciferase-PCR (lower column). Results are shown as a mean of three independent experiments and SD. The values corresponding to transfections with Luc construct together with empty pSG5 vector was set to one. A representative anti-Flag western blot of p100-Flag expression is shown (bottom panel). (B) SN-like domains increase the activity of Luc (1–300) construct. COS-1 cells were transfected with Luc(1–300) construct together with empty pSG5 vector, or expression constructs for p100-Flag, SN-Flag or TD-Flag together with renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. Value corresponding to transfection with Luc(1–300) construct together with empty pSG5 vector was set to one. An anti-Flag western blot shows the relative expression levels of p100, SN-like domains and TD.
Downregulation of p100 expression decreases AT1R expression
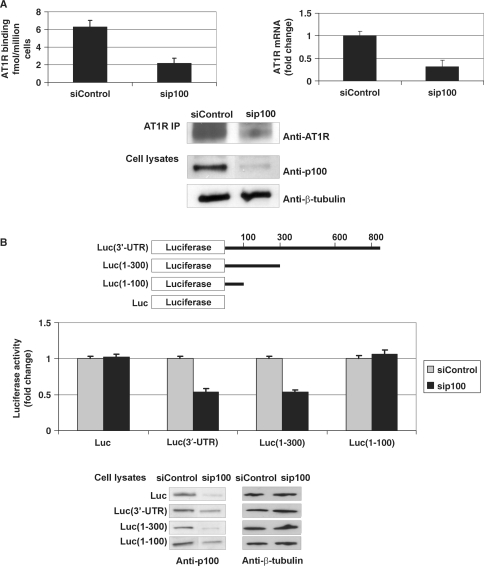
Next we used a siRNA-based intervention to reduce p100 expression levels in early-passage coronary artery VSMC. We found that p100 downregulation leads to a decline in endogenous AT1R levels as measured by ligand binding and western blot (Figure 6A). We also saw that sip100 reduced mRNA of endogenous AT1R in VSMCs. Consistent with these results, we observed that silencing of p100 caused about 50% reduction in the luciferase activity of Luc(3′-UTR) and Luc(1–300) constructs in COS-1 cells. Silencing of p100 had no effect on the 3′-UTR-less or Luc(1–100) constructs lacking the interaction with p100. Taken together, this data further implies that p100 increases AT1R expression through an interaction with 3′-UTR. These data also demonstrated that functional effects of p100 are similar in COS-1 cells and in VSMCs. At this point we tested whether Ago2 had functional effects on AT1R expression in the absence of miR-155. We observed that Ago2 depletion had no effect on receptor expression measured at the protein and mRNA level (Figure 6C and D). This suggests that Ago2 has no function of its own related to the regulation of AT1R expression but that the effects mediated by Ago2 require the presence of miR-155. In addition, these results indicate that the stabilizing effect on AT1R expression exerted by p100 does not require Ago2.
Figure 6.
Silencing of p100 expression decreases AT1R expression. (A) The effect of p100 silencing on AT1R expression in VSMCs. VSMCs were transfected with 30 nM siRNA for p100 (sip100) or equal amount of control siRNA (siControl). AT1R expression was then quantified by ligand binding (left column). Transfected VSMCs were also subjected to RNA extraction, followed by RT–PCR and quantitative AT1R-PCR (right column). Results are shown as a mean of three independent experiments and SD. The mean mRNA value of siControl-transfected cells was set to one. Endogenous AT1R was immunoprecipitated from VSMCs and detected by western blotting (upper panel). The expression levels of p100 and β-tubulin in VSMC lysates are also shown (lowest two panels). (B) Silencing of p100 reduces the activity of Luc(3′-UTR) and Luc(1–300) constructs. A schematic representation of different luciferase constructs used is shown. COS-1 cells were transfected with luciferase constructs with or without regions of 3′-UTR, as indicated, and co-transfected with 30 nM sip100 or siControl, together with plasmid expressing renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The mean value of each luciferase construct transfected together with siControl was set to one. Below are the western blots demonstrating the efficiency of the silencing of p100 (left panels) and β-tubulin control blots (right panels). (C) Silencing of Ago2 expression has no effect on luciferase constructs. This experiment was done as 6B but 30 nM siAgo2 was used instead of sip100. Results are shown as a mean of three independent experiments and SD. The mean value of each luciferase construct transfected together with siControl was set to one. Below are the representative western blots of Ago2 (upper panel) and β-tubulin (lower panel) expression. (D) The effect of Ago2 silencing on AT1R mRNA. COS-1 cells were transfected with 30 nM siAgo2 or equal amount of siControl. The cells were subjected to RNA extraction, followed by RT–PCR and quantitative AT1R-PCR. Results are shown as a mean of three independent experiments and SD. The mean mRNA value of siControl-transfected cells was set to one. The representative expression levels of Ago2 and β-tubulin in cell lysates are shown (lowest two panels).
Identification of the p100-binding site
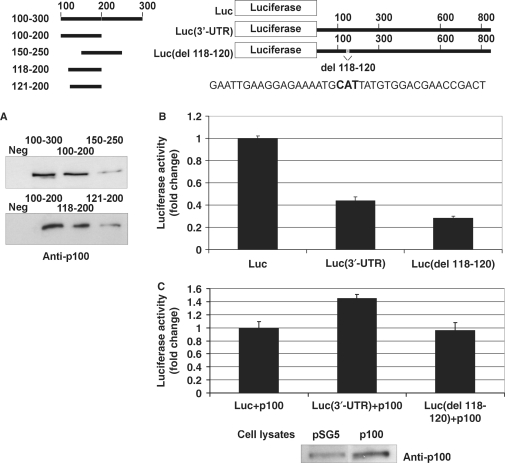
To test whether the effect of p100 depends on p100 binding to 3′-UTR, we mapped the binding site in more detail. For this purpose, we generated shorter probes of the 100–300 area and used them in affinity purification assay. We found that p100 requires 3′-UTR residues 118–120 for its binding (Figure 7A). The importance of this site for the function of 3′-UTR itself was tested in luciferase assays. Loss of p100 binding resulted in reduced luciferase activity (Figure 7B) and the construct became unresponsive to p100 overexpression (Figure 7C). These results further underline the importance of p100 binding for its functional effects.
Figure 7.
Mapping and deletion of the p100-binding site. A schematic of the probes used in the affinity purification and luciferase constructs used in luciferase assays is shown. The schematic also shows the sequence around 118–120 (118–120 shown in bold). (A) Nucleotides 118–120 of AT1R 3′-UTR are critical for p100 interaction. A negative control probe (from luciferase coding region; Neg) or different 3′-UTR probes were incubated with cell lysates and the binding products were analysed by western blot with anti-p100 antibodies. (B) Deletions of p100-binding site lead to reduced luciferase activity. COS-1 cells were transfected either with a luciferase coding region only (Luc), with Luc(3′-UTR), or with Luc(del 118–120) constructs, as indicated, and co-transfected with renilla luciferase. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The luciferase activity corresponding to transfection with Luc construct was set to one. (C) Deletion of p100 binding site (del 118–120). COS-1 cells were transfected with the same set of luciferase constructs as in (B), and co-transfected with empty pSG5 vector, or pSG5 plasmid expressing p100-Flag, together with renilla luciferase, as indicated. Cells were lysed and assayed for firefly and renilla luciferase activities. Results are shown as a mean of three independent experiments and SD. The value corresponding to transfection with Luc construct together with empty pSG5 vector was set to one. Below is the representative control western blot of p100 expression.
p100 stabilizes AT1R mRNA
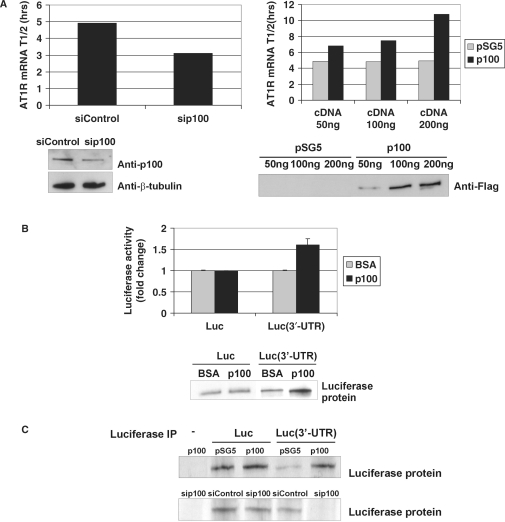
To ascertain if the increase in AT1R mRNA levels was due to changes in mRNA stability, the AT1R half-life was analysed following actinomycin D treatment to inhibit de novo transcription. The half-life of AT1R mRNA with 3′-UTR was 4.9 h, respectively, whereas the silencing of p100 expression resulted in the reduction of the mRNA half-life to 3.1 h (Figure 8A). We also studied the effect of p100 overexpression to AT1R mRNA half-life as a function of p100 protein level. The half-life of AT1R mRNA was increased to 6.8 h with 50 ng, to 7.5 h with 100 ng and to 10.8 h with the transfection of 200 ng of p100 cDNA. The half-life of the UTR-less AT1R or β-actin mRNA was not altered by depletion or by overexpression of p100 (data not shown). Thus, p100 affected AT1R mRNA levels by regulating its stability through specific binding to AT1R 3′-UTR.
Figure 8.
p100 regulates positively AT1R mRNA stability and translation. (A) p100 silencing and overexpression. COS-1 cells were transfected with AT1R 3′-UTR construct with 30 nM of siControl or sip100 (left column) or with increasing amounts of pSG5 vector or p100-Flag (right column). The half-life of the AT1R mRNA in each p100 expression level condition was first plotted as linear fit and the resulting half-lives are shown in columns. Below are the control western blots for p100 silencing, β-tubulin expression and p100-Flag overexpression. (B) p100 enhances translation. In vitro translation level of luciferase coding region only (Luc) or of Luc(3′-UTR) transcript were compared in the presence or absence of affinity-purified p100. BSA was used as a control protein. Luciferase activity was measured from the in vitro translation reaction mixture. The results represent the means and SD of an average of three independent experiments. The mean luciferase value of the Luc transcript incubated with BSA was set to one. Biotin-labelled translation products were also detected by blotting with streptavidin-HRP antibodies (lower panel). (C) 35S incorporation. COS-1 cells were transfected with Luc or Luc(3′-UTR) constructs (except sample in the first lane) and co-transfected with pSG5 vector, p100-Flag expression plasmid, siControl or sip100, as indicated. After incubation with 35S-methionine, cells were lysed and subjected to IP with anti-luciferase antibody. The immunoprecipitated proteins were detected by autoradiography. Results are shown both after p100 overexpression (upper panel) and p100 silencing (lower panel).
p100 increases AT1R mRNA translation
To examine the effect of p100 on the 3′-UTR-regulated translation, in vitro translation reactions were performed in the presence of BSA control or immunoaffinity-purified p100. When the luciferase activities of the translation products were measured, we found that the addition of p100 into the reaction together with RNA transcript in which luciferase coding region was fused with 3′-UTR, Luc(3′-UTR), we saw a clear up-regulation of luciferase translation by ∼50% (Figure 8B, upper panel). In contrast, when p100 was added to the reaction with 3′-UTR-less transcript (Luc), there was no effect, consistent with the interpretation that p100 interaction with the mRNA is essential for its translational effects. Biotin-labelled translation products were also separated by SDS–PAGE, electroblotted and detected by streptavidin-HRP with similar results (Figure 8B, lower panel).
A pulse incubation with 35S, followed by luciferase IP, revealed a decrease in luciferase translation from the Luc(3′-UTR) mRNA after p100 silencing and increase in translation after p100 overexpression (Figure 8C). Translation of mRNA lacking 3′-UTR was unaffected by the overexpression or silencing of p100 expression. These findings provide an additional demonstration that p100 enhances AT1R mRNA translation. Together, our findings support the notion that the posttranscriptional regulation of AT1R expression by p100 is elicited through altered stability and translation, respectively.
DISCUSSION
We found a novel RISC-independent mRNA regulatory mechanism for p100. Our findings support a model in which the SN-like domains of p100 enhance mRNA translation and repress mRNA degradation by binding to the 3′-UTR of AT1R. The effect of p100 on AT1R is mediated by specific 3′-UTR interaction as the deletion of p100-binding site and subsequent decrease in p100 binding removes p100 effects. Calreticulin (20), AUF1 (21) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; J.L. and K.P., unpublished results) bind either the proximal or the distal region of AT1R 3′-UTR, and p100 is thus so far the only protein described to bind to the 100–300 region. The p100 effect is independent of either Ago2 or miR-155 consistent with the RISC-independent mRNA regulation by p100. Several lines of evidence indicate that p100 function on AT1R 3′-UTR depends on the level of p100 binding. The effect of p100 increased when p100 was overexpressed, whereas a decrease in p100 binding to AT1R 3′-UTR either by p100 silencing or by the deletion of p100 binding site led to a decreased receptor expression. Thus, our results are consistent with p100 being a positive regulator of AT1R expression.
p100 was first identified as a co-activator for EBNA-2, and subsequently discovered as co-regulator for Pim-1, Signal transducer and activator of transcription 6 (STAT6) in interleukin-4-mediated gene regulation, and STAT5 in prolactin (PRL) signalling (25,28,31,38,39). The four SN-like domains of p100 resemble staphylococcal nucleases, which are small calcium-dependent enzymes that hydrolyze both DNA and RNA, and also SN-like domains of p100 have been shown to possess nuclease activity (23,24). The SN-like domains have been implicated also in protein interactions, and the SN-like domains of p100, for example, recruit CREB-binding protein and RNA polymerase II to STAT6 and facilitate the formation of STAT6 enhanceosome (25). In line with the function of SN-like domains in protein interactions, we found that these domains mediated the p100 interaction with Ago2. In accordance with recently published findings showing the ability of SN-like domains to mediate RNA binding (37), we identified that the p100 interaction with 3′-UTR and its functional effects related to interaction were mediated by SN-like domains. Whether the interaction between SN-like domains and 3′-UTR mRNA is direct or indirect remains uncertain. The REMSA experiment with GST fusion proteins did not show any binding suggesting that the interaction of p100 with AT1R 3′-UTR might be indirect. Recently, it was found that the TD mediates the interaction between p100 and small nuclear RNPs that function in pre-mRNA splicing, and that the TD accelerates the kinetics of the spliceosome assembly (40). Thus p100 is a novel regulator of gene expression that participates via distinct domains in both transcription and splicing. In our assays, we did not see any functional effect specific for TD. The TD appears to modulate the specificity of p100–3′-UTR interaction, since SN-like domains alone showed a slightly different pattern of interaction with 3′-UTR compared to the full-length p100.
As a further evidence for its multifunctional role, p100 has been indicated in degradation of edited primary transcripts of some miRNAs (41–43). p100 has been found also to be a part of RISC with yet unknown function (24,29). RISC is a multiprotein assembly consisting of proteins with different functions and is induced by miRNA binding to 3′-UTR (44,45). Since it is possible that protein–protein interactions might mediate p100–AT1R 3′-UTR interaction, we pursued the hypothesis that the binding of p100 to AT1R mRNA 3′-UTR would be mediated by RISC. Ago2, the central engine of RISC and a known partner of p100, did not mediate this interaction, since the depletion of Ago2 did not influence the p100 binding to 3′-UTR. The consequence of p100–Ago2 complex formation at 3′-UTR remains elusive. We probed the blots containing affinity purification reactions with antibodies against other well-known RISC components (46,47). We did not detect fragile-X-mental-retardation-related protein 1 or GW182 protein in the same complex with p100 and Ago2 (data not shown), implying that p100–Ago2 complex may no be a functional RISC. The silencing of Ago2 expression did not have any effect on AT1R expression at protein or mRNA level, indicating that Ago2 does not mediate the p100-induced effects on AT1R mRNA. Interaction of p100 with 3′-UTR or the p100-mediated effects do not depend on RISC as endogenous miR-155 was not present in COS-1 or HEK293 cells and that miR-155-induced RISC function did not require p100.
The p100 protein is an evolutionarily conserved protein present in the genomes of various eukaryotes (28,39,48–51). It appears that p100 is regulated at posttranscriptional level and that this regulation has also functional consequences. In our experiments p100 did not mediate the posttranscriptional regulation of AT1R in response to estrogen, angiotensin II, atorvastatin, or insulin. The increase in p100 protein expression during lactation is a common feature among mammals (52). Accordingly, our results showing that p100 protein levels are increased by PRL in mouse mammary epithelial cells further related increased p100 expression to lactation (38). Interestingly, rat p100 has been shown to increase in VSMCs in response to oxidative stress (53). This suggests that reactive oxygen species-activated expression of p100 may contribute to the VSMC growth and proliferation. In view of the upregulation of AT1R expression by both oxidative stress and by p100 overexpression, it will be interesting to study the relationship of p100 and the increase in AT1R expression induced by oxidative stress.
p100 is localized both in nucleus and cytoplasm and it regulates RNA processing in both these compartments. p100 regulates mRNA maturation, and as shown in this study, also mRNA stability and translation. Thus, p100 could potentially have a role in coordinating mRNA metabolism. Many other RBPs, such as embryonic lethal abnormal vision-like RNA-binding protein HuR, and alpha-globin poly(C)-binding protein have similar, multilayered effects on RNA (54–57). Linking nuclear and cytoplasmic controls by the action of a particular RBP may be of general importance and underscore the intricate connections that exist between RNA processing in different stages of mRNA life cycle, as we seek a better understanding of the molecular underpinnings of mRNA regulation.
ACKNOWLEDGEMENTS
We thank Ms Susanna Tverin, Ms Saara Nyqvist, Ms Hanna Nieminen and Ms Gunilla Rönnholm for expert technical assistance. This work was supported by grants from the Finnish Academy, the Finnish Foundation for Cardiovascular Research, the Sigrid Juselius Foundation (to K.K.) as well as the Finnish Medical Foundation, and Instrumentarium Research Foundation (to J.L.). Funding to pay the Open Access publication charges for the article was provided by Finnish Foundation for Cardiovascular Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 2.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol. Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 5.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 7.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 10.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 13.de Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol. Rev. 2000;52:415–472. [PubMed] [Google Scholar]
- 14.Nickenig G, Murphy TJ. Down-regulation by growth factors of vascular smooth muscle angiotensin receptor gene expression. Mol. Pharmacol. 1994;46:653–659. [PubMed] [Google Scholar]
- 15.Nickenig G, Murphy TJ. Enhanced angiotensin receptor type 1 mRNA degradation and induction of polyribosomal mRNA binding proteins by angiotensin II in vascular smooth muscle cells. Mol. Pharmacol. 1996;50:743–751. [PubMed] [Google Scholar]
- 16.Nickenig G, Sachinidis A, Michaelsen F, Bohm M, Seewald S, Vetter H. Upregulation of vascular angiotensin II receptor gene expression by low-density lipoprotein in vascular smooth muscle cells. Circulation. 1997;95:473–478. doi: 10.1161/01.cir.95.2.473. [DOI] [PubMed] [Google Scholar]
- 17.Nickenig G, Baumer AT, Grohe C, Kahlert S, Strehlow K, Rosenkranz S, Stablein A, Beckers F, Smits JF, Daemen MJ, et al. Estrogen modulates AT1 receptor gene expression in vitro and in vivo. Circulation. 1998;97:2197–2201. doi: 10.1161/01.cir.97.22.2197. [DOI] [PubMed] [Google Scholar]
- 18.Nickenig G, Roling J, Strehlow K, Schnabel P, Bohm M. Insulin induces upregulation of vascular AT1 receptor gene expression by posttranscriptional mechanisms. Circulation. 1998;98:2453–2460. doi: 10.1161/01.cir.98.22.2453. [DOI] [PubMed] [Google Scholar]
- 19.Nickenig G, Bohm M. Interaction between insulin and AT1 receptor. Relevance for hypertension and arteriosclerosis. Basic Res. Cardiol. 1998;93(Suppl. 2):135–139. doi: 10.1007/s003950050240. [DOI] [PubMed] [Google Scholar]
- 20.Nickenig G, Michaelsen F, Muller C, Berger A, Vogel T, Sachinidis A, Vetter H, Bohm M. Destabilization of AT(1) receptor mRNA by calreticulin. Circ. Res. 2002;90:53–58. doi: 10.1161/hh0102.102503. [DOI] [PubMed] [Google Scholar]
- 21.Pende A, Giacche M, Castigliola L, Contini L, Passerone G, Patrone M, Port JD, Lotti G. Characterization of the binding of the RNA-binding protein AUF1 to the human AT(1) receptor mRNA. Biochem. Biophys. Res. Commun. 1999;266:609–614. doi: 10.1006/bbrc.1999.1862. [DOI] [PubMed] [Google Scholar]
- 22.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J. Biol. Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 23.Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem. J. 1997;321(Pt 1):125–132. doi: 10.1042/bj3210125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 25.Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J. Biol. Chem. 2005;280:14989–14996. doi: 10.1074/jbc.M410465200. [DOI] [PubMed] [Google Scholar]
- 26.Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 27.Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 28.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Grifoni SC, Gannon KP, Stec DE, Drummond HA. ENaC proteins contribute to VSMC migration. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H3076–3086. doi: 10.1152/ajpheart.00333.2006. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. Embo J. 2002;21:4950–4958. doi: 10.1093/emboj/cdf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuda H, Fukabori Y, Nakano K, Shimizu N, Yamanaka H. Expression of bone morphogenetic protein-7 (BMP-7) in human prostate. Prostate. 2004;59:101–106. doi: 10.1002/pros.20030. [DOI] [PubMed] [Google Scholar]
- 33.Minami M, Daimon Y, Mori K, Takashima H, Nakajima T, Itoh Y, Okanoue T. Hepatitis B virus-related insertional mutagenesis in chronic hepatitis B patients as an early drastic genetic change leading to hepatocarcinogenesis. Oncogene. 2005;24:4340–4348. doi: 10.1038/sj.onc.1208628. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 35.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 36.Akishita M, Ito M, Lehtonen JY, Daviet L, Dzau VJ, Horiuchi M. Expression of the AT2 receptor developmentally programs extracellular signal-regulated kinase activity and influences fetal vascular growth. J. Clin. Invest. 1999;103:63–71. doi: 10.1172/JCI5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 2008;36:3579–3589. doi: 10.1093/nar/gkn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 39.Leverson JD, Koskinen PJ, Orrico FC, Rainio EM, Jalkanen KJ, Dash AB, Eisenman RN, Ness SA. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, Frilander MJ, Silvennoinen O. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 2007;35:4485–4494. doi: 10.1093/nar/gkm470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connell MA, Keegan LP. Drosha versus ADAR: wrangling over pri-miRNA. Nat. Struct. Mol. Biol. 2006;13:3–4. doi: 10.1038/nsmb0106-3. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 44.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda K, Satoh M, Pauley KM, Fritzler MJ, Reeves WH, Chan EK. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J. Immunol. Methods. 2006;317:38–44. doi: 10.1016/j.jim.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchiya N, Ochiai M, Nakashima K, Ubagai T, Sugimura T, Nakagama H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007;67:9568–9576. doi: 10.1158/0008-5472.CAN-06-2707. [DOI] [PubMed] [Google Scholar]
- 49.Keenan TW, Winter S, Rackwitz HR, Heid HW. Nuclear coactivator protein p100 is present in endoplasmic reticulum and lipid droplets of milk secreting cells. Biochim. Biophys. Acta. 2000;1523:84–90. doi: 10.1016/s0304-4165(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 50.Porta A, Colonna-Romano S, Callebaut I, Franco A, Marzullo L, Kobayashi GS, Maresca B. An homologue of the human 100-kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem. Biophys. Res. Commun. 1999;254:605–613. doi: 10.1006/bbrc.1998.9894. [DOI] [PubMed] [Google Scholar]
- 51.Palacios L, Ochoa B, Gomez-Lechon MJ, Castell JV, Fresnedo O. Overexpression of SND p102, a rat homologue of p100 coactivator, promotes the secretion of lipoprotein phospholipids in primary hepatocytes. Biochim. Biophys. Acta. 2006;1761:698–708. doi: 10.1016/j.bbalip.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Broadhurst MK, Wheeler TT. The p100 coactivator is present in the nuclei of mammary epithelial cells and its abundance is increased in response to prolactin in culture and in mammary tissue during lactation. J. Endocrinol. 2001;171:329–337. doi: 10.1677/joe.0.1710329. [DOI] [PubMed] [Google Scholar]
- 53.Sakamoto K, Yamasaki Y, Kaneto H, Fujitani Y, Matsuoka T, Yoshioka R, Tagawa T, Matsuhisa M, Kajimoto Y, Hori M. Identification of oxidative stress-regulated genes in rat aortic smooth muscle cells by suppression subtractive hybridization. FEBS Lett. 1999;461:47–51. doi: 10.1016/s0014-5793(99)01419-2. [DOI] [PubMed] [Google Scholar]
- 54.Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji X, Kong J, Carstens RP, Liebhaber SA. The 3′ untranslated region complex involved in stabilization of human alpha-globin mRNA assembles in the nucleus and serves an independent role as a splice enhancer. Mol. Cell. Biol. 2007;27:3290–3302. doi: 10.1128/MCB.02289-05. [DOI] [PMC free article] [PubMed] [Google Scholar]