Abstract
The WW domain-containing oxidoreductase (WWOX) gene encodes a tumor suppressor. We have previously shown that targeted ablation of the Wwox gene in mouse increases the incidence of spontaneous and chemically induced tumors. To investigate WWOX function in vivo, we examined Wwox-deficient (Wwox-/-) mice for phenotypical abnormalities. Wwox-/- mice are significantly reduced in size, die at the age of 2-3 weeks, and suffer a metabolic disorder that affects the skeleton. Wwox-/- mice exhibit a delay in bone formation from a cell autonomous defect in differentiation beginning at the mineralization stage shown in calvarial osteoblasts ex vivo and supported by significantly decreased bone formation parameters in Wwox-/- mice by microcomputed tomography analyses. Wwox-/- mice develop metabolic bone disease, as a consequence of reduced serum calcium, hypoproteinuria, and hypoglycemia leading to increased osteoclast activity and bone resorption. Interestingly, we find WWOX physically associates with RUNX2, the principal transcriptional regulator of osteoblast differentiation, and on osteocalcin chromatin. We show WWOX functionally suppresses RUNX2 transactivation ability in osteoblasts. In breast cancer MDA-MB-242 cells that lack endogenous WWOX protein, restoration of WWOX expression inhibited Runx2 and RUNX2 target genes related to metastasis. Affymetrix mRNA profiling revealed common gene targets in multiple tissues. In Wwox-/- mice, genes related to nucleosome assembly and cell growth genes were down-regulated, and negative regulators of skeletal metabolism exhibited increased expression. Our results demonstrate an essential requirement for the WWOX tumor suppressor in postnatal survival, growth, and metabolism and suggest a central role for WWOX in regulation of bone tissue formation.
WW domain-containing oxidoreductase (WWOX)3 is a 46-kDa protein that contains two N-terminal WW domains and a central short-chain dehydrogenase/reductase domain (1, 2). WWOX was identified as a putative tumor suppressor in cancer cells because it lies in a genomic region that is frequently altered in pre-neoplastic and neoplastic lesions (1, 2). Indeed, expression of WWOX is deregulated in several types of cancer, including breast, prostate, lung, stomach, and pancreatic carcinomas (3, 4). Ectopic expression of WWOX in cancer cells lacking expression of endogenous WWOX results in significant growth inhibition and prevents the development of tumors in athymic nude mice (5, 6). Recently, we generated a mouse carrying a targeted deletion of the Wwox gene (7). We reported that loss of both alleles of Wwox resulted in the formation of frequent juvenile osteosarcomas, whereas loss of one allele increased the incidence of spontaneous and chemically induced tumors (7, 8) thus confirming that Wwox is a bona fide tumor suppressor.
The identification of WWOX-interacting proteins has provided insights into the potential roles of WWOX in cell signaling and its impact on cell fate. WWOX cytosolic interactions, through its first WW domain that binds PPXY ligand-containing proteins, regulate the transactivation activity of several transcription factors. For example, we recently showed that WWOX interacts with p73 and suppresses its transactivation activity (9). However, this association contributes to the increased rate of WWOX pro-apoptotic activity. We also reported that the association of WWOX with the AP2γ transcription factor and the ErbB4 intracellular domain might contribute to breast carcinogenesis (10, 11). In addition, WWOX antagonizes the function of Yes-associated protein (YAP), another WW domain-containing protein, and suppresses its co-activation ability of ErbB4-dependent transcription. WWOX physically associates with c-Jun following ultraviolet radiation and functionally suppresses its transcriptional ability (12). Other research groups have shown that WWOX associates with proline-rich motifs of SIMPLE (13) and EZRIN (14) and with proteins lacking the PPXY motif such as p53 and c-Jun N-terminal kinase (JNK) (15). Thus, although WWOX is involved in specific protein complexes that define its function as an important suppressor of transactivator functions, its specific function in vivo is not clearly defined.
To investigate in vivo requirements for WWOX, we examined the Wwox-null mice for phenotypical abnormalities. Wwox-deficient mice display many postnatal defects that include growth retardation, postnatal lethality, and abnormalities of bone metabolism.
EXPERIMENTAL PROCEDURES
Mice—C57Bl/6J × 129/SvJ-F1,-F2,-F3,-F4, and -F5 mice (B6-129 F1-F5; (7)) were produced at Ohio State University animal facility. Animals were sacrificed; tissues of all organs were removed, fixed in 10% buffered formalin, and examined for histological abnormalities by two pathologists after hematoxylin and eosin staining. Bones were cleaned by separation from soft tissue (skin, muscle) leaving the periosteum intact for radiograph and μCT studies.
Histology and LacZ Staining—Tissue from different organs were processed, embedded, sectioned (4 μm), and hematoxylin and eosin-stained according to standard methods. Bones were dissected for fixation in 4% paraformaldehyde for 48 h and either demineralized in 18% EDTA for paraffin embedding or embedded in methyl methacrylate for examination of sections of mineralized tissues. Sections were stained for mineral and matrix using the von Kossa 3% silver nitrate stain combined with toluidine blue to distinguish cartilage and bone. Alkaline phosphatase and tartrate-resistant acid phosphatase enzyme detection for demonstrating osteoblast and osteoclast activities, respectively, was performed using reagent kits from Sigma. Because our targeting construct included the lacZ gene (7), whole mount β-galactosidase staining was performed using standard procedures (16). Prior to preparing frozen sections from E17.5 embryos, limbs (femur with tibia) were removed for separate embedment. Frozen sections of stained embryos and limbs were refixed in 0.5% glutaraldehyde and restained in X-gal solution and serial sections counterstained in 0.5% eosin after dehydration (50% alcohol). Immunochemistry for WWOX was performed using WT limb sections of newborn mice with WWOX polyclonal anti-rabbit antibody (a gift from Dr. Kay Huebner, Ohio State University) at 1:1000 dilution (7).
Bone Analyses—Skeletons were prepared for visualization of cartilage and bone using Alcian blue and alizarin red stains, respectively. Briefly, eviscerated embryos were fixed in 100% ethanol, stained overnight in a solution containing 4 parts ethanol, 1 part glacial acetic acid, and 0.3 mg/ml Alcian blue 8GX (Sigma). Soft tissues were dissolved for 6 h in a 2% KOH followed by an overnight staining in a 1% KOH solution with 75 μg/ml alizarin red S (Sigma). Skeletons are destained in 20% glycerol, 1% KOH for several days and stored in 50% glycerol, 50% ethanol. Radiography of dissected limbs at ages 7, 12, 17, and 20 days was performed after fixation in 4% paraformaldehyde at 4 °C under vacuum 2 days and rinsing in PBS using a Faxitron MX-20 specimen radiography system. Microcomputed tomography studies were performed on limbs fixed in 70% ethanol for scanning. Qualitative and quantitative three-dimensional analysis of femurs was carried out using micro-CT imaging (μCT 40, Scanco Medical AG, Bassersdorf, Switzerland) at the University of Connecticut Health Science Center μCT Facility by Dr. Douglas J. Adams.
Calvarial Osteoblast Differentiation—Primary calvarial osteoblasts were isolated from postnatal day 3 wild-type and homozygous KO pups following collagenase P digestion, and cultured in α-minimal essential medium supplemented with 10% FBS, as described previously (17). For osteogenic differentiation of both primary osteoblast and MC3T3 cells, stimulus was provided at confluence by subsequent feedings with the BGJb medium containing 10 mm β-glycerol phosphate (BGP04) and 50 μg/ml ascorbic acid. For histochemical staining, cell layers were fixed in 2% paraformaldehyde before staining, and reagents used were from Sigma. Briefly, cells were stained with 0.05% naphthol AS-Mx phosphate disodium salt, 2.8% dimethyl formamide,. and 0.1% Fast Red TR salt in 0.1 m Tris maleate buffer (pH 8.4) at 37 °C for 30 min.
Osteoclast Differentiation—Bone marrow cells were prepared from the long bones of 7-9-week-old BALB/c mice. Marrow was flushed from bones with a 30-gauge syringe, and cells were dispersed through a metal filter. 1 × 105 cells were plated in each well of a 12-well plate and incubated at 37 °C in α-minimum Eagle's medium supplemented with 10% FBS, 2 mm l-glutamine, 1% (v/v) penicillin/streptomycin, and 25 ng/ml macrophage colony-stimulating factor (R & D Systems, Minneapolis, MN) for 3 days before addition of recombinant RANKL (5 ng/ml). RAW264.7 cells (a kind gift from Philip Osdoby (Washington University, St. Louis, MO) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mm l-glutamine, and 1% (v/v) penicillin/streptomycin and plated in 6-well (2-4 × 105 cells/well) or 100-mm plates (1 × 106 cells). Cells were allowed to settle 2 days after plating before addition of RANKL (5 ng/ml). After differentiated osteoclasts were observed, mononuclear cells were shaken off and cell layers rinsed in PBS. Mononuclear cells (controls in which no RANKL was added) and the enriched multinucleated osteoclasts on the plates were harvested in TRIzol for mRNA isolation according to manufacturers' procedure (Invitrogen).
Cell Lines, Transfection, and Reporter Assays—NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium and MC3T3 in a growth medium of α-minimum Eagle's medium (Invitrogen) supplemented with 10% FBS (Sigma). Transient transfections were performed at subconfluency using FuGENE 6 transfection reagent (Roche Applied Science). The RUNX2 and WWOX expression vector (pCMV-MYC-WWOX; pCMV-MYC-WWOXY33R; pcDNA3.1-HA-RUNX2) were used in this study. For control of expression, vector pcDNA3.1 was transfected according to the experimental conditions. Previously described constructs (18) containing the rat osteocalcin (Oc) (-1097/+23 or -208/+23) promoter fused to the chloramphenicol acetyltransferase (CAT), as well as the Oc promoter with all three RUNX sites mutated (-1.1-kb rOc mABC-CAT) were co-transfected with expression plasmids.
Real Time PCR—RNA was isolated from the different tissues followed by homogenizing in TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Bones were cut at the mid-diaphysis and flushed free of marrow by PBS after removing the epiphyses from either end. The bone halves were placed and frozen in TRIzol for RNA isolation by homogenization. cDNA was synthesized with oligo(dT) primers using the Super-Script first strand synthesis kit (Invitrogen) according to the manufacturer's protocol. Gene expression was assessed by semi-quantitative and quantitative real time PCR (Cyber Green and TaqMan) using primers listed in Table 1.
TABLE 1.
Nucleotide sequence of primers used for quantitative reverse transcription-PCR detection
For indicates forward and Rev indicates reverse.
| Gene | Primer Sequence |
|---|---|
| Runx2 | For 5′ CGC CCC TCC CTG AAC TCT 3′ |
| Rev 5′ TGC CTG CCT GGG ATC TGT A 3′ | |
| BSP | For 5′ GCA CTC CAA CTG CCC AAG A 3′ |
| Rev 5′ TTT TGG AGC CCT GCT TTC TG 3′ | |
| Col I | For 5′ GTA TCT GCC ACA ATG GCA CG 3′ |
| Rev 5′ CTT CAT TGC ATT GCA CGT CAT 3′ | |
| Osteocalcin | For 5′ CTG ACA AAG CCT TCA TGT CCA A 3′ |
| Rev 5′ GCG CCG GAG TCT GTT CAC TA 3′ | |
| Alkaline phosphatase | For 5′ TTG TGC GAG AGA AAG GAG A 3′ |
| Rev 5′ GTT TCA GGG CAT TTT TCA AGG T 3′ | |
| Histone H4 | For 5′ CCA GCT GGT GTT TCA GAT TAC A 3′ |
| Rev 5′ ACC CTT GCC TAG ACC CTT TC 3′ | |
| Wwox | For 5′ TCA CAC TGA GGA GAA GAC CCA 3′ |
| Rev 5′ CCT ATT CCC GAA TTT GCT CCA 3′ | |
| Osteocalcin (human) | For 5′ GGC AGC GAG GTA GTG AAG AG 3′ |
| Rev 5′ CGA TAG GCC TCC TGA AAC TC 3′ | |
| VEGF (human) | For 5′ CCT TGC TGC TCT ACC TCC AC 3′ |
| Rev 5′ CCA TGA ACT TCA CCA CTT CG 3′ | |
| RUNX2 (human) | For 5′-CGG CCC TCC CTG AAC TCT-3′ |
| Rev 5′-TGC CTG CCT GGG GTC TGT A-3′ |
Immunoprecipitation and Western Blot Analysis—Immunoprecipitation was performed as described previously (9). Antibodies used were monoclonal anti-WWOX, monoclonal or polyclonal anti-RUNX-2, anti-MYC and anti-HA (Santa Cruz Biotechnology), anti-actin (Santa Cruz), and Lamin B1 (Zymed).
Chromatin IP (ChIP) Assay—For identifying association of RUNX2 and WWOX on chromatin of a RUNX2 target gene osteocalcin, the mouse MC3T3-E1 osteoblastic cells at 30 to 50% confluence were transfected using Oligofectamine (Invitrogen) with small interfering RNA (siRNA) duplexes specific for murine Runx2 gene-specific siRNA ((5′-r(UGC CUC UGC UGU UUG AAA) d(TT)-3′) and nonspecific siRNA (5′-r(UUC UCC GAA CGU GUC ACG U) dTdT-3′) duplexes for 48 h, obtained from Qiagen Inc. (Stanford, CA). Total proteins from the specific siRNA oligonucleotide-treated and nonspecific-oligonucleotide-treated cells were analyzed by Western blotting to evaluate the RUNX2 knockdown profile. After Runx2 siRNA treatment, cells were cross-linked with 1% form-aldehyde, and soluble chromatin fractions were immunoprecipitated with RUNX2 rabbit polyclonal (3 μg) and WWOX rabbit polyclonal (3 μg) antibody. The procedure for ChIP in osteoblasts has been described (19). DNA fragments from immunoprecipitates were analyzed by quantitative real time PCR using 2× SYBR Green Master Mix (Eurogentec) and a two-stage cycling protocol (60 °C annealing and extension, 94 °C denaturation, 40 cycles). The following primers were used to amplify the proximal mouse osteocalcin (mOC) promoter (mOC-F1 5′-CCC TCA GGG AAG AGG TCT G-3′ and mOC-R1 5′-CTA ATT GGG GGT CAT GTG CT-3′).
Affymetrix Chip Analysis—mRNAs from 26 samples (including kidney, spleen, brain, pituitary gland, femur, and calvarial bones) were hybridized with Affymetrix mouse gene-chip 430 2.0 arrays. Normalization was performed by using GC robust multiarray average. Genes showing minimal variation across the set of arrays were excluded from the analysis. Genes whose expression differed by at least 1.5-fold from the median in at least 20% of the arrays were retained. For class comparison, we identified genes that were differentially expressed between KO and WT mice by using a multivariate permutation test (www.linus.nci.nih). In comparing the genotypes we controlled for the analyzed tissues, we used the multivariate permutation test to provide 90% confidence that the false discovery rate was less than 10%. The false discovery rate is the proportion of the list of genes claimed to be differentially expressed that are false positives. The test statistics used are random variance F-statistics for the effect of genotype on each gene. Although F-statistics were used, the multivariate permutation test is nonparametric and does not require the assumption of a gaussian distribution. The summarized cDNAs presented in supplemental Table 4 represent those up- and down-regulated genes that were common to all analyzed tissues.
RESULTS
Wwox Is Essential for Postnatal Survival—To characterize the role of the Wwox gene in vivo, B6-129 F1-F5 hybrid mice were examined for phenotypical abnormalities. Genotype analysis of newborns obtained from a Wwox+/- intercross demonstrated the presence of all three genotypes with ratios consistent with the Mendelian distribution (7). Wwox heterozygous (HET) pups were indistinguishable from wild-type (WT) animals at all stages of development and postnatal life. At birth, homozygous Wwox-null pups were indistinguishable from their WT or HET littermates up to 3 days postpartum; subsequently, homozygous (KO) pups were easily recognizable by their smaller size (Fig. 1A). KO pups continued to grow more slowly than their littermates, and 100% of the KO mice died by 3 weeks after birth (Fig. 1B). Although homozygous mice were runted, they did not exhibit any abnormal behavior or impaired motor skills. All KO pups showed normal suckling behavior, and milk was found in their stomachs. At 2 or 3 days prior to their death, Wwox-null pups became lethargic and showed signs of wasting.
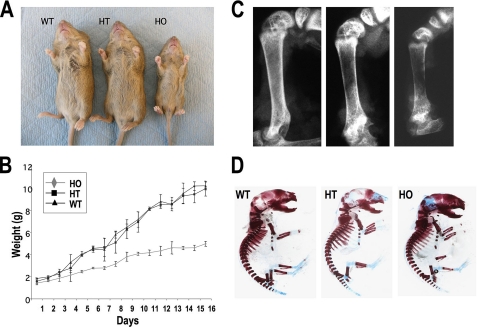
FIGURE 1.
Phenotypic analysis of Wwox null mice. Impaired growth is shown in A. Three-week-old mice showing significant small size of KO mouse compared with WT and HET mice. B, growth curves of WT, HET, and KO mice. Newborn pups were weighed every day from day 1. Mice were then genotyped, and weights were plotted on growth curve. The weights of KO mice diverge after day 3 from that of the WT and HET mice. HT, heterozygous. C, radiography of femurs of 3-week-old mice showing smaller but also less dense bone of the homozygous (HO) Wwox-/- mouse. D, alizarin red/Alcian blue staining of newborn skeletons.
Consistent with growth retardation, key organs, including spleen, thymus, brown adipose tissue, heart, and liver, weighed less in both female and male KO pups compared with wild-type littermates as measured on postnatal day 14 (supplemental Table 2). Interestingly, the brain, adrenal and the pituitary gland of KO pups were heavier compared with WT littermates. Macroscopic and histological examination of the organs confirmed the atrophy of many organs in KO animals without significant microscopic lesions, though KO mice are born with gonadal abnormalities and displayed impaired steroidogenesis.4 Serum chemistry analysis showed marked hypoproteinemia, hypoalbuminemia, hypoglycemia, hypocalcemia, hypotriglyceridemia, and hypocholesterolemia (supplemental Table 3). Reduced serum levels of proteins, carbohydrates, and lipids concurrently without significant histological lesions in liver, pancreas, intestinal tract, and kidney indicate that KO pups most likely suffered from severe metabolic defect.
To further determine the in vivo functions of Wwox, we studied the differential expression of mRNAs in the Wwox-deficient mice compared with their WT and HET littermates. mRNAs extracted from spleen, kidney, brain, pituitary, femur and calvarial bone were analyzed using mouse Affymetrix microarray gene-chip 430 2.0 arrays. Chromatin structure, cell cycle, and other related genes were among the most differentially expressed genes in KO mice tissues as compared with control mice (supplemental Table 4).
A Bone Metabolic Disorder in Wwox-null Mice—Limbs of the KO mice revealed not only a size difference proportional to the animal weight, but radiography revealed that KO limbs exhibited decreased density of trabeculae bone and a thinner cortex compared with WT beginning at day 7 (data not shown). Fig. 1C shows the striking differences in bone density at 3 weeks age. To determine whether the observed osteopenia was a consequence of a metabolic disorder or related to cell autonomous defects in bone cell populations, we first examined skeletal development using alizarin red/Alcian blue staining of newborn pups. As shown in Fig. 1D, no apparent size differences or abnormalities related to skeletal patterning were observed. Some variability in ossification of the calvarium was found at birth among HET and KO Wwox mice, but radiography showed no differences in craniofacial bones on day 7 (data not shown).
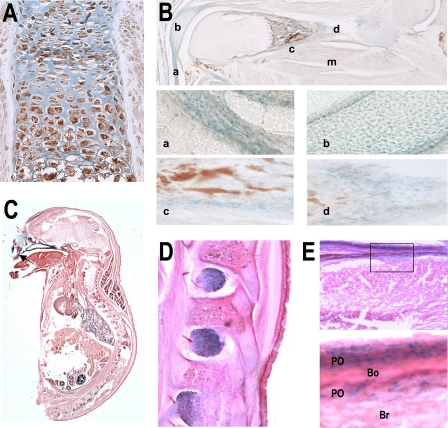
Because our targeting vector included the lacZ gene (7), we next examined expression of Wwox promoter in the skeleton. In our previous report, lacZ staining of Wwox-/- mice revealed Wwox expression throughout the skeleton of whole embryos in craniofacial bones, vertebrae, and limb bones (7). To address the activity of Wwox promoter in specific cell populations of the skeleton, we performed in situ immunohistochemistry (Fig. 2A, newborn) and sectioning of X-gal-stained embryos at E17.5 (Fig. 2, B-E). In the embryo, WWOX protein is expressed in chondrocytes and osteoblasts of the developing WT limb (Fig. 2A, newborn). Wwox promoter activity was identified in limb sections in chondrocytes, osteoblasts, and tendon fibroblasts in long bone (Fig. 2B). Chondrocytes and osteoblasts in vertebral bodies of the spine were also positive (Fig. 2, C and D). In the developing calvarium (Fig. 2E), we found robust expression of Wwox promoter activity in early differentiating osteoblasts that are forming the bone matrix.
FIGURE 2.
Wwox promoter activity in cartilage and bone of Wwox-null mice. A, in situ immunohistochemistry of newborn femur showing expression of WWOX protein in epiphysis, growth plates, chondrocytes, and osteoblasts in primary spongiosa. B, lacZ staining in bone section of KO femur at E17.5 (no counterstain). The four lower panels of B show higher magnification (×20) of tissues for visualization of Wwox-positive cells in tendon (a), chondrocytes (b), osteoblasts on the endosteal surface (c), and osteoblasts forming bone in the periosteal side of the limb (d). Muscle fibers (m) surrounding the bone do not show β-galactosidase activity. C, lacZ and eosin counterstain of E17.5 embryo section with higher magnifications in D showing vertebral body (VB) at ×20. E, calvarium at ×10 (top panel) and ×40 (lower panel). Intense staining of cells on the bone (Bo)-forming surfaces but not in mature osteocytes in the bone center. Br, brain tissue; Po, periosteum.
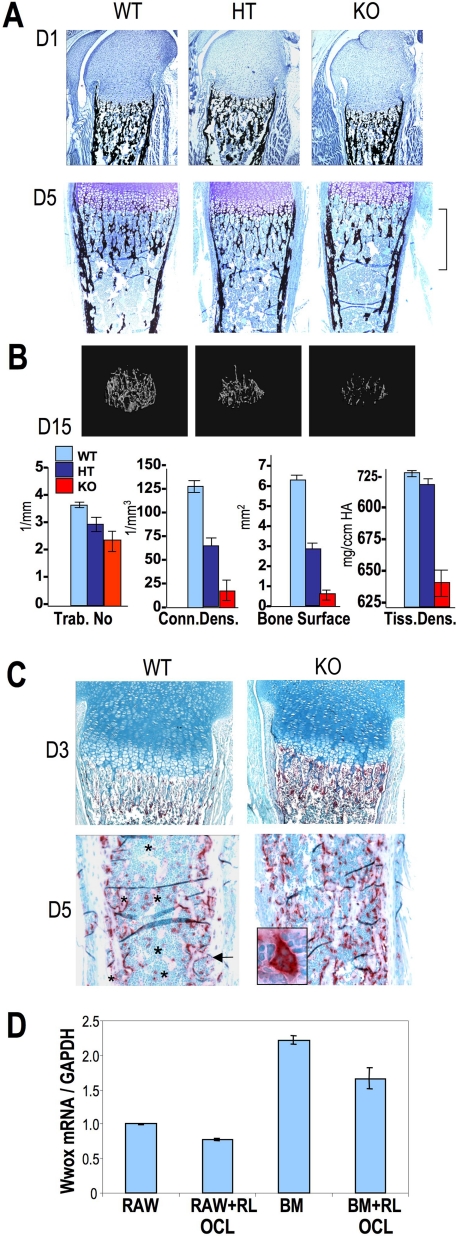
To identify modifications in bone cell populations and tissue organization to account for the apparent reduced bone formation revealed by radiography (less dense bone) in the Wwox-deficient mice, histologic sectioning of limbs was performed at several ages. Normal organization of zones of chondrocyte maturation at the growth plate was observed, and mineralization of trabecular and cortical bone tissue appeared normal on day 1 (Fig. 3A), but the KO mouse bone size was slightly smaller (∼8-10%). By day 3 impaired bone growth was visually evident, and as shown at day 5, the KO mouse bones clearly have fewer trabeculae compared with WT. The diminished volume of trabecular bone in the KO mice continued until the mice died. The three-dimensional μCT images of femur metaphysis in mice at day 15 (Fig. 3B) show reduced trabecular member connectivity and bone surface area in both the HET and KO. However, mineral content (tissue density parameter) of the trabecular bone in HET is not significantly changed from WT.
FIGURE 3.
Metabolic bone disease phenotype of the Wwox null mouse. A, histologic sectioning of long bone of postnatal mouse day 1 (D1) and day 5 (D5) shows normal organization of the epiphysis and the growth plate and mineralization of bone tissue. By day 5 fewer trabeculae were in the KO mouse compared with WT and HT (bracket). Sections were stained with toluidine blue and von Kossa silver stain (for detection of mineralized tissues; black). B, μCT image of the metaphysis region of WT and KO femur at day 15 showing less volume of trabecular (Trab.) bone with accompanying quantitation of trabecular bone loss. HT, heterozygous; Conn Dens, connectivity density; Tiss Dens, tissue density. C, high rate of bone resorption in Wwox null mice is evident by tartrate-resistant acid phosphatase histochemical staining (red) with fast green counterstain of femurs from wild-type (WT) and homozygous (HO) mice. Note increased osteoclast activity in the primary spongiosa under the growth plate on day 3 and osteoclasts on nearly all trabeculae on day 5, consistent with decreased trabeculae number on day 15 in B. Inset in KO panel shows ×63 magnification of osteoclast. On day 5, asterisk shows bone spicules in WT without osteoclasts. D, WWOX expression/GAPDH by Q-PCR in the RAW274.6 monocyte cell line and bone marrow cells compared with osteoclasts (+Rank ligand-treated 4 days).
Given the reduced serum values of 50% lower calcium and 20% increased phosphate in the KO mouse (supplemental Table 3), as well as loss of trabecular bone, these findings suggest a metabolic bone disease contributing to the osteopenic phenotype in KO mouse. We therefore examined bone tissue for the number and activity of osteoclasts (bone resorbing cells) by tartrate-resistant acid phosphatase histochemical staining at several postnatal ages (Fig. 3C). Higher acid phosphatase activity was consistently observed in femur bone sections of KO mice compared with the WT by day 3 in the primary spongiosa (Fig. 3C, upper panel). The day 5 (Fig. 3C, lower panel) sections of diaphysis are shown to illustrate the less osteoclast activity around mature cortical and trabeculae bone (indicated by *) compared with the KO.
We next addressed if the stimulated osteoclast activity was solely because of the physiologic response to low serum calcium in Wwox-deficient mice or if WWOX deficiency directly influenced stimulation of osteoclast activity. Because of organ wasting, it was not possible to isolate viable progenitors from bone marrow or spleen from the Wwox-/- mice for ex vivo osteoclast differentiation. Therefore, WWOX expression was examined in two in vitro osteoclast differentiation systems (Fig. 3D). Both the RAW264.7 cell line and bone marrow mouse mononuclear precursors express WWOX with higher expression in ex vivo marrow-derived cells. Differentiation to osteoclasts (induced by RANKL) did not result in significant changes in WWOX expression. In conclusion, WWOX is expressed in normal osteoclasts, but we cannot ascertain if WWOX deficiency in the osteoclast contributed to stimulated activity of the osteoclast. However, it is clear that functional osteoclasts in Wwox null mice respond to the metabolic bone disease of reduced serum calcium with no apparent defects in osteoclast activity per se as HET mice have normal calcium and phosphorous serum levels.
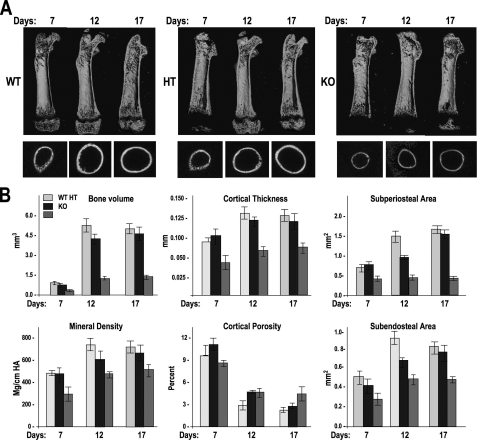
Quantitative changes in bone growth, density, formation, and resorption characteristics in the Wwox knock-out mice were determined by high resolution μCT imaging of limb bones for the WT, HET, and KO mouse groups at three postnatal ages days 7, 12, and 17 (the latter age group did not yet exhibit signs of wasting) (Fig. 4). Longitudinal mid-sections of the femur are shown in Fig. 4A (top panel), which emphasizes a delay in postnatal bone formation in the KO, reflected by reduced appearance of the secondary center of ossification in the epiphysis from days 7 to 17. The cross-section of the diaphysis (Fig. 4A, middle panel) reveals a thinner cortical bone in KO mice compared with WT. Selected μCT parameters are shown in Fig. 4B to confirm the decrease in bone volume and mineral density, which reflects calcified trabecular and cortical bone. The decrease in cortical thickness (day 7-17) and the higher cortical porosity in the Wwox null mouse during the post-natal growth period (day 12 and day 17) reflect a slow rate of bone formation. Both the periosteal and endosteal bone forming surfaces are significantly reduced in the Wwox KO mouse, parameters that also indicate reduced bone formation. Taken together, these results identify a bone phenotype contributed by a decrease in bone formation, as well as increased bone resorption from a metabolic disorder.
FIGURE 4.
Delayed bone formation in the Wwox KO mouse. A, microcomputed tomography (μCT) three-dimensional images of WT, HET (HT), and KO genotypes at the indicated ages. Top panel, image from the midsection of femur that shows less trabecular bone and a delay in the secondary center of ossification in the KO; middle panel, cross-section at mid-diaphysis shows thin cortical bone of KO. B, quantitation of selected bone formation parameters, including bone volume, mineral density, area of bone formation surfaces, and cortical bone quality from μCT data analyses as indicated.
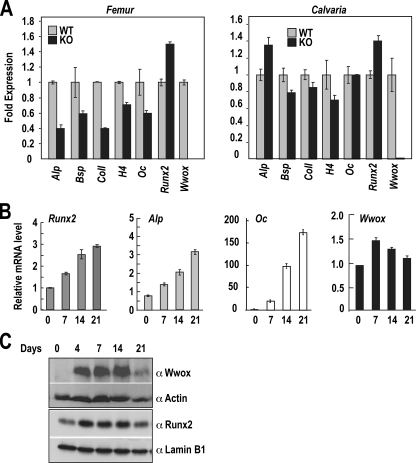
WWOX Plays an Essential Role during Osteoblast Differentiation—To uncover the mechanism contributing to the delayed bone growth, we first evaluated the consequences of Wwox deletion on the expression of essential bone gene markers. For analysis of bone formation on femurs and calvaria from age day 7, KO and WT were used in performing real time PCR (Fig. 5A). Our data demonstrate that expression of Runx2, an essential transcription factor bone formation, is increased ∼25% in KO compared with WT littermates in both tissues. This change in Runx2, which is a marker of committed osteoprogenitors, may reflect a stimulated number of cells recruited into the osteoblast lineage to compensate for bone loss. In contrast, the osteoblastic genes representing newly synthesized bone matrix and osteoblast differentiation (alkaline phosphatase (Ap), collagen type I (ColI), bone sialoprotein (Bsp), osteocalcin (Oc)) were expressed at a 50% lower level in femur. In calvaria, both RUNX2 and Alp were slightly elevated, and Oc was unchanged compared with femur. This result reflects that intramembranous calvarial bone is not as involved in metabolic bone turnover compared with long bone. However, in femur the rate of bone resorption exceeds the rate of bone formation. It is also noteworthy that histone H4, which reflects DNA synthesis, is decreased in femur and calvaria of KO bone tissue, further indicating a metabolic wasting of tissues in the absence of the WWOX tumor suppressor.
FIGURE 5.
Gene expression markers of bone formation in Wwox WT and KO mice and during MC3T3 osteoblast differentiation. A, quantitative RT-PCR for expression of key genes in osteoblast-related genes in femur and calvarial bones from day 7 postnatal mice (n = 3). Expression of genes was normalized to Gapdh and plotted as relative expression in KO compared with WT set to 1.0. Alp, alkaline phosphatase; Bsp, bone sialoprotein; ColI, collagen-I; H4, histone (proliferation marker); Oc, osteocalcin. B, total RNA was prepared at the indicated day from differentiating MC3T3 calvarial osteoblasts (day 0, osteoprogenitors; day 7, growth; day 14, matrix maturation; and day 21, mineralization stages). C, Western blot analyses from cell lysates from the same experiment in which quantitative-PCR was performed (shown in Fig. 4B).
We next obtained a profile of WWOX expression during osteoblast growth and/or differentiation to identify specific stages for its function. Using the mouse MC3T3 osteoprogenitor cell line (Fig. 5, B and C), we find Wwox mRNA is found expressed in osteoblasts from proliferating to mature osteoblasts. A 1.5-fold increase occurs from undifferentiated confluent cells on day 0 to differentiated cells on day 7. Wwox mRNA is detected in the undifferentiated exponentially growing MC3T3 cells (day 0), but protein (Fig. 5C) does not appear until the cells are confluent or differentiated (day 4 is 24 h after confluency). The protein levels remain constitutive until the mineralization stage (day 21) when both mRNA and protein decline. At this time point osteoblast marker genes reach peak expression (Fig. 5B). Thus WWOX expression exhibits changes at the early stage of osteoprogenitor commitment and in mature osteoblasts.
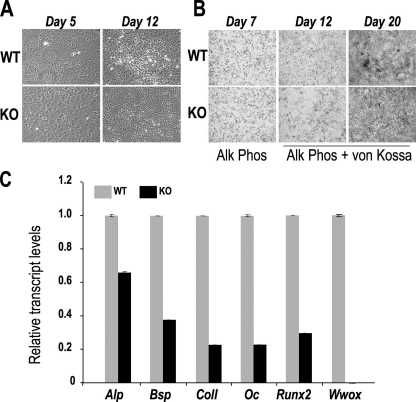
To verify if WWOX may be indispensable for osteoblast differentiation, we performed an ex vivo osteoblast differentiation study with cells derived from calvaria of 3-day-old Wwox-/- and wild-type mice (Fig. 6). Although cells were plated at equivalent density, phase contrast micrographs show that Wwox-/- cells reached confluency and exhibited the typical cuboidal morphology earlier than WT (Fig. 6A, day 5). However, by day 12 the production of multilayered nodules arising from osteoprogenitors is greatly reduced (Fig. 6A, day 12). Fig. 6B shows that WT and KO-derived cells come to monolayer confluency (day 7) and exhibit similar alkaline phosphatase activity. However, there is less representation of alkaline phosphatase-positive cells and a decrease in mineralization of the cell layer (day 20) in the KO cell layers. Cells were harvested on day 12 for detection of osteoblast markers by quantitative RT-PCR (Fig. 6C). Expression of the markers of early stage commitment (Runx2, Alp), the matrix production period (Bsp, ColI), and mineralization stage (Oc) are significantly decreased. The decreased Alp and Runx2 gene expression on day 12 is consistent with delayed/reduced formation of mineralized bone tissue. These findings suggest that WWOX is an important coregulatory factor for proteins that influence osteogenic differentiation. Taken together, these findings confirm that an impaired differentiation of osteoblast lineage cells in the Wwox-/- mice also contributes to the observed bone phenotype.
FIGURE 6.
Cell autonomous defects in osteoblast differentiation in Wwox null mice. Ex vivo osteoblasts from day 3 calvaria postnatal of Wwox WT and KO mice are shown. A, phase contrast micrographs at the indicated days showing an increased proliferation with Wwox null cells reaching confluency sooner (day 5), but unable to form multilayered nodules (day 12). B, alkaline phosphatase (Alk Phos) and von Kossa histochemical staining for mineral shows cells reach the first stage of differentiation (alkaline phosphatase-positive), but have reduced mineralized nodules. C, quantitative reverse transcription-PCR expression of phenotypic genes on day 12. Alp, alkaline phosphatase; Bsp, bone sialoprotein; ColI, collagen-I; Oc, osteocalcin.
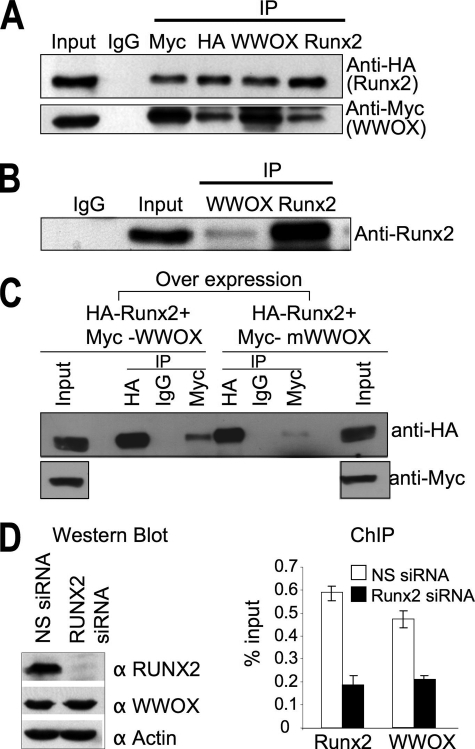
WWOX Regulates Activity of RUNX2—To further understand WWOX functions in osteoblast, we addressed physical and functional interactions of WWOX with RUNX2. Based on the knowledge of a functional PPXY motif in RUNX proteins that interact with several WW domain proteins (20-24), NIH3T3 cells were co-transfected with vectors expressing MYC-WWOX and HA-RUNX2. Cell lysates were immunoprecipitated with anti-HA or anti-MYC antibodies followed by immunoblotting with horseradish peroxidase-conjugated secondary antibody against HA or MYC. Our results demonstrate that WWOX physically associates with RUNX2 (Fig. 7A). As a control, there were no detectable complexes with normal IgG immunoprecipitates. Furthermore, we demonstrate that interaction of the endogenous proteins occurs in osteoblasts (Fig. 7B). We have previously shown that mutation in the first WW domain of WWOX (WWOX Y33R, tyrosine at position 33 replaced by arginine) abolishes interaction of WWOX with its partners (9-12). Therefore, we examined whether WWOXY33R can co-immunoprecipitate RUNX2. As shown in Fig. 7C, the Y33R point mutation significantly reduced WWOX association with RUNX2. These results indicate that WWOX, via its first WW domain, associates with RUNX2. This interaction was confirmed by chromatin immunoprecipitation on the osteocalcin promoter (Fig. 7D). Both factors associate with the Oc gene, but the binding of WWOX is decreased to the same extent as RUNX2 in osteoblasts treated with siRUNX2. The Western blot shows a near complete knockdown of RUNX2, without affecting WWOX which validates that WWOX association with Oc chromatin is dependent on RUNX2 binding.
FIGURE 7.
WWOX physically binds RUNX2 and together will associate with osteocalcin chromatin. A, physical interaction between RUNX2 and WWOX. Co-immunoprecipitation studies were carried out in non-osseous NIH3T3 cells transfected with expression vectors encoding MYC-WWOX and HA-RUNX2, followed by Western blot analysis using anti-HA and anti-Myc antibodies. B, co-immunoprecipitation of endogenous RUNX2 and WWOX by pulldown with anti-RUNX2 and anti-WWOX antibodies in ROS17/2.8 osteoblasts. The immunoprecipitates (IP) were detected by monoclonal anti-RUNX2 antibody. C, WWOX associates with RUNX2 via its first WW domain. HEK293 cells were co-transfected with HA-RUNX2 and MYC-WWOX or MYC-WWOXY33R plasmids. 24 h later, whole cell lysates were immunoprecipitated using anti-Myc and anti-HA antibodies followed by immunoblotting with anti-HA antibody. D, WWOX association with osteocalcin chromatin is dependent on RUNX2 occupancy of the Oc gene promoter. Chromatin immunoprecipitation was performed on endogenous mouse osteocalcin promoter in MC3T3 cells. Cells were treated with RUNX2 gene-specific and nonspecific siRNA duplexes for 48 h. Western blot with anti-RUNX2 and anti-WWOX antibodies demonstrate RUNX2 protein knockdown, but WWOX protein is not affected. Actin protein was used as loading control. ChIP-soluble chromatins from siRNA-treated cells were immunoprecipitated with anti-RUNX2 and anti-WWOX antibody. DNA fragments from immunoprecipitates were analyzed by quantitative real time.
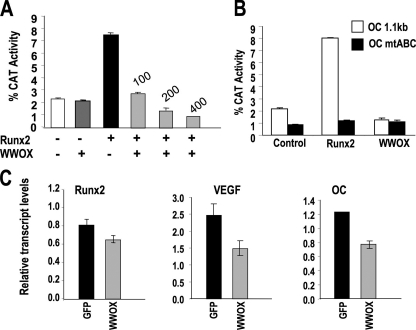
The interaction of WWOX and RUNX2 suggests that WWOX may be an important regulator of RUNX2-mediated transcriptional activation of bone-related genes. We therefore performed a functional assay using the osteocalcin gene promoter, known to be responsive to RUNX2-WW domain coregulatory protein interactions (20-22). NIH3T3 cells were used for transient transfection studies with the full-length osteocalcin promoter (rOc-CAT) containing three Runx2 regulatory elements. RUNX2 expression by itself resulted in a 4-4.5-fold stimulation (Fig. 8A). Although WWOX alone had no significant affect on Oc promoter activity, a dose-dependent inhibition of Oc-CAT induced by RUNX2 was observed by WWOX (Fig. 8A). To establish that WWOX inhibition of basal Oc promoter activity is mediated solely by protein-protein interactions with RUNX2, we compared the effect of WWOX on activation of Oc wild type and an Oc promoter in which all the three RUNX sites (A, B, and C) are mutated (25). This mutation in the Oc gene promoter reduced basal activity by ∼50% of WT Oc-CAT (Fig. 8B). We observed no significant activation of the mABC-Oc promoter by RUNX2 in contrast to a 4-fold activation of WT Oc-CAT. WWOX expression down-regulated WT Oc promoter activity, abolishing RUNX2-mediated activation, but had no effect on Oc mutant ABC-CAT activity. These data demonstrate that WWOX functions as a suppressor of osteogenic genes activated by RUNX2.
FIGURE 8.
WWOX suppresses RUNX2-transcriptional activity. A, dose-dependent suppression of RUNX2-mediated activation of the osteocalcin gene by WWOX is shown. NIH3T3 cells were transiently transfected with 1 μg of -1.1-kb rOc-CAT, with empty vector (100 ng of pcDNA, bar graph 1), WWOX (200 ng), RUNX2 (100 ng), and indicated increased amounts of WWOX. The Oc promoter activity (percent CAT conversion activity) is the average of six replicates. Values were calculated from the ratios of converted mono or diacetylchloramphenicol versus the total input chloramphenicol (S.E. of n = 6 wells). B, evidence that WWOX repression is mediated via RUNX2 regulatory elements. Comparison of inhibition on WT-Oc promoter and Oc promoter in which all three RUNX sites mutated (mABC-Oc). RUNX2 (100 ng) were co-transfected with WWOX (200 ng). C, WWOX suppresses RUNX2 target genes in metastatic breast cancer cells. MDA-MB-231 cells were transduced with adenovirus expressing green fluorescent protein (GFP) or WWOX. The relative expression of RUNX2, vascular endothelial growth factor (VEGF), and OC normalized to GAPDH is shown.
Because RUNX2 levels are elevated in metastatic breast cancer cells (26) and absence of WWOX in these and other tumor cells is known (5), we tested a functional relationship between RUNX2 and WWOX in MDA-MB-231 cells (Fig. 8C). It is well documented that this metastatic breast cancer cell line MDA-MB-231 causes osteolytic disease in bone through RUNX2 activation of metastatic related genes (26). We therefore tested WWOX function in these cells on RUNX2 target genes, osteocalcin, and vascular endothelial growth factor. Interestingly, we found that WWOX expression resulted in a 40-50% decrease in mRNA levels of these genes. These data suggest that both in normal osteoblasts as well as in cancer cells, WWOX tumor suppressor functions affect RUNX2-mediated transcription.
DISCUSSION
Mice lacking Wwox die by 3 weeks after birth and display defects in growth rate, steroid metabolism, and bone growth. The complexity of the mutant phenotype, together with postnatal survival lethality, supports multiple roles for WWOX in vivo. In general, the Wwox-null mice do not appear to have embryonic problems because all pups look indistinguishable at birth, and histological examination of homozygous embryos did not reveal any major developmental defects in soft tissue organs and the skeleton. Instead, our results reveal a vital requirement for WWOX for postnatal survival and maintenance of proper cell growth and metabolism. A key finding of these studies is the metabolic bone phenotype from consequences of WWOX deficiency in many tissues that contribute to the regulation of normal homeostasis.
We show that WWOX is expressed in osteoblasts and osteoclasts. The complex skeletal phenotype we observe is contributed by multiple factors. First, LacZ staining in embryo sections identified robust Wwox expression in numerous skeletal cell populations, including tendon fibroblasts, osteochondroprogenitor cells, and mature osteoblasts and chondrocytes (Fig. 2). Second, after birth Wwox KO pups exhibit a retarded bone growth (Fig. 3). This finding indicates that in utero nutrition of the embryo could contribute to normal growth and development until the perinatal period. The WWOX KO phenotype is further emphasized during postnatal growth with delayed formation of the epiphyseal bone and minimal increase of overall skeletal growth. Third, the KO mice exhibit low serum calcium that triggers the calcitrophic hormone axis (1,25(OH)2D3 and parathyroid hormone) to resorb bone. Interestingly, our Affymetrix gene profile identified the up-regulation of thyroid hormone just below the cutoff value of 1.5. Thyroid hormone increases are known to contribute to metabolic bone disease, as hyperthyroidism is well documented to increase bone turnover (27, 28). By 7 days postnatally, fewer trabeculae are observed in null mice compared with WT and HET mice because of activated osteoclasts. Although WWOX is expressed in osteoclasts, osteoclast activity is not impaired in vivo with WWOX deficiency. Osteoclasts retain apparent normal function and respond to low serum calcium by increasing bone resorption in an attempt to normalize mineral homeostasis. Analyses of bone formation markers show that on day 7, osteoblasts are functioning at a near normal level in intramembranous bone (calvaria), but in the trabecular bone of the femur in the KO mouse, their activity is very diminished in the environment of increased bone resorption. The turnover from bone resorption to bone formation is not coupled and net bone loss occurs.
Our previous and current analyses of the Wwox knock-out mouse phenotype demonstrated several important aspects of WWOX function in the occurrence of bone tumors, and herein in metabolic bone defects and a cell autonomous osteoblast defect. Thirty one percent of KO mice develop very early chondroid osteosarcomas (7). Thus, the tumor suppressor activity of WWOX is important for regulating proliferation and maturation of osteochondroprogenitor cells during endochondral bone formation. At E17.5 and at birth, bone formation is apparently normal and may be related to systemic factors from the dam. We focused our studies on the postnatal delayed bone formation phenotype (Fig. 4) and show osteoblast differentiation is impaired at the mature osteoblast stage (beginning on day 12 in vitro). This may arise from an insufficient number of cells competent to form the multilayered nodule and/or reduced expression of bone matrix producing genes at the mineralization stage. The in vitro defects in osteoblast maturation and decreased expression of markers of the mineralization stage are consistent with the in vivo μCT and osteoblast gene expression analyses in the KO. These findings suggest cell autonomous osteoblast defects with loss-of-function of WWOX. Trabecular bone mineral content is significantly reduced, suggesting reduced numbers of fully functional differentiated osteoblasts. There is adequate RUNX2 for initiation of osteoblastogenesis, but the final stage of differentiation is compromised. These are likely the properties of the osteoblasts that are present during the 3-week survival period, which cannot produce sufficient bone matrix (e.g. ColI, Bsp, and Oc) to compensate for bone loss by resorption. Therefore, we conclude that the decreased skeletal growth and failure to survive reflect both an inherent defect in osteoblast activity as well as a metabolic disease in the KO mice.
Our findings demonstrate that WWOX, in part, contributes to bone formation through regulation of RUNX2 activity in osteoblasts. This transcription factor is essential for the control of osteoblast growth by promoting exit from the cell cycle (29) and for differentiation by activation of bone matrix genes (30). Expression of Runx2, one of the earliest markers of mesenchymal stem cell commitment for bone formation, is slightly increased in both calvarial and femoral bone of WWOX KO mice. RUNX2 is known to be autoregulated having at least seven RUNX2 regulatory elements in the gene (30). Because WWOX inhibits RUNX2 transcriptional activity (shown in Fig. 8), then in the absence of WWOX repressor function, RUNX2 expression could be elevated.
WWOX is a co-regulatory protein that interacts with transcription factors, growth regulatory factors, and signal transduction proteins (e.g. p73, Ap2) through PPXY motifs in the interacting protein (4). WWOX appears to regulate RUNX2 at two levels as follows: first, an indirect effect that leads to decreased RUNX2 expression in bone and in isolated osteoblasts; second, as a suppressor of RUNX2 transcriptional activity. WWOX protein contains two WW domains predicted to mediate protein-protein interaction. Indeed, the first WW domain of WWOX belongs to class I of WW domains (9, 13). We have shown that WWOX binds PPXY motifs of several transcription factors, sequesters them in the cytoplasm, and reduces their transactivation abilities (9-12). Here, we further confirm WWOX transcriptional co-repressor activity as demonstrated by WWOX-RUNX2 physical and functional interaction. Furthermore, the association of WWOX with OC chromatin is dependent on RUNX2 binding as revealed by chromatin immunoprecipitation studies in control and RUNX2 siRNA-treated osteoblasts. The RUNX2 PPXY motif is a target of other WW domain-containing proteins that modify RUNX2 levels and/or transcriptional activity by either repression, such as YAP (20), activation by TAZ (32), or by targeting RUNX2 to the proteosome through SMURF1 (21) or a Schnurri-3-WWP1 complex (22) through recruitment of the E3 ubiquitin ligases. Therefore, in the absence of WWOX and RUNX2 interaction, the balance between the different WW domain-adaptor proteins may determine the functional outcome of RUNX2 and therefore the cell autonomous defects in osteoblast differentiation. Our data suggest that certain WWOX functions in the skeleton may not be compensated for by redundant functions of related proteins of the WW domain-containing proteins or proteins containing the short-chain dehydrogenase/reductase domain. However, only an osteoblast-specific in vivo deletion of WWOX can establish a direct role for WWOX in bone formation. Taken together, our findings identify a transcriptional mechanism involving WWOX regulation of bone formation through the transcription factor RUNX2 in all skeletal elements, as well as WWOX function as a tumor suppressor of osteochondroprogenitor and RUNX2-expressing tumor cells.
It is noteworthy that genetic abnormalities in RUNX factors are associated with tumor progression (33). RUNX2 is present at very low levels in normal breast and prostate tissue but is overexpressed in breast and prostate cancer metastases to bone (26, 33). Increased activity of Runx2 in cancer cells appears to be a contributing factor to the growth of tumors in bone (26, 34). It is therefore possible that WWOX suppression of RUNX2 transactivation ability contributes to the tumor suppressor function of WWOX (7). Our data suggest that the absence of WWOX in cancer cells is contributing to increased RUNX2 activity and likely a significant factor to the tumor growth in bone and the accompanying bone disease (26).
The low level of glucose, protein, and lipid in Wwox KO mice are indicative of a severe metabolic defect that may explain the postnatal lethality of Wwox KO mice. Recently, Aldaz and co-workers (35) described the generation of Wwox hypomorphic mice that display reduced levels of WWOX expression. Interestingly, these mice survived, although they had a shorter life span than WT. These data suggest that low levels of WWOX protein might be enough to overcome the postnatal lethality we observed in our Wwox-null mice. Of note, Wwox hypomorphic mice had higher incidence of B-cell lymphoma similar to Wwox heterozygous mice (7). Our microarray profiling data provide insight into the spectrum of cellular activities that are modified by Wwox deficiency that leads to postnatal lethality and the bone defect. Targeted deletion of Wwox resulted in down-regulation of genes involved in cell cycle and chromatin structure (supplemental Table 4), suggesting that WWOX may be involved in regulation of cell growth. In addition, we find from our microarray studies, increased expression of systemic growth factors that are implicated as negative regulators of bone formation. One example is IGFBP-3, a multifunctional regulatory peptide of cell growth and survival. Changes in IGFBP-3 levels affect glucose homeostasis and bone turnover (36) and because IGFBP-3 has high affinity for insulin-like growth factors, it is positively correlated with bone metabolism in healthy children (37). However, overexpression of IGBP-3 in transgenic mice increases osteoclast number and attenuates the osteoblast response to insulin-like growth factor-I (36). PDGF-2 is mitogenic for mesenchymal cells and is secreted by osteoblasts (38) but is associated with tumor progression in osteosarcomas (39). LTBP-3 contributes to transforming growth factor-β activity, and Ltbp-3 null mice exhibit bone abnormalities contributed by decreased osteoclast function (40, 41). Thus the elevated LTBP-3 levels may be related to stimulated bone resorption in the Wwox-/- mice. Regulation of FGF-18 levels in bone and cartilage tissues is required for normal cell proliferation and differentiation of chondrocytes and osteoblasts (31). These elevated growth factors in Wwox null mice may be contributing to both impaired osteogenesis and the observed delay in the secondary center of ossification as well as stimulated bone resorption during the 3-week postnatal survival period of the Wwox null mice. Such changes in gene expression reflect the complexity of the Wwox KO phenotype.
In conclusion, our phenotypic analysis of the Wwox knock-out mice demonstrates an important role of WWOX in fundamental cellular processes, including survival, growth, bone metabolism, and neoplastic transformation.
Supplementary Material
Acknowledgments
We thank Dr. Valerie Bergdall and Dr. Dona Kusewitt from School of Veterinary-Ohio State University for advice and assistance in mouse phenotyping. We are also grateful to Nicola Zanesi, Dean Marshall, Chris Lemmon, Muareen Mork, Eugenio Gaudio, and Valentina Donati for technical assistance. We thank Dr. Kay Hubener (Ohio State University, Columbus) for providing WWOX polyclonal antibody, Dr. Douglas Adams (Director of the University of Connecticut Imaging Core) for discussions of the μCT data, and Dr. Philip Osdoby (Washington University, St. Louis, MO) for providing RAW264.7 cells. We thank Judy Rask and Charlene Baron (UMass Medical School) for manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 CA082834 and P01 AR048818 (to G. S. S. and J. B. L.). This work was also supported by Ohio Cancer Research Associates and Kimmel Scholar Award (to R. I. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 2-4.
Footnotes
The abbreviations used are: WWOX, WW domain-containing oxidoreductase; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; KO, knock-out; WT, wild type; HET, heterozygous; FBS, fetal bovine serum; PBS, phosphate-buffered saline; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interfering RNA; RANKL, receptor activator of NF-κB ligand; μCT, microcomputed tomography; HA, hemagglutinin.
R. Aqeilan, unpublished data.
References
- 1.Bednarek, A. K., Laflin, K. J., Daniel, R. L., Liao, Q., Hawkins, K. A., and Aldaz, C. M. (2000) Cancer Res. 60 2140-2145 [PubMed] [Google Scholar]
- 2.Ried, K., Finnis, M., Hobson, L., Mangelsdorf, M., Dayan, S., Nancarrow, J. K., Woollatt, E., Kremmidiotis, G., Gardner, A., Venter, D., Baker, E., and Richards, R. I. (2000) Hum. Mol. Genet. 9 1651-1663 [DOI] [PubMed] [Google Scholar]
- 3.Paige, A. J., Taylor, K. J., Taylor, C., Hillier, S. G., Farrington, S., Scott, D., Porteous, D. J., Smyth, J. F., Gabra, H., and Watson, J. E. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11417-11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aqeilan, R. I., and Croce, C. M. (2007) J. Cell Physiol. 212 307-310 [DOI] [PubMed] [Google Scholar]
- 5.Bednarek, A. K., Keck-Waggoner, C. L., Daniel, R. L., Laflin, K. J., Bergsagel, P. L., Kiguchi, K., Brenner, A. J., and Aldaz, C. M. (2001) Cancer Res. 61 8068-8073 [PubMed] [Google Scholar]
- 6.Fabbri, M., Iliopoulos, D., Trapasso, F., Aqeilan, R. I., Cimmino, A., Zanesi, N., Yendamuri, S., Han, S. Y., Amadori, D., Huebner, K., and Croce, C. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15611-15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aqeilan, R. I., Trapasso, F., Hussain, S., Costinean, S., Marshall, D., Pekarsky, Y., Hagan, J. P., Zanesi, N., Kaou, M., Stein, G. S., Lian, J. B., and Croce, C. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3949-3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aqeilan, R. I., Hagan, J. P., Aqeilan, H. A., Pichiorri, F., Fong, L. Y., and Croce, C. M. (2007) Cancer Res. 67 5606-5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aqeilan, R. I., Pekarsky, Y., Herrero, J. J., Palamarchuk, A., Letofsky, J., Druck, T., Trapasso, F., Han, S. Y., Melino, G., Huebner, K., and Croce, C. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4401-4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aqeilan, R. I., Palamarchuk, A., Weigel, R. J., Herrero, J. J., Pekarsky, Y., and Croce, C. M. (2004) Cancer Res. 64 8256-8261 [DOI] [PubMed] [Google Scholar]
- 11.Aqeilan, R. I., Donati, V., Palamarchuk, A., Trapasso, F., Kaou, M., Pekarsky, Y., Sudol, M., and Croce, C. M. (2005) Cancer Res. 65 6764-6772 [DOI] [PubMed] [Google Scholar]
- 12.Gaudio, E., Palamarchuk, A., Palumbo, T., Trapasso, F., Pekarsky, Y., Croce, C. M., and Aqeilan, R. I. (2006) Cancer Res. 66 11585-11589 [DOI] [PubMed] [Google Scholar]
- 13.Ludes-Meyers, J. H., Kil, H., Bednarek, A. K., Drake, J., Bedford, M. T., and Aldaz, C. M. (2004) Oncogene 23 5049-5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin, C., Ge, L., Ding, X., Chen, Y., Zhu, H., Ward, T., Wu, F., Cao, X., Wang, Q., and Yao, X. (2006) Biochem. Biophys. Res. Commun. 341 784-791 [DOI] [PubMed] [Google Scholar]
- 15.Chang, N. S., Hsu, L. J., Lin, Y. S., Lai, F. J., and Sheu, H. M. (2007) Trends Mol. Med. 13 12-22 [DOI] [PubMed] [Google Scholar]
- 16.Kim, I. S., Otto, F., Zabel, B., and Mundlos, S. (1999) Mech. Dev. 80 159-170 [DOI] [PubMed] [Google Scholar]
- 17.Pratap, J., Galindo, M., Zaidi, S. K., Vradii, D., Bhat, B. M., Robinson, J. A., Choi, J. Y., Komori, T., Stein, J. L., Lian, J. B., Stein, G. S., and van Wijnen, A. J. (2003) Cancer Res. 63 5357-5362 [PubMed] [Google Scholar]
- 18.McCabe, L. R., Banerjee, C., Kundu, R., Harrison, R. J., Dobner, P. R., Stein, J. L., Lian, J. B., and Stein, G. S. (1996) Endocrinology 137 4398-4408 [DOI] [PubMed] [Google Scholar]
- 19.Hassan, M. Q., Tare, R. S., Lee, S. H., Mandeville, M., Morasso, M. I., Javed, A., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2006) J. Biol. Chem. 281 40515-40526 [DOI] [PubMed] [Google Scholar]
- 20.Zaidi, S. K., Sullivan, A. J., Medina, R., Ito, Y., van Wijnen, A. J., Stein, J. L., Lian, J. B., and Stein, G. S. (2004) EMBO J. 23 790-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao, M., Qiao, M., Oyajobi, B. O., Mundy, G. R., and Chen, D. (2003) J. Biol. Chem. 278 27939-27944 [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. C., Wein, M. N., Oukka, M., Hofstaetter, J. G., Glimcher, M. J., and Glimcher, L. H. (2006) Science 312 1223-1227 [DOI] [PubMed] [Google Scholar]
- 23.Yamashita, M., Ying, S. X., Zhang, G. M., Li, C., Cheng, S. Y., Deng, C. X., and Zhang, Y. E. (2005) Cell 121 101-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong, J. H., Hwang, E. S., McManus, M. T., Amsterdam, A., Tian, Y., Kalmukova, R., Mueller, E., Benjamin, T., Spiegelman, B. M., Sharp, P. A., Hopkins, N., and Yaffe, M. B. (2005) Science 309 1074-1078 [DOI] [PubMed] [Google Scholar]
- 25.Javed, A., Gutierrez, S., Montecino, M., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (1999) Mol. Cell. Biol. 19 7491-7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Javed, A., Barnes, G. L., Pratap, J., Antkowiak, T., Gerstenfeld, L. C., van Wijnen, A. J., Stein, J. L., Lian, J. B., and Stein, G. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1454-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassett, J. H., and Williams, G. R. (2003) Trends Endocrinol. Metab. 14 356-364 [DOI] [PubMed] [Google Scholar]
- 28.Sun, L., Davies, T. F., Blair, H. C., Abe, E., and Zaidi, M. (2006) Ann. N. Y. Acad. Sci. 1068 309-318 [DOI] [PubMed] [Google Scholar]
- 29.Galindo, M., Pratap, J., Young, D. W., Hovhannisyan, H., Im, H. J., Choi, J. Y., Lian, J. B., Stein, J. L., Stein, G. S., and van Wijnen, A. J. (2005) J. Biol. Chem. 280 20274-20285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lian, J. B., Javed, A., Zaidi, S. K., Lengner, C., Montecino, M., van Wijnen, A. J., Stein, J. L., and Stein, G. S. (2004) Crit. Rev. Eukaryotic Gene Expression 14 1-41 [PubMed] [Google Scholar]
- 31.Ohbayashi, N., Shibayama, M., Kurotaki, Y., Imanishi, M., Fujimori, T., Itoh, N., and Takada, S. (2002) Genes Dev. 16 870-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui, C. B., Cooper, L. F., Yang, X., Karsenty, G., and Aukhil, I. (2003) Mol. Cell. Biol. 23 1004-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blyth, K., Cameron, E. R., and Neil, J. C. (2005) Nat. Rev. Cancer 5 376-387 [DOI] [PubMed] [Google Scholar]
- 34.Pratap, J., Lian, J. B., Javed, A., Barnes, G. L., van Wijnen, A. J., Stein, J. L., and Stein, G. S. (2006) Cancer Metastasis Rev. 25 589-600 [DOI] [PubMed] [Google Scholar]
- 35.Ludes-Meyers, J. H., Kil, H., Nunez, M. I., Conti, C. J., Parker-Thornburg, J., Bedford, M. T., and Aldaz, C. M. (2007) Genes Chromosomes Cancer 46 1129-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silha, J. V., Mishra, S., Rosen, C. J., Beamer, W. G., Turner, R. T., Powell, D. R., and Murphy, L. J. (2003) J. Bone Miner. Res. 18 1834-1841 [DOI] [PubMed] [Google Scholar]
- 37.Leger, J., Mercat, I., Alberti, C., Chevenne, D., Armoogum, P., Tichet, J., and Czernichow, P. (2007) Eur. J. Endocrinol. 157 685-692 [DOI] [PubMed] [Google Scholar]
- 38.Centrella, M., McCarthy, T. L., Kusmik, W. F., and Canalis, E. (1992) J. Clin. Investig. 89 1076-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulzbacher, I., Birner, P., Trieb, K., Traxler, M., Lang, S., and Chott, A. (2003) Mod. Pathol. 16 66-71 [DOI] [PubMed] [Google Scholar]
- 40.Dabovic, B., Chen, Y., Colarossi, C., Zambuto, L., Obata, H., and Rifkin, D. B. (2002) J. Endocrinol. 175 129-141 [DOI] [PubMed] [Google Scholar]
- 41.Dabovic, B., Levasseur, R., Zambuto, L., Chen, Y., Karsenty, G., and Rifkin, D. B. (2005) Bone (N. Y.) 37 25-31 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.