Abstract
O-Linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) catalyzes the addition of O-linked β-N-acetylglucosamine (O-GlcNAc) onto serine and threonine residues in response to stimuli or stress analogous to phosphorylation by Ser/Thr-kinases. Like protein phosphatases, OGT appears to be targeted to myriad specific substrates by transiently interacting with specific targeting subunits. Here, we show that OGT is activated by insulin signaling. Insulin treatment of 3T3-L1 adipocytes stimulates both tyrosine phosphorylation and catalytic activity of OGT. A subset of OGT co-immunoprecipitates with the insulin receptor. Insulin stimulates purified insulin receptor to phosphorylate OGT in vitro. OGT is a competitive substrate with reduced and carboxyamidomethylated lysozyme (RCAM-lysozyme), a well characterized insulin receptor substrate. Insulin stimulation of 3T3-L1 adipocytes results in a partial translocation of OGT from the nucleus to the cytoplasm. The insulin activation of OGT results in increased O-GlcNAc modification of OGT and other proteins including, signal transducer and activator of transcription 3 (STAT3). We conclude that insulin stimulates the tyrosine phosphorylation and activity of OGT.
O-linked β-N-acetylglucosamine (O-GlcNAc)3 is a sugar modification that is found on serine and threonine residues throughout the nucleocytoplasm of metazoans (1, 2). O-GlcNAc is on myriad proteins and cycles like phosphorylation. On several proteins, O-GlcNAc is attached at or adjacent to phosphorylation sites, suggesting that the saccharide also plays a competitive role in cell signaling pathways (3). O-GlcNAc attachment and detachment (cycling) is mediated by O-GlcNAc transferase (OGT) and O-GlcNAc specific β-d-N-acetylglucosaminidase (O-GlcNAcase), respectively (4, 5). A global reciprocal relationship between O-phosphorylation and O-GlcNAcylation has been observed (6, 7), as well as a dynamic interplay at specific sites (6, 8–11). Unlike phosphorylation, which is orchestrated by hundreds of kinases, in mammals, there is predominantly only the 110-kDa catalytic subunit of OGT and a few other isoforms that result from alternative promoter usage and splicing of a single gene (4, 12, 13).
Studies of OGT protein structure (14), protein interactions, and post-translational modifications are beginning to reveal some of the mechanisms that regulate OGT. Mammalian OGT contains multiple tetratricopeptide repeats consisting of 34-residue motifs in an elongated superhelix with a phylogenetically conserved inner surface that is similar in structure to importin-α and the armadillo repeats of β-catenin. This structural similarity and recent data suggest that a wide range of physiological protein-protein interactions with the tetratricopeptide repeats help target OGT to its many substrates in a highly specific manner (14–17). OGT also has a catalytic domain related to glycogen phosphorylase and is itself modified by O-GlcNAc and tyrosine phosphorylation, suggesting that the enzyme is regulated by signal transduction cascades (4, 15). OGT is responsive to morphogens (18), cellular stress (19), cell cycle (20), and changes in glucose metabolism (3).
Recently, OGT was found to play an important role in nematode insulin signaling (21). An OGT-1 deletion allele in nematodes suppresses dauer formation induced by the partial loss of daf-2 (insulin-like receptor) and results in defective macronutrient storage analogous to mammalian insulin resistance (21). Deletion of OGT resulted in elevated glycogen storage and decreased fat storage, suggesting that OGT and O-GlcNAc are important for the regulation of the insulin-like pathway in worms. OGT is involved in the liver's insulin signaling pathway and contributes to AKT-mediated signaling at the level of IRS-1 in a phosphatidylinositol 3,4,5-trisphosphate-dependent manner (22). In mammals, deletion of OGT is lethal in embryonic stem cells and in single cells in cell culture (23, 24). OGT's regulation by nutrients is largely due to its extraordinary sensitivity to intracellular UDP-GlcNAc concentrations. UDP-GlcNAc is the end product of the hexosamine biosynthetic pathway and is the direct donor sugar nucleotide for OGT (25). Virtually all nutrients including fatty acids, glucose, and amino acids dramatically affect the levels of UDP-GlcNAc in cells, which in-turn affects dynamic O-GlcNAcylation (26–28). We hypothesize that post-translational modification of OGT may also affect OGT's activity and O-GlcNAcylation of substrates. In this study, we show that insulin signaling stimulates tyrosine phosphorylation of OGT, increases its catalytic activity, affects OGT localization, and results in the O-GlcNAc modification of STAT3, a key kinase and transcription factor in 3T3-L1 adipocytes (29, 30).
EXPERIMENTAL PROCEDURES
Reagents—Antibodies used in this study are against STAT3 (Cell Signaling), α-tubulin (T5168, Sigma), AL-28 OGT (Hart laboratory (31)), anti-O-GlcNAc CTD 110.6 (Hart laboratory and Covance), and the polyclonal insulin receptor β-chain antibody (M. Daniel Lane laboratory (32)). Horseradish peroxidase-conjugated secondary antibodies were from Amersham Biosciences, and anti-IgM was from Sigma. Fluorescently labeled secondary antibodies Alexa Fluor 647 and 488 were from Molecular Probes. Blots were developed with ECL reagent and Hyper-film (GE Healthcare). Recombinant human insulin was from Roche Diagnostics. All other reagents used were from Sigma unless otherwise indicated.
Cell Culture—3T3-L1 adipocytes were grown and differentiated into adipocytes, as described with minor modifications (33). Preadipocytes were grown to confluency in Dulbecco's modified Eagle's medium 10% calf serum containing penicillin-streptomycin and biotin. Two days after confluency, preadipocytes were differentiated in Dulbecco's modified Eagle's medium, 10% fetal bovine serum with dexamethasone (390 ng/ml), insulin (1 μg/ml), and methylisobutylxanthine (115 μg/ml) for 48 h and then treated with insulin (1 μg/ml) for an additional 48 h. 3T3-L1 adipocytes were used for experiments 8–11 days after differentiation commenced.
Western Blotting and Immunoprecipitation—Whole cell lysates were harvested by washing 3T3-L1 three times in ice-cold phosphate-buffered saline and scraping into Nonidet P-40 buffer (1% Nonidet P-40, 15 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, protease inhibitor mixture 1, 40 mm GlcNAc (GlcNAc prevents O-GlcNAcase from removing O-GlcNAc from proteins during cell lysis), and 1 mm NaVO4). For Western blot analysis, the cell lysate was sonicated 2 × 10 s with a 30-s pause between on ice. An equal volume (1:1) of ice-cold Freon (1,1,2-trichloro-trifluoroethane) was added and mixed to remove the lipids and centrifuged at 15, 000 × g, and the protein concentration was determined by Bio-Rad protein reagent. For immunoprecipitations, cell lysates were rocked at 4 °C for 2 h to gently lyse cells and solubilize proteins in cell membranes before treating samples in a manner similar to Western blot samples, as stated above. Immunoprecipitations where then performed with the indicated antibodies overnight at 4 °C, captured with protein A/G-Sepharose (Santa Cruz Biotechnology), and washed four times in phosphate-buffered saline. In some cases stated in the text, shorter incubations of 4 h were necessary when trying to preserve post-translational modifications of proteins. Proteins were eluted by boiling Sepharose beads in modified Laemmli buffer. SDS-PAGE was performed with precast Criterion gels (Bio-Rad), transferred to polyvinylidene difluoride membrane (Millipore), blocked with 3% bovine serum albumin (w/v, Sigma) in Tris-buffered saline 0.1% Tween 20, and probed with the indicated antibodies overnight at 4 °C. The respective horseradish peroxidase-conjugated antibodies were incubated with the blots, and ECL detection was used. When necessary, blots were stripped with 100 mm glycine, pH 2.8, for 30 min at room temperature and reprobed with antibodies.
Confocal Microscope—Essentially, all microscopy was conducted as described by Slawson et al. (20). 3T3-L1 fibroblasts were plated onto Lab-Tek II chamber slides (Nalge Nunc) coated with poly-d-lysine (Sigma). Cells were fixed with 3% paraformaldehyde, immunohistochemistry was performed with the indicated antibodies overnight, and cells were incubated with secondary antibody for 1 h in the dark and treated with propidium iodide for nuclear staining, and slides were mounted before fluorescent images were obtained using a Zeis Ultra view 2 confocal microscope at The Johns Hopkins University Microscope Core Facility.
Overexpression of OGT in Insect Cells—Essentially overexpression of OGT in insect cells was conducted as indicated in Kreppel and Hart (15) with minor modifications. High Five (Invitrogen) insect cells were grown in Graces's insect medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum to 70–80% confluency (15-cm dishes) and infected for 40–48 h before harvesting. Cell pellets were frozen at -20 °C. Infected cell pellets were resuspended in Buffer A (20 mm Tris, pH 7.9, 0.5 m NaCl) and lysed by sonication (two pulses each lasting 10 s, power 3.5). Lysates were centrifuged for 30 min at 30,000 × g, filtered through a 0.45-μm filter, and loaded on a fast protein liquid chromatography (Amersham Biosciences) at a flow rate of 1 ml/min (onto a 5-ml chelating column precharged with NiSO4 and pre-equilibrated with buffer A). The column was washed with 10 column volumes of buffer A followed by a 10-ml wash at 5% buffer B and then eluted with a 25-min linear gradient of 5–100% Buffer B (20 mm Tris, pH 7.9, 0.5 m NaCl, 100 mm imidazole) at 1 ml/min. Fractions (1 ml) were collected throughout the separation and subjected to SDS-PAGE and analyzed for purity by Coomassie G250 staining and Western blot analysis. Fractions containing OGT were pooled and concentrated using a Vivaspin6 concentrator (MW 10, 000 cutoff) as per the manufacturer's directions (Vivascience) and buffer-exchanged into 12 ml of Buffer C (20 mm Tris, pH 7.5, 20% glycerol, 1 mm dithiothreitol) and concentrated. The pooled concentrate was brought to 40% glycerol, 1 mm dithiothreitol and stored at -20 °C.
OGT Activity Assay—Samples treated as indicated were immunoprecipitated with covalently attached AL-28 to Sepharose A beads for 4 h at 4 °C. Samples were washed three times with phosphate-buffered saline and one time with OGT assay buffer (10×: 0.5 m sodium cacodylate, pH 6.5, 10 mg/ml bovine serum albumin, 0.35 m NaF), divided in half for treatment with or without the acceptor substrate CK2 peptide (PGGSTPVS-SANMM) in UDP-[3H]GlcNAc (0.5 μCi/μl), 25 mm AMP, 0.5 μl, 10× buffer, 5 μl, alkaline phosphatase, 0.25 μl, 10 mm CK2 peptide, 5 μl, and H2O, 10 μl on ice. Samples were incubated at 24 °C for 30 min, and the reaction was stopped with 450 μl of 50 mm formate, 0.5 m NaCl. CK-2 peptides were cleaned up with C18 columns and eluted with methanol, and radioactive incorporation was measured by a scintillation counter.
Kinase Assay—Insulin receptor kinase assays were performed essentially as outlined in Kohanski et al. (34). Insulin receptor was immunoprecipitated from ±100 nm insulin-treated 3T3-L1 adipocytes and restimulated with 1 μm insulin in vitro for 10 min in kinase buffer (20 μm [γ-32P]ATP, 5 mm Mn(CH3CO2)2, 50 mm HEPES, 0.1% Triton X-100, pH 6.9) before purified OGT substrate was added. Five μg of OGT was added to each reaction and incubated with [γ-32P]ATP for 15 min at 30 °C. Where indicated commercially produced insulin receptor (IR) (Sigma) was incubated in 1 μm insulin for 10 min before 5 μg of OGT or RCAM-lysozyme was added to the reaction for 15 min at 30 °C. Sigma protocol was followed (product I 9266).
β-1,4-Galactosyltransferase Labeling—Immunoprecipitated proteins were rinsed twice with phosphate-buffered saline and then twice with 1× galactostransferase (GalT) labeling buffer (10×: 100 mm HEPES, pH 7.5, 100 mm Gal, 50 mm MnCl2) and then incubated in GalT buffer with 1 μCi/reaction of UDP-[3H]Gal, 1 unit of calf intestinal alkaline phosphatase at 4 °C overnight (35). Reactions were stopped with modified Laemmli buffer.
RESULTS
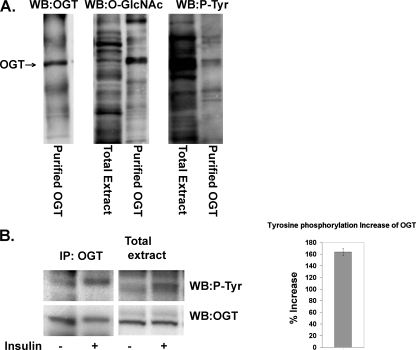
Insulin Increases Tyrosine Phosphorylation of OGT in 3T3-L1 Adipocytes—OGT is modified with both O-GlcNAc and tyrosine phosphorylation when overexpressed in insect cells (Fig. 1A) (15). We examined whether increased tyrosine phosphorylation of OGT occurs in insulin-stimulated 3T3-L1 adipocytes. OGT was immunoprecipitated from insulin-stimulated (10 min ± 100 nm insulin) 3T3-L1 adipocytes and immunoblotted with a phospho-tyrosine antibody. Tyrosine phosphorylation of OGT increased significantly after a 10-min insulin treatment (Fig. 1B). Whole cell extract in adjacent lanes was also examined for OGT and tyrosine phosphorylation, as a control.
FIGURE 1.
Insulin increases OGT tyrosine phosphorylation in 3T3-L1 adipocytes. A, OGT overexpressed in insect cells was purified by nickel chromatography, electrophoresed on SDS-PAGE, and Western blotted (WB) with OGT (AL28), CTD 110.6 O-GlcNAc, and phospho-tyrosine antibody (P-Tyr) (PY20, Santa Cruz Biotechnology). B, OGT was immunoprecipitated (IP) from ±100 nm insulin-treated 3T3-L1 adipocytes, electrophoresed on SDS-PAGE with whole cell extract, and Western blotted with phospho-tyrosine antibody. The same blots were stripped and blotted for OGT (AL28) to demonstrate equal OGT protein levels. Data are representative of experiments done in triplicate. The increase in OGT phosphorylation was quantified by densitometer readings of Western blots using the software program ImageJ from the National Institutes of Health. Insulin-induced tyrosine phosphorylation increased about 160% over basal levels.
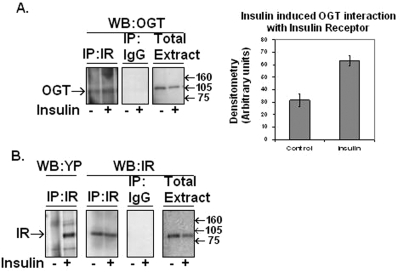
A Portion of OGT Co-immunoprecipitates with the Insulin Receptor—Insulin receptor was immunoprecipitated from 3T3-L1 adipocytes ±100 nm insulin. Immunoprecipitates were immunoblotted for the presence of OGT (Fig. 2A). Whole cell extract was used as a positive control for the presence of OGT. There was a slight increase in the amount of OGT interacting with the insulin receptor upon insulin treatment. We also confirmed the presence of IR- and insulin-induced phosphorylation of the IR in the immunoprecipitation and IR in whole cell extracts (Fig. 2B).
FIGURE 2.
Insulin receptor immunoprecipitation reveals OGT association via Western blot analysis. A, OGT (AL28) Western (WB) blot of immunoprecipitated (IP) IR, rabbit IgG control immunoprecipitation, and whole cell extract from 3T3-L1 adipocytes pretreated stimulated with ±100 nm insulin. B, the same samples as in A were Western blotted with insulin receptor antibody with the addition of IR immunoprecipitation Western blotted with tyrosine phosphorylation (YP) antibody. Data are representative of experiments done in triplicate. Data were quantified by densitometer readings of Western blots using the software program ImageJ from the National Institutes of Health.
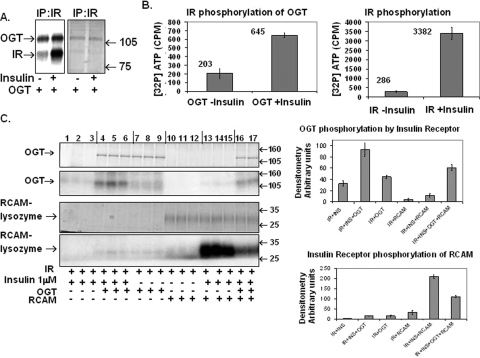
Isolated Insulin-stimulated IR Complex Phosphorylates OGT in Vitro—To test the hypothesis that OGT is a direct substrate of the insulin receptor and/or interacting with the insulin receptor proteins, we immunoprecipitated insulin receptor from ±100 nm insulin-stimulated 3T3-L1 adipocytes at 10 min and then restimulated the isolated IR complex with insulin in vitro for 10 min to maximize IR activity. Insulin stimulation of 3T3-L1 adipocytes increased the autophosphorylation of the insulin receptor and concomitantly caused increased phosphorylation of OGT (Fig. 3A). The incorporation of [γ-32P]ATP into OGT was measured by scintillation counting of excised Coomassie G250-stained OGT bands from SDS-PAGE (Fig. 3B). Insulin induced a 3-fold increase in OGT phosphorylation and over an 11-fold increase in insulin receptor autophosphorylation. Experiments were performed in triplicate.
FIGURE 3.
Insulin-stimulated IR phosphorylates OGT. A, purified OGT is incubated with immunoprecipitated (IP) insulin receptor from ±100 nm insulin (10 min)-treated 3T3-L1 adipocytes in kinase reaction buffer and stimulated with 1 μm insulin for 15 min. The reaction was stopped with the addition of modified Laemmli buffer, boiled, loaded onto 10% SDS-PAGE, stained with Coomassie G-250, and incubated with film. mw, molecular weight. B, OGT and insulin receptor phosphorylated in vitro similar to the samples in A are cut out of SDS-PAGE and [32P]ATP incorporation counted by a scintillation counter. Data are an average of three reactions and representative of three separate experiments. C, OGT is incubated with ±1 μm insulin-treated (10 min) insulin receptor in [32P]ATP kinase reaction buffer without and with a known insulin receptor substrate, RCAM-lysozyme, for 20 min in triplicate. The reaction was stopped with modified Laemmli buffer, electrophoresed on SDS-PAGE, stained with Coomassie Blue G-250, and autoradiographed onto film. Data are representative of triplicate experiments on two separate occasions. Data were quantified by densitometer readings of Western blots using the software program ImageJ from the National Institutes of Health.
We then tested whether OGT could compete with a well characterized insulin receptor substrate, carboxyamidomethylated lysozyme (RCAM-lysozyme), in an in vitro IR kinase assay. Purified OGT was incubated with non-stimulated (Fig. 3C, lanes 7–12) and insulin-stimulated insulin receptor (lanes 1–6 and lanes 13–17), with and without RCAM-lysozyme (Fig. 3C). OGT becomes phosphorylated upon incubation with activated insulin receptor, and OGT phosphorylation is competitively inhibited by the addition of RCAM-lysozyme (Fig. 3C, upper panel, lanes 10–17). Conversely, RCAM-lysozyme phosphorylation (Fig. 3C, lower panel, lanes 16 and 17) is inhibited by the presence of OGT, indicating that both substrates are competing in vitro for insulin-stimulated insulin receptor phosphorylation. Data represent triplicate experiments conducted on two separate occasions.
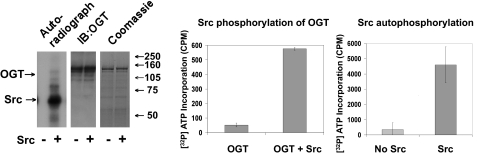
We also tested and found that a well known tyrosine kinase, Src, could phosphorylate OGT similar to the IR (Fig. 4). Purified OGT was incubated with ±Src in Src kinase buffer containing [γ-32P]ATP for 15 min. Gel bands of OGT incubated with either +Src or -Src from triplicate experiments were cut out of SDS-PAGE, and [32P]ATP was counted via a scintillation counter.
FIGURE 4.
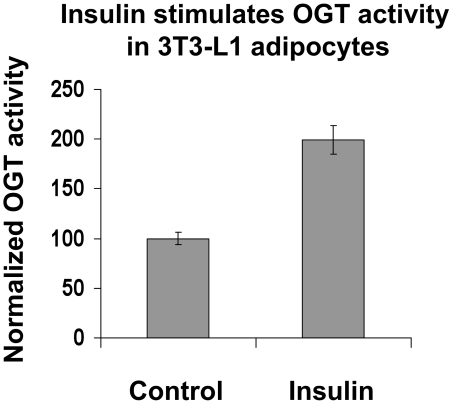
Insulin stimulates OGT activity in 3T3-L1 adipocytes. OGT was immunoprecipitated with OGT (AL28) antibody covalently attached to Sepharose A beads from ±100 nm insulin-stimulated 3T3-L1 adipocytes. The affinity bound OGT (AL28) was washed and incubated in OGT buffer containing UDP-[3H]GlcNAc and 1 mm CK2 peptide (OGT peptide substrate). The reaction was conducted at 22 °C for 30, min and radioactive [3H]GlcNAc incorporation into CK2 was measured by a scintillation counter. The graphs are an average of three experiments, and the graph represents three similar experiments on separate occasions. IB, immunoblot.
Insulin Stimulates OGT Activity in 3T3-L1 Adipocytes—Insulin stimulation of 3T3-L1 adipocytes activates an entire cascade of insulin signaling proteins. Insulin-stimulated tyrosine phosphorylation of IRS-1 and IRS-2, resulting in their activation of phosphatidylinositol 3-kinase, represents one of the earliest steps in the insulin signaling cascade. Using 100 nm insulin, the same concentration previously used to maximally stimulate IRS-1 and IRS-2 protein activation in 3T3-L1 adipocytes, we found that insulin stimulation (10 min) of 3T3-L1 adipocytes significantly increased OGT activity when compared with control adipocytes (Fig. 5). To further determine whether tyrosine phosphorylation correlates with OGT activity, we treated 3T3-L1 cells with the tyrosine phosphatase inhibitor sodium vanadate (supplemental Fig. 1). Treatment of 3T3-L1 cells with vanadate also resulted in increased OGT activity. When insulin was added to 3T3-L1 cells preincubated with sodium vanadate, there was a further increase in OGT activity versus inhibitor alone. Results shown in Fig. 5 are an average of three experiments, and data represent three independent experiments on separate occasions.
FIGURE 5.
Tyrosine kinase Src in vitro phosphorylation of OGT. OGT overexpressed from insect cells was incubated with ±Src in Src kinase buffer containing [32P]ATP for 15 min. Reactions were stopped with modified Laemmli buffer, electrophoresed on 10% SDS-PAGE, and either stained with Coomassie Blue G250 and underwent an audioradiogram or was transferred to polyvinylidene difluoride for Western blot analysis with OGT (AL-28). Gel bands of OGT incubated with ±Src were cut out of SDS-PAGE, and [32P]ATP was counted via a scintillation counter. Data are representative of experiments done in triplicate.
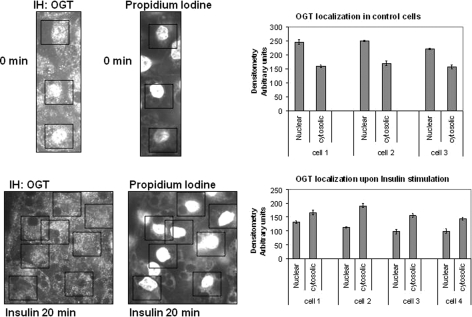
OGT Localizes to the Cytoplasm upon 20 Min of Insulin Stimulation of 3T3-L1 Adipocytes—3T3-L1 adipocytes were starved overnight in serum-free media, stimulated with ±100 nm insulin for 20 min, and fixed, and immunohistochemistry was performed with an OGT polyclonal antibody. The nuclei of the 3T3-L1 adipocytes were stained with propidium iodine to locate and differentiate the nucleus from lipid droplets within the cells. The high concentration of OGT within the nuclei of cells has been established (4, 36). After about 20 min of insulin stimulation, OGT becomes less concentrated within the nucleus, and a portion of the enzyme translocates to the cytoplasm (Fig. 6). We also subfractionated ±100 nm insulin-treated NIH 3T3 fibroblasts that overexpress the insulin receptor and analyzed the insulin-dependent concentration of OGT in cytoplasmic or nuclear fractions via Western blot analysis. In these cells, at 20 min of insulin stimulation, the proportion of OGT within the nucleus decreased, whereas that in the cytoplasm increased (supplemental Fig. 2 and cell fractionation protocol).
FIGURE 6.
OGT localizes to the cytoplasm upon 20 min of insulin treatment. 3T3-L1 adipocytes were starved overnight in serum-free media, stimulated with ±100 nm insulin for 20 min, and fixed, and immunohistochemistry (IH) was performed with AL-28 OGT antibody. Nuclear staining was performed with propidium iodine to locate the nucleus. The boxes outline the nucleus of the adipocyte in which OGT is present in the control and is absent in the insulin-treated cells. Data were quantified by densitometer readings of three different locations within the nucleus and the average when compared with the average of densitometer readings of three different locations within the cytoplasm of each cell listed using the software program ImageJ from the National Institutes of Health.
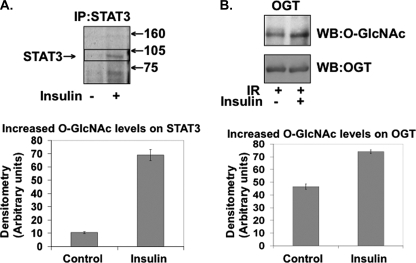
Insulin Stimulates O-GlcNAc Modification on STAT3 and OGT in 3T3-L1 Adipocytes—Since insulin stimulates OGT activity, we analyzed whether OGT's increased activity resulted in it modifying other O-GlcNAcylated proteins present. Insulin-stimulated 3T3-L1 adipocytes resulted in O-GlcNAc modification of STAT3 within 10 min (Fig. 7A). When OGT was incubated in the presence of insulin-activated IR and UDP-[3H]GlcNAc, Western blot analysis showed an increase in O-GlcNAc modification of OGT (Fig. 7B). These data support the hypothesis that OGT is activated by insulin signaling through the IR and that insulin stimulates specific O-GlcNAc modification substrates in a dynamic and transient fashion similar to phosphorylation.
FIGURE 7.
Insulin stimulation results in increased O-GlcNAc modification of OGT substrates. A, STAT3 was immunoprecipitated (IP) from ±insulin-treated 3T3-L1 adipocytes and labeled with UDP-[3H]Gal by GalT, electrophoresed on SDS-PAGE, enhanced, and exposed by autoradiography. B, OGT substrate was incubated with insulin-stimulated insulin receptor in the presence of UDP-[3H]GlcNAc, electrophoresed on SDS-PAGE, and Western blotted (WB) for O-GlcNAc and OGT. Data are representative of experiments done in triplicate.
DISCUSSION
The role of O-GlcNAc cycling in cell signaling, cell cycle control, transcriptional regulation, cellular stress, and protein degradation, as well as many other cellular processes, is just beginning to be revealed (1–3, 37–41). Increased O-GlcNAc has been shown to directly inhibit insulin signaling in several cell lines (42–44). OGT itself is known to be modified by both O-GlcNAc and tyrosine phosphorylation (15). The complexity of OGT's regulation, the stimuli that regulate OGT's activity, and specificity, which in turn control the enzyme's actions on its myriad substrates, may be the combinatorial result of: 1) intricate protein-protein interactions at OGT's tetratricopeptide repeats that allow it to interact with potentially thousands of proteins; 2) a number of post-translational modifications of OGT, such as O-GlcNAc, tyrosine, serine, and threonine phosphorylation; 3) OGT's quaternary structure, and; 4) OGT's response to nutrient sensing through different levels of UDP-GlcNAc. OGT has a number of potential tyrosine, serine, and threonine phosphorylation motif sites that may play a role in the regulation of OGT activity.4
Here, we show that OGT is actively involved in the insulin signaling pathway. Insulin stimulation increases OGT tyrosine phosphorylation. The insulin receptor phosphorylates OGT directly, and other down-stream tyrosine kinases may also be involved, such as Src, a known tyrosine kinase. Interestingly, there are potentially 15 Src tyrosine phosphorylation motifs on OGT.4 Insulin-stimulated tyrosine phosphorylation correlates with increased OGT activity. Inhibition of tyrosine phosphatases by vanadate also increases OGT activity. Upon insulin stimulation, a portion of the OGT translocates from the nucleus to the cytoplasm. Finally, insulin stimulation results in increased O-GlcNAcylation of several endogenous proteins. Since insulin is capable of stimulating OGT activity and increases O-GlcNAc modification, it is plausible that hyperinsulinemia, in conjunction with hyperglycemia, which increases UDP-GlcNAc pools, may be synergistically activating OGT, leading to abnormally increased O-GlcNAc levels in cells and contributing to insulin resistance. In addition, insulin-stimulated self-GlcNAcylation by OGT may also contribute to the activity of OGT.
Post-translational Modification of OGT Regulates Its Activity—Western blot analyses of overexpressed OGT in insect cells initially revealed the presence of O-GlcNAc and tyrosine phosphorylation on OGT. Elevations in O-GlcNAc caused by high glucose, glucosamine, insulin, and the inhibition of O-GlcNAcase inhibit the insulin responsiveness of adipocytes, muscle, and endothelial cells (42–44). Treating 3T3-L1 cells with 100 nm insulin resulted in increased tyrosine phosphorylation of OGT.
OGT has a number of predicted Grb2 SH2 binding motifs that may play a role in OGT's interactions with the insulin receptor. We found that OGT co-immunoprecipitates with the insulin receptor in ±insulin-treated cells, with slightly more OGT bound in the insulin-stimulated samples. We found that insulin-dependent tyrosine phosphorylation of OGT increased 3-fold after 15 min of incubation. Analyses of substrate competition between OGT and RCAM-lysozyme phosphorylation, a well studied IR substrate, are competitive substrates for the IR.
Insulin Stimulation of 3T3-L1 Adipocytes Also Increased OGT Activity—Insulin stimulation of 3T3-L1 adipocytes not only increased tyrosine phosphorylation but also increased OGT activity as well. Consistent with the increased OGT activity, OGT O-GlcNAc modification increased upon incubation with insulin-induced activated IR. Inhibition of tyrosine phosphatases also increased OGT activity. The finding that Src tyrosine kinase is also capable of phosphorylating OGT implies that there may be many kinases in different signaling pathways that are capable of phosphorylating and regulating OGT function. OGT may also be regulated, or in turn, regulating insulin signaling at a number of check points in the insulin signaling pathway. It remains to be seen whether insulin also stimulates the interaction of OGT with other insulin signaling proteins, but OGT has 17 potential GSK3β serine kinase motifs and two potential phosphatidylinositol 3-kinase p85 tyrosine binding motifs.4
Insulin Affects the Localization of OGT—In most cells, OGT is mostly localized to the nucleus, with smaller amounts within the cytoplasm (36). This localization may be important in the regulation of gene transcription since nearly every transcription factor analyzed to date is modified by O-GlcNAc. Significant amounts of OGT translocated from the nucleus at around 20 min after insulin stimulation and relocalized back to the nucleus after about 40 min. Although OGT translocation correlates with tyrosine phosphorylation, we do not claim this is the cause and effect of tyrosine phosphorylation but rather a result of insulin signaling possibly affecting a multitude of mechanisms downstream including kinases, scaffolding proteins, and/or nuclear pore proteins, resulting in a number of possible post-translation modifications on OGT and protein-protein interactions.
Insulin Stimulates the O-GlcNAc Modification of Known O-GlcNAc Protein STAT3—Insulin stimulation of adipocytes resulted in the increased O-GlcNAc modification of Stat3. Stat3 is important in adipogenesis (30) and leptin response in the hypothalamus, regulating food intake and energy expenditure (45, 46), and is involved in the activation of suppressor of cytokine signaling 3 (SOCS3), which in turn is responsible for the serine phosphorylation of IRS-1, resulting in its ubiquitination (47, 48). Prolactin-induced O-GlcNAcylation of Stat5 was found to be an important post-translational modification for Stat5 binding to the coactivator of transcription cAMP-response element-binding protein binding protein (49). Since this is an essential interaction for Stat5-mediated gene transcription, the insulin-induced OGT O-GlcNAcylation of Stat3 may also have a crucial role in its function in insulin signaling. Further studies would be interesting to see whether O-GlcNAc modification has any effect on SOCS3 and IRS-1 ubiquitination. The complex regulation of OGT is just beginning to be understood (4, 15, 21, 31, 50–52).
We have demonstrated that a major and a well characterized signaling pathway affects the activity of OGT in the insulin-responsive 3T3-L1 cell line. A growing number of groups are only beginning to elucidate the role that OGT and O-GlcNAc plays in a multitude of cellular functions. This study documents that insulin signaling affects OGT activity, tyrosine phosphorylation, localization, and downstream Stat3 O-GlcNAc modification, which may have a role in the etiology of diabetes.
Supplementary Material
Acknowledgments
We especially thank Chad Slawson for critical reading and suggestions of this manuscript and the Hart laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK61671 and National Institutes of Health Contract N01-HV-28180. G. W. H. receives a share of royalty received by the university on sales of the CTD 110.6 antibody. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures and supplemental text.
Footnotes
The abbreviations used are: O-GlcNAc, O-linked β-N-acetylglucosamine; OGT, O-GlcNAc transferase; O-GlcNAcase, O-GlcNAc specific β-d-N-acetylglucosaminidase; GalT, galactostransferase; IR, insulin receptor; IRS, insulin receptor substrate; STAT3, signal transducer and activator of transcription 3; RCAM-lysozyme, carboxyamidomethylated lysozyme.
Akilesh Pandey, personal communication.
References
- 1.Whelan, S. A., and Hart, G. W. (2003) Circ. Res. 93 1047-1058 [DOI] [PubMed] [Google Scholar]
- 2.Wells, L., Vosseller, K., and Hart, G. W. (2001) Science 291 2376-2378 [DOI] [PubMed] [Google Scholar]
- 3.Zachara, N. E., and Hart, G. W. (2006) Biochim. Biophys. Acta 1761 599-617 [DOI] [PubMed] [Google Scholar]
- 4.Kreppel, L. K., Blomberg, M. A., and Hart, G. W. (1997) J. Biol. Chem. 272 9308-9315 [DOI] [PubMed] [Google Scholar]
- 5.Lubas, W. A., Frank, D. W., Krause, M., and Hanover, J. A. (1997) J. Biol. Chem. 272 9316-9324 [DOI] [PubMed] [Google Scholar]
- 6.Comer, F. I., and Hart, G. W. (2001) Biochemistry 40 7845-7852 [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre, T., Alonso, C., Mahboub, S., Dupire, M. J., Zanetta, J. P., Caillet-Boudin, M. L., and Michalski, J. C. (1999) Biochim. Biophys. Acta 1472 71-81 [DOI] [PubMed] [Google Scholar]
- 8.Chou, T. Y., Hart, G. W., and Dang, C. V. (1995) J. Biol. Chem. 270 18961-18965 [DOI] [PubMed] [Google Scholar]
- 9.Kelly, W. G., Dahmus, M. E., and Hart, G. W. (1993) J. Biol. Chem. 268 10416-10424 [PubMed] [Google Scholar]
- 10.Arnold, C. S., Johnson, G. V., Cole, R. N., Dong, D. L., Lee, M., and Hart, G. W. (1996) J. Biol. Chem. 271 28741-28744 [DOI] [PubMed] [Google Scholar]
- 11.Tao, G. Z., Kirby, C., Whelan, S. A., Rossi, F., Bi, X., MacLaren, M., Gentalen, E., O'Neill, R. A., Hart, G. W., and Omary, M. B. (2006) Biochem. Biophys. Res. Commun. 351 708-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haltiwanger, R. S., Holt, G. D., and Hart, G. W. (1990) J. Biol. Chem. 265 2563-2568 [PubMed] [Google Scholar]
- 13.Love, D. C., Kochan, J., Cathey, R. L., Shin, S. H., and Hanover, J. A. (2003) J. Cell Sci. 116 647-654 [DOI] [PubMed] [Google Scholar]
- 14.Jinek, M., Rehwinkel, J., Lazarus, B. D., Izaurralde, E., Hanover, J. A., and Conti, E. (2004) Nat. Struct. Mol. Biol. 11 1001-1007 [DOI] [PubMed] [Google Scholar]
- 15.Kreppel, L. K., and Hart, G. W. (1999) J. Biol. Chem. 274 32015-32022 [DOI] [PubMed] [Google Scholar]
- 16.Lamb, J. R., Tugendreich, S., and Hieter, P. (1995) Trends Biochem. Sci. 20 257-259 [DOI] [PubMed] [Google Scholar]
- 17.Lubas, W. A., and Hanover, J. A. (2000) J. Biol. Chem. 275 10983-10988 [DOI] [PubMed] [Google Scholar]
- 18.Slawson, C., Shafii, S., Amburgey, J., and Potter, R. (2002) Biochim. Biophys. Acta 1573 121-129 [DOI] [PubMed] [Google Scholar]
- 19.Zachara, N. E., O'Donnell, N., Cheung, W. D., Mercer, J. J., Marth, J. D., and Hart, G. W. (2004) J. Biol. Chem. 279 30133-30142 [DOI] [PubMed] [Google Scholar]
- 20.Slawson, C., Zachara, N. E., Vosseller, K., Cheung, W. D., Lane, M. D., and Hart, G. W. (2005) J. Biol. Chem. 280 32944-32956 [DOI] [PubMed] [Google Scholar]
- 21.Hanover, J. A., Forsythe, M. E., Hennessey, P. T., Brodigan, T. M., Love, D. C., Ashwell, G., and Krause, M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11266-11271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, X., Ongusaha, P. P., Miles, P. D., Havstad, J. C., Zhang, F., So, W. V., Kudlow, J. E., Michell, R. H., Olefsky, J. M., Field, S. J., and Evans, R. M. (2008) Nature 451 964-969 [DOI] [PubMed] [Google Scholar]
- 23.Shafi, R., Iyer, S. P., Ellies, L. G., O'Donnell, N., Marek, K. W., Chui, D., Hart, G. W., and Marth, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5735-5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell, N., Zachara, N. E., Hart, G. W., and Marth, J. D. (2004) Mol. Cell Biol. 24 1680-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haltiwanger, R. S., Blomberg, M. A., and Hart, G. W. (1992) J. Biol. Chem. 267 9005-9013 [PubMed] [Google Scholar]
- 26.Wells, L., Vosseller, K., and Hart, G. W. (2003) CMLS Cell Mol. Life Sci. 60 222-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zachara, N. E., and Hart, G. W. (2004) Trends Cell Biol. 14 218-221 [DOI] [PubMed] [Google Scholar]
- 28.Hawkins, M., Barzilai, N., Liu, R., Hu, M., Chen, W., and Rossetti, L. (1997) J. Clin. Investig. 99 2173-2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cernkovich, E. R., Deng, J., Bond, M. C., Combs, T. P., and Harp, J. B. (2008) Endocrinology 149 1581-1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng, J., Hua, K., Lesser, S. S., and Harp, J. B. (2000) Endocrinology 141 2370-2376 [DOI] [PubMed] [Google Scholar]
- 31.Iyer, S. P., Akimoto, Y., and Hart, G. W. (2003) J. Biol. Chem. 278 5399-5409 [DOI] [PubMed] [Google Scholar]
- 32.Modan-Moses, D., Janicot, M., McLenithan, J. C., Lane, M. D., and Casella, S. J. (1998) Biochem. J. 333 825-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Student, A. K., Hsu, R. Y., and Lane, M. D. (1980) J. Biol. Chem. 255 4745-4750 [PubMed] [Google Scholar]
- 34.Kohanski, R. A., Frost, S. C., and Lane, M. D. (1986) J. Biol. Chem. 261 12272-12281 [PubMed] [Google Scholar]
- 35.Whelan, S. A., and Hart, G. W. (2006) Methods Enzymol. 415 113-133 [DOI] [PubMed] [Google Scholar]
- 36.Akimoto, Y., Kreppel, L. K., Hirano, H., and Hart, G. W. (1999) Diabetes 48 2407-2413 [DOI] [PubMed] [Google Scholar]
- 37.Kudlow, J. E. (2006) J. Cell Biochem. 98 1062-1075 [DOI] [PubMed] [Google Scholar]
- 38.Hanover, J. A. (2001) FASEB J. 15 1865-1876 [DOI] [PubMed] [Google Scholar]
- 39.Love, D. C., and Hanover, J. A. (2005) Sci's. STKE 2005 re13. [DOI] [PubMed] [Google Scholar]
- 40.Wells, L., Whelan, S. A., and Hart, G. W. (2003) Biochem. Biophys. Res. Commun. 302 435-441 [DOI] [PubMed] [Google Scholar]
- 41.Slawson, C., Housley, M. P., and Hart, G. W. (2006) J. Cell Biochem. 97 71-83 [DOI] [PubMed] [Google Scholar]
- 42.Patti, M. E., Virkamaki, A., Landaker, E. J., Kahn, C. R., and Yki-Jarvinen, H. (1999) Diabetes 48 1562-1571 [DOI] [PubMed] [Google Scholar]
- 43.Vosseller, K., Wells, L., Lane, M. D., and Hart, G. W. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5313-5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Federici, M., Menghini, R., Mauriello, A., Hribal, M. L., Ferrelli, F., Lauro, D., Sbraccia, P., Spagnoli, L. G., Sesti, G., and Lauro, R. (2002) Circulation 106 466-472 [DOI] [PubMed] [Google Scholar]
- 45.Buettner, C., Pocai, A., Muse, E. D., Etgen, A. M., Myers, M. G., Jr., and Rossetti, L. (2006) Cell Metabol. 4 49-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bates, S. H., Stearns, W. H., Dundon, T. A., Schubert, M., Tso, A. W., Wang, Y., Banks, A. S., Lavery, H. J., Haq, A. K., Maratos-Flier, E., Neel, B. G., Schwartz, M. W., and Myers, M. G., Jr. (2003) Nature 421 856-859 [DOI] [PubMed] [Google Scholar]
- 47.Rui, L., Yuan, M., Frantz, D., Shoelson, S., and White, M. F. (2002) J. Biol. Chem. 277 42394-42398 [DOI] [PubMed] [Google Scholar]
- 48.Ishizuka, K., Usui, I., Kanatani, Y., Bukhari, A., He, J., Fujisaka, S., Yamazaki, Y., Suzuki, H., Hiratani, K., Ishiki, M., Iwata, M., Urakaze, M., Haruta, T., and Kobayashi, M. (2007) Endocrinology 148 2994-3003 [DOI] [PubMed] [Google Scholar]
- 49.Gewinner, C., Hart, G., Zachara, N., Cole, R., Beisenherz-Huss, C., and Groner, B. (2004) J. Biol. Chem. 279 3563-3572 [DOI] [PubMed] [Google Scholar]
- 50.Lazarus, B. D., Love, D. C., and Hanover, J. A. (2006) Glycobiology 16 415-421 [DOI] [PubMed] [Google Scholar]
- 51.Okuyama, R., and Marshall, S. (2004) Biol. Pharm. Bull 27 1293-1296 [DOI] [PubMed] [Google Scholar]
- 52.Marshall, S., Okuyama, R., and Rumberger, J. M. (2005) Biochem. Biophys. Res. Commun. 332 263-270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.