Abstract
Using a multiple alignment of 175 cytochrome P450 (CYP) family 2 sequences, 20 conserved sequence motifs (CSMs) were identified with the program PCPMer. Functional importance of the CSM in CYP2B enzymes was assessed from available data on site-directed mutants and genetic variants. These analyses suggested an important role of the CSM 8, which corresponds to187RFDYKD192 in CYP2B4. Further analysis showed that residues 187, 188, 190, and 192 have a very high rank order of conservation compared with 189 and 191. Therefore, eight mutants (R187A, R187K, F188A, D189A, Y190A, K191A, D192A, and a negative control K186A) were made in an N-terminal truncated and modified form of CYP2B4 with an internal mutation, which is termed 2B4dH/H226Y. Function was examined with the substrates 7-methoxy-4-(trifluoromethyl)coumarin (7-MFC), 7-ethoxy-4-(trifluoromethyl)coumarin (7-EFC), 7-benzyloxy-4-(trifluoromethyl)coumarin (7-BFC), and testosterone and with the inhibitors 4-(4-chlorophenyl)imidazole (4-CPI) and bifonazole (BIF). Compared with the template and K186A, the mutants R187A, R187K, F188A, Y190A, and D192A showed ≥2-fold altered substrate specificity, kcat, Km, and/or kcat/Km for 7-MFC and 7-EFC and 3- to 6-fold decreases in differential inhibition (IC50,BIF/IC50,4-CPI). Subsequently, these mutants displayed 5-12 °C decreases in thermal stability (Tm) and 2-8 °C decreases in catalytic tolerance to temperature (T50) compared with the template and K186A. Furthermore, when R187A and D192A were introduced in CYP2B1dH, the P450 expression and thermal stability were decreased. In addition, R187A showed increased activity with 7-EFC and decreased IC50,BIF/IC50,4-CPI compared with 2B1dH. Analysis of long range residue-residue interactions in the CYP2B4 crystal structures indicated strong hydrogen bonds involving Glu149-Asn177-Arg187-Tyr190 and Asp192-Val194, which were significantly-reduced/abolished by the Arg187→Ala and Asp192→Alasubstitutions, respectively.
One of the most intriguing recent discoveries about mammalian cytochromes P450 (CYP)3 is the remarkable conformational plasticity exhibited by a number of the enzymes (1-4). This plasticity allows a single P450 to adapt its ligand binding site to a wide variety of compounds of different size, shape, and chemistry. The rabbit CYP2B4 provides some of the most striking examples of an adaptable active site as inferred from x-ray crystal structures of ligand-free, 4-(4-chlorophenyl)imidazole (4-CPI)-bound, 1-(4-chlorophenyl)imidazole-bound, and bifonazole (BIF)-bound x-ray crystal structures as well as solution thermodynamics derived from isothermal titration calorimetry (5-10). In particular, comparisons of three structures identified five plastic regions in CYP2B4 (8), which account for almost one-third of the protein and contribute to broad substrate specificity by allowing different conformations in response to ligands. These results suggest an important role of non-active site regions/residues in ligand-induced conformational transitions and differential substrate binding and catalysis. However, predicting the role of non-active site regions/residues is currently very difficult.
Recently, directed evolution of CYP2B1 has been carried out to locate important non-active site amino acid residues. Residues Val187, Phe202, Lys236, Asp257, His295, and Ser334 were found to contribute to enzyme catalysis and/or stability (11, 12). As a complement to directed evolution, amino acid sequences were compared among CYP2B6 (low heterologous expression in bacteria) and CYP2B1, CYP2B4, and CYP2B11 (high P450 expression), which led to the design of CYP2B6 L264F. This mutant showed enhanced expression and stability compared with the wild type (13).
CYP2B6 is one of the most polymorphic P450 enzymes in humans, with 28 alleles described to date. Several genetic variants are linked to altered plasma levels and/or in vitro metabolism of bupropion (14 -17). Mutations in all the variants are located in non-active site regions. Two of the non-synonymous changes in particular, Q172H and K262R, are found in multiple haplotypes. Frequencies of the three most common variants range from 14 to 49% for Q172H, 17 to 63% for K262R, and 0 to 14% for R487C depending on the ethnicity of the population studied (17). At present, the structural basis for the altered function of P450 2B6 variants or for species differences relative to CYP2B1, CYP2B4, or CYP2B11 with regard to the oxidation of steroids (18), or inhibition by imidazoles (16) is largely unknown.
Based on the x-ray crystal structure of bacterial P450cam, Osamu Gotoh in 1992 analyzed 52 P450 family 2 sequences (CYP2), and identified 6 substrate recognition sites (SRSs) (19). These SRSs have been used extensively to guide site-directed studies in CYP2B enzymes (20). Subsequent x-ray crystal structures have verified most of the active site residues inferred from mutagenesis studies (1, 7, 10, 21). However, to investigate the role of non-active site regions/residues we need an additional approach. One such approach is conserved sequence motifs (CSM) analysis. In previous work, a multiple sequence alignment from representative alphaviruses was used to determine physical chemical property motifs (likely functional areas) with our PCPMer program. Information on residue variability, propensity to be in protein interfaces, and surface exposure on the model was combined to predict surface clusters likely to interact with other viral or cellular proteins. Mutagenesis of these clusters indicated that the predictions accurately detected areas crucial for virus infection (22). In addition, we successfully used this approach to locate regions far from the active site that modulate substrate binding and processivity in apurinic/apyrimidinic endonuclease (APE1) and related nucleases (23, 24). We also showed that CSM as defined by the software package PCPMer can find functionally important residues in surface-exposed regions of viral proteins (25, 26). In the present study, we examined a set of 175 P450 sequences from family 2 and identified 20 CSMs. Based on existing structural and functional information on the various CSMs, the role of CSM 8 (187RFDYKD192 in CYP2B4) in enzyme catalysis, inhibition, and stability was studied by site-directed mutagenesis.
EXPERIMENTAL PROCEDURES
Materials—7-Methoxy-4-(trifluoromethyl)coumarin (7-MFC), 7-ethoxy-4-(trifluoromethyl)coumarin (7-EFC), and 7-benzyloxy-4-(trifluoromethyl)coumarin (7-BFC) were purchased from Invitrogen. [4-14C]Testosterone was obtained from Amersham Biosciences. 4-CPI, BIF, NADPH, and H2O2 were bought from Sigma. Recombinant NADPH-cytochrome P450 reductase (CPR) and cytochrome b5 (b5) from rat liver were prepared as described previously (27). Oligonucleotide primers for PCR were obtained from Sigma Genosys (Woodlands, TX). The QuikChange XL site-directed mutagenesis kit was obtained from Stratagene (La Jolla, CA). Nickel-nitrilotriacetic acid affinity resin was purchased from Qiagen. All other chemicals were of the highest grade available and were obtained from standard commercial sources.
Identification of Conserved Sequence Motifs—We selected 175 CYP2 sequences from the species human, mouse, rat, dog, fugu, and zebrafish, and generated a multiple sequence alignment with ClustalW (28), as presented in supporting data (supplemental Table S1). This multiple sequence alignment was further analyzed with our program PCPMer. In PCPMer each of the 20 natural amino acids was represented by a 5-dimensional vector. The basis vectors of this 5-dimensional space were derived by multidimensional scaling of 237 dimensional physicochemical property (PCP) space (23). The eigenvector that had the highest eigenvalue correlated very well (r = 0.95) with the hydrophilicity scale. However, every eigenvector was a linear combination of different PCP of the 237 dimensional spaces. PCPMer generated a profile for the alignment at every position, which included the standard deviation and relative entropy (30) for each position and component of the 5-dimensional space. PCPMer then used these profiles to identify high relative entropy clusters (highly conserved regions) CSM.
Mutagenesis, Expression, and Purification of P450 Enzymes—All the site-directed single mutants in CSM 8 and a negative control K186A were created using CYP2B4dH/H226Y (H226Y) as the template and appropriate forward and reverse primers as presented in supplemental Table S2. In addition, R187A and D192A were created in CYP2B1dH. To confirm the desired mutation and verify the absence of unintended mutations all constructs were sequenced at the University of Texas Medical Branch Protein Chemistry Laboratory (Galveston, TX). H226Y and CSM 8 mutants were expressed as His-tagged proteins in Escherichia coli TOPP3 and purified using a nickel-affinity column as described previously (7). The P450 content was measured by reduced CO-difference spectra. Protein concentrations were determined using the Bradford protein assay kit (Bio-Rad, Hercules, CA).
Enzyme Assays—The standard NADPH-dependent enzyme assay with 7-MFC, 7-EFC, 7-BFC, and testosterone was essentially carried out as described previously (12, 31). To determine the substrate specificity, 200 μm of 7-MFC, 7-EFC, and testosterone and 75 μm of 7-BFC in 2% methanol were used. For steady-state kinetic analysis a concentration range of 10-200 μm of 7-MFC and 7-EFC was used. Above 200 μm concentrations the substrates were not soluble in 2% methanol. The reconstituted system contained P450 (0.05 μm), CPR, and b5 at a molar ratio of 1:4:2. Cumene hydroperoxide (CuOOH)-supported reactions were carried out at 1 mm CuOOH and were devoid of NADPH, CPR, and b5. Steady-state kinetic parameters were determined by regression analysis using SigmaPlot (Jandel, San Rafel, CA). The kcat and Km values were determined using the Michaelis-Menten equation. Each kinetic experiment included H226Y, K186A, and the CSM 8 mutants simultaneously for more accurate comparison of the data.
Inhibition Studies—Enzyme inhibition was measured using the 7-EFC O-deethylation assay in a final reaction volume of 100 μl at 0.005-0.1 μm 4-CPI and 0.1-2.5 μm BIF concentrations as described previously (31). BIF and 4-CPI inhibition studies for all the mutants utilized 0.025 μm P450. Nonlinear regression analysis was performed to fit the data using a four-parameter logistic function to derive the IC50 values. Each experiment included H226Y, K186A, and the CSM 8 mutants simultaneously for more accurate comparison of the data.
Thermal Stability—Inactivation of P450 was essentially carried out as described (11, 13). The reaction mixture contained 1 μm protein in 100 mm Hepes buffer, pH 7.4, in a 1-ml semi-micro spectrophotometric cell with constant stirring using a Shimadzu-2600 spectrophotometer. Thermal inactivation was carried out by measuring a series of absorbance spectra in the 340-700 nm range as a function of temperature between 30 and 70 °C with the interval of 2.5-5 °C and a 3-min equilibration at each temperature range. Determination of the total concentration of the heme protein was done by non-linear least square approximation of the spectra using a linear combination of spectral standards of CYP2B4 low spin, high spin, and P420 states. All data treatment and fitting of the titration curves were performed with our SpectraLab software as described (32). Fitting of the temperature profile curves was performed by regression analysis using SigmaPlot (Jandel, San Rafel, CA). The inactivation profiles were fit to a sigmoidal curve using four-parameter logistic function to obtain the mid-point of the thermal transition temperature (Tm) as described (11, 13).
Catalytic Tolerance to Temperature—The catalytic tolerance to temperature was studied by incubating enzyme (20 pmol in 20 μl dialysis buffer for two assays) at different temperatures (30-70 °C) with the interval of 2.5-5 °C for 10 min. The samples were then chilled in ice for 15 min and then brought to room temperature prior to measuring enzyme activity using a 7-EFC O-deethylation assay as described earlier (13). The temperature at which the enzyme retains 50% of the activity (T50) was calculated by fitting the data to a sigmoidal curve using a four-parameter function by regression analysis using SigmaPlot.
RESULTS
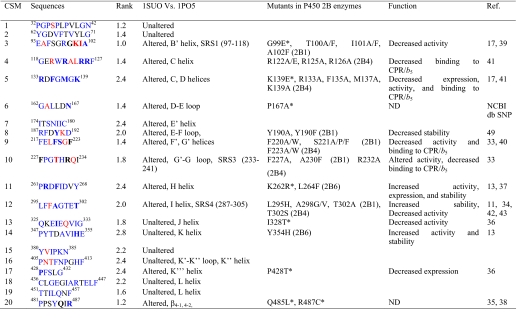
Identification of CSM in P450 Family 2—The program PCPMer identified 20 CSM from the multiple alignments of the selected 175 CYP2 sequences (supplemental Table S1). Because we included sequences from diverse subfamilies, we did not expect to pick up motifs that are specific for subfamilies but rather CSMs that are important and common for the whole CYP2 enzymes. These CSMs were conserved structurally, functionally, and dynamically because of the general features of the family, and they were spread throughout the sequences (Fig. 1). Furthermore, individual motifs and residues within the motifs were analyzed from 175 sequences to rank the level of conservation (Table 1). The rank order for conservation as defined by the relative entropy level of the motifs is presented in column 3, whereas the rank order of the residues within the motifs are in the order blue > gray > red.
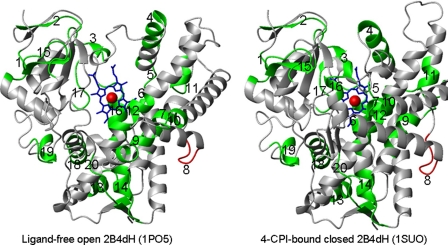
FIGURE 1.
Structure of ligand-free CYP2B4 (1PO5) and CYP2B4-CPI (1SUO). The segment in green shows all the CSM and in red is the CSM 8, which was studied experimentally.
TABLE 1.
PCPMer motifs of CYP2 enzymes generated using 175 different sequences
Residue numbers correspond to the rabbit CYP2B4 sequence.
In column 2 the colors of the residues in the motifs represent the rank order of sequence conservation as a function of relative entropy; red is the least conserved, black is intermediate, and blue represents the most conserved residues. The residues in bold represent ones for which genetic variants or site-directed mutants are known. In column 3 the rank order for conservation of the motifs is presented. In column 4 “altered” stands for either change in secondary structure or displacement of the backbone by ≥ 1 Å when open and closed structures are compared. In column 5, the asterisks represent genetic variants in the human CYP2B6, and “P450” represents the P450 in which the mutants, indicated prior to parenthesis, were created. Please note that some of the residues in column 4 are different from column 2 because the sequence differences among the CYP2B enzymes. SRS: substrate recognition site. ND: not determined. Activity is shown for 7-BR, 7-EFC, benzphetamine, testosterone, progesterone, PCB, CPA, IFA, or specific CYP2B6 substrate(s).
Structural and Functional Analyses of the CSM—Using the reported data detailed structural and functional analyses of the CSMs were performed. Structural analysis was performed by comparing CYP2B4 ligand-free (open, 1PO5) and 4-CPI-bound (closed, 1SUO) structures, which differ in their secondary structures and backbone movements (1-8 Å) (6, 7), referred to as enzyme plasticity (8). We analyzed each CSM, and the results describing altered regions of the CYP2B4 are presented in Table 1 and Fig. 1. It was not surprising that regions that interact directly with the heme are conserved, such as CSM 12 in the middle of helix I or CSM 18 at the beginning of helix L. CSM 9 and 10 (helix F′,G′, and beginning of helix G) are likely involved in enzyme opening/closing and alter their conformation drastically upon ligand binding. On the other hand, CSM 1, a loop before helix A′, and CSM 2, a β strand after helix A, are also conserved without any visible involvement in function.
A probable functional role of these CSMs was assessed by reviewing the reported mutagenesis data on CYP2B enzymes, including genetic variants of human CYP2B6. Detailed observations are presented in Table 1 (1, 6, 13, 17, 33-45). It was intriguing to see that many of these CSMs were subjected previously to experimental studies, which showed effects on structure, function, expression, and/or stability (Table 1). Based on the literature, many residues in helices C and D are involved in CPR and/or b5 binding. Similarly, the role of SRS/active site and substrate access channels mainly through B/C, F/G, H, and I loops/helices has been studied extensively, especially in CYP2B1 (13, 33-34, 39). CSMs 3, 10, and 12 are part of SRS1, SRS3, and SRS4, respectively. In contrast, CSMs 1, 2, 7, 15, 16, 18, and 19 have not been studied earlier and have the least dynamic structures. The most intriguing finding was that the CYP2B6 variants, whose altered functions cannot be explained based on existing structure-function information, are found in the CSM. For example, K262R*, which is the most frequent variant (17-63%) and shows altered activity, is located in motif 11 (37). Interestingly, a mutant in the same CSM, L264F, showed increased stability and expression (13).
A significant correlation was found in rank orders of the residues within the motifs and their known functions (Table 1). This correlation suggests that the CSM can be used as one of the approaches to identify the functional role of the residues within motifs. Two interesting regions to test this concept were CSM 8 (residues 187-192 in the E-F loop) and CSM 10 (residues 227-234 in the F-G or G′-G loop), because these loops are juxtaposed to the most dynamic regions of the protein, the region involving helices F/G (6, 7) (Table 1). In addition, these loops are within regions (residues 165-300) that harbor 80% of the beneficial mutations in CYP1A1, CYP2A6, CYP2B1, CYP2B6, CYP2B11, and CYP3A4 enzymes (11-13, 46, 47, 49-53, 60) identified by directed evolution or rational mutagenesis of non-active site residues. Although the rank orders of conservation CSM 8 and 10 are not very high, they contain an appropriate blend of rank orders of conservation of the residues within the motifs (Table 1). For further functional analysis we selected CSM 8, because it is present between plastic regions 3 (177-188) and 4 (203-298) (8). In CSM 8 Arg187, Phe188, Tyr190, and Asp192 have the highest rank order of conservation with ≤10 non-identical residues (blue), whereas Asp189 has an intermediate rank order (gray) and Lys191 has the lowest rank order (red) of conservation (data not shown). Next we tested the hypothesis that the rank orders of conservation of residues within motif 8 are related to their importance in CYP2B4 structure and function.
Role of CSM 8 in CYP2B4 Substrate Specificity—We created R187A, R187K, F188A, D189A, Y190A, K191A, and D192A in addition to K186A, which serves as a negative control. The mutants were then characterized using coumarin substrates (7-MFC, 7-EFC, and 7-BFC) of variable sizes (Fig. 2). Changes ≥2-fold in substrate specificity, kcat, Km, and/or kcat/Km compared with H226Y are considered significant in the discussion below. K186A, R187K, D189A, and K191A showed no significant changes in the ratio of dealkylation of 7-EFC/7-MFC or 7-BFC/7-MFC at a single substrate concentration (Table 2). R187A showed a ∼2-fold higher 7-EFC/7-MFC activity ratio than H226Y. In contrast, R187A, Y190A, and D192A showed >4-fold lower 7-BFC/7-MFC ratios.
FIGURE 2.
Structures of 7-MFC, 7-EFC, 7-BFC, 4-CPI, and BIF.
TABLE 2.
O-deethylation of 7-MFC, 7-EFC, and 7-BFC by H226Y, K186A, and CSM mutants
Results are representative of two or three independent determinations carried out simultaneously for more accurate comparison between the template and the mutants. The variation between the experiments is approximately ± 20%.
|
P450
|
Turnover
|
Substrate specificity
|
|||||
|---|---|---|---|---|---|---|---|
| 7-MFC | 7-EFC | 7-BFC | 7-EFC/7-MFC | 7-BFC/7-MFC | |||
| nmol/min/nmol P450 | |||||||
| H226Y | 8.5 | 8.1 | 2.9 | 0.95 | 0.38 | ||
| K186A | 12 | 11 | 3.8 | 0.95 | 0.31 | ||
| R187A | 5.1 | 11 | 0.34 | 2.2 | 0.07 | ||
| R187K | 5.2 | 7.5 | 2.8 | 1.4 | 0.54 | ||
| F188A | 2.1 | 3.7 | 0.51 | 1.7 | 0.24 | ||
| D189A | 8.5 | 7.2 | 3.0 | 0.84 | 0.35 | ||
| Y190A | 8.2 | 9.3 | 0.08 | 1.2 | 0.01 | ||
| K191A | 8.5 | 8.3 | 3.1 | 0.99 | 0.37 | ||
| D192A | 4.7 | 3.0 | 0.51 | 0.62 | 0.11 | ||
Role of CSM 8 in CYP2B4 Steady-state Kinetics—In steady-state kinetic analysis, R187A showed an unchanged kcat/Km with 7-MFC but >2-fold increase in kcat/Km (0.20 versus 0.09 min-1, μm-1) with 7-EFC (Table 3). In contrast to the template, R187K and F188A showed ∼2.5-fold lower kcat and Km with 7-MFC than 7-EFC. In addition, whereas Y190A showed ∼2-fold decrease in the Km for 7-MFC, D192A showed >2- and >3-fold decreases in kcat/Km for 7-MFC and 7-EFC, respectively (0.04 and 0.025 versus 0.08 and 0.09, respectively), compared with H226Y. K186A, D189A, and K191A did not show significant changes in the kcat, Km, or kcat/Km values compared with H226Y (Table 3).
TABLE 3.
Steady-state kinetic analysis of the oxidation of coumarin substrates by H226Y, K186A, and CSM 8 mutants
|
P450
|
7-MFC
|
7-EFC
|
||||
|---|---|---|---|---|---|---|
| kcat | Km | kcat/Km | kcat | Km | kcat/Km | |
| min−1 | μM | min−1 | μM | |||
| H226Y | 11 (0.8)a | 134 (18) | 0.081 | 11 (1) | 127 (26) | 0.09 |
| K186A | 17 (1.6) | 143 (24) | 0.12 | 16 (1.0) | 170 (18) | 0.09 |
| R187A | 6 (0.3) | 88.6 (10) | 0.07 | 11 (0.6) | 54.4 (8.4) | 0.20 |
| R187K | 5.5 (0.3) | 64.9 (8.2) | 0.08 | 12 (2) | 173 (45) | 0.07 |
| F188A | 2.7 (0.1) | 114 (11) | 0.021 | 8.9 (2) | 355 (222) | 0.025 |
| D189A | 14 (1.6) | 176 (25) | 0.08 | 11 (1.8) | 170 (4.8) | 0.06 |
| Y190A | 7.7 (0.2) | 72.3 (2.5) | 0.10 | 11 (0.4) | 81.2 (7.2) | 0.13 |
| K191A | 11 (0.5) | 110 (11) | 0.10 | 10 (0.5) | 91.4 (19) | 0.11 |
| D192A | 7.8 (1.3) | 185 (52) | 0.04 | 4.9 (0.35) | 194 (23) | 0.025 |
Standard errors for fit to the Michaelis-Menten equation. The variation between two or three experiments was ≤15%.
Role of CSM 8 in CYP2B4 Inhibition—To examine the effect of the mutations on binding of inhibitors of different sizes, 4-CPI and BIF were used, and 7-EFC O-deethylation was monitored (31) (Table 4). Of the eight mutants generated, the most interesting results were observed with R187A, R187K, F188A, Y190A, and D192A. Compared with H226Y, these mutants showed >6-, >4-, >3, ∼4-, and ∼4-fold decreased differential inhibition (IC50,BIF/IC50,4-CPI), respectively, (Table 4). Consistent with the lack of changes in steady-state kinetics of alkoxy-coumarin oxidation, K186A, D189A, and K191A showed < 2-fold changes in differential inhibition.
TABLE 4.
Inhibition by 4-CPI or BIF of 7-EFC O-Deethylation by H226Y, K186A, and CSM 8 mutants
|
P450
|
IC50
|
||
|---|---|---|---|
| BIF | 4-CPI | BIF/4-CPI | |
| μM | μM | ||
| H226Y | 0.80 (0.09)a | 0.010 (0.004) | 80 |
| K186A | 0.70 (0.02) | 0.014 (0.002) | 50 |
| R187A | 0.41 (0.02) | 0.033 (0.004) | 12 |
| R187K | 0.46 (0.02) | 0.024 (0.002) | 19 |
| F188A | 0.18 (0.03) | 0.007 (0.001) | 26 |
| D189A | 0.64 (0.16) | 0.009 (0.001) | 71 |
| Y190A | 0.55 (0.11) | 0.025 (0.005) | 22 |
| K191A | 1.0 (0.16) | 0.008 (0.001) | 125 |
| D192A | 0.56 (0.10) | 0.024 (0.006) | 23 |
Standard errors for fit to the sigmoidal curve using four-parameter logistic function are shown in parentheses. The variation between two or three experiments is ≤15%.
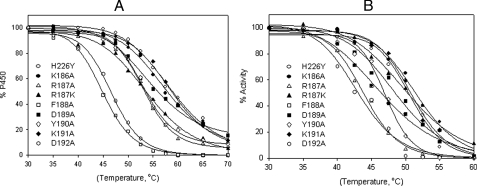
Role of the CSM 8 in Protein Stability—R187A, Y190A, and D192A showed ∼8-, 3-, and 3-fold lower expression, and higher P420, respectively, than H226Y (data not shown), suggesting decreased protein stability. Therefore, all the CSM 8 mutants along with H226Y and K186A were studied for thermal stability (Fig. 3A and Table 5). The Tm values for R187A, R187K, F188A, D189A, Y190A, and D192A were 5.2, 4.6, 13.4, 3.6, 4.6, and 11.6 °C, respectively, lower than H226Y. Similarly, the T50 of the CSM mutants and K186A was measured and compared with H226Y (Fig. 3B and Table 5). The T50 values for R187A, R187K, F188A, D189A, Y190A, and D192A were 7.4, 1.5, 4.2, 3.0, 4.4, and 8.5 °C, respectively, lower than H226Y. Consistent with the functional assays reported above, the Tm and T50 of K186A and K191A were similar to H226Y.
FIGURE 3.
A, thermal inactivation of CYP2B4dH mutants (1 μm) monitored by a decrease in the amount of total heme protein as a function of temperature as described under “Experimental Procedures.” Determination of the total concentration of the heme protein was done by non-linear least square approximation of the spectra by a linear combination of spectral standards of CYP2B4 low spin, high spin, and P420 states. The data were fit to a sigmoidal curve to obtain Tm. B, catalytic tolerance to temperature monitored by a decrease in the enzyme activity with 7-EFC as a function of temperature as described under “Experimental Procedures.” The data were fit to a sigmoidal curve to obtain T50.
TABLE 5.
Thermal stability of H226Y, K186A, and CSM 8 mutants
Standard errors for fit to the sigmoidal curve using four-parameter logistic function are shown in parentheses.
| P450 | Tm | T50 | |
|---|---|---|---|
| °C | |||
| H226Y | 58.4 (0.7)a | 51.5 (0.7)b | |
| K186A | 57.3 (0.6) | 50.7 (0.6) | |
| R187A | 53.2 (0.3) | 43.9 (0.3) | |
| R187K | 53.8 (0.6) | 50.0 (0.7) | |
| F188A | 45.0 (0.2) | 47.3 (0.9) | |
| D189A | 55.3 (0.7) | 48.5 (0.8) | |
| Y190A | 53.8 (0.6) | 47.1 (0.4) | |
| K191A | 58.4 (0.5) | 50.7 (0.6) | |
| D192A | 46.8 (0.3) | 43.0 (0.7) | |
Variations between two or three experiments are ≤0.75 °C.
Variations between two or three experiments are ≤1.0 °C.
Functional Role of CSM 8 in CYP2B1—To investigate whether the functional role of CSM 8 is conserved in other CYP2B enzymes, we created R187A and D192A in CYP2B1dH and characterized the mutants for P450 expression, substrate specificity, differential inhibition, and thermal stability. R187A and D192A showed ∼10-fold lower expression and higher P420 than CYP2B1dH (data not shown), suggesting decreased protein stability. The purified D192A showed no holo P450 based on CO-difference spectra, and therefore was omitted from further studies. R187A showed a 1.6-fold higher 7-EFC/7-MFC activity ratio than CYP2B1dH (Table 6). Compared with CYP2B1dH, R187A showed ∼3-fold decreased differential inhibition (IC50,BIF/IC50,4-CPI). Finally, the Tm values for R187A and D192A were 11.4 and 9 °C, respectively, lower than 2B1dH, and the T50 of R187A was 3.2 °C lower than 2B1dH.
TABLE 6.
Characterization of 2B1dH and CSM 8 mutants R187A and D192A
|
P450
|
Activitya
|
Specificity of 7-EFC/7-MFC
|
IC50
|
Tm
|
T50
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 7-MFC | 7-EFC | BIF | 4-CPI | BIF/4-CPI | ||||||
| nmol/min/nmol P450 | °C | |||||||||
| 2B1dH | 6.2 | 7.7 | 1.2 | 0.47 (0.09) | 0.034 (0.003) | 14 | 57.0 (1.0) | 49.6 (0.6) | ||
| K187A | 6.1 | 11.6 | 1.9 | 0.42 (0.07) | 0.081 (0.010) | 5.2 | 45.6 (0.5) | 46.4 (0.5) | ||
| D192A | NDb | ND | ND | ND | ND | ND | 48.1 (0.7) | ND | ||
The activity was determined at 150 μm substrate. Inhibition and catalytic tolerance to temperature were carried out using 150 μm 7-EFC. Standard errors for fit to the sigmoidal curve using four-parameter logistic function are shown in parentheses. The variations between two experiments in the case of activity and inhibition determinations were ≤20% and in the case of thermal stability determination was ≤1.0 °C.
ND, not determined because there was no holo P450 in D192A when measured using CO difference spectra.
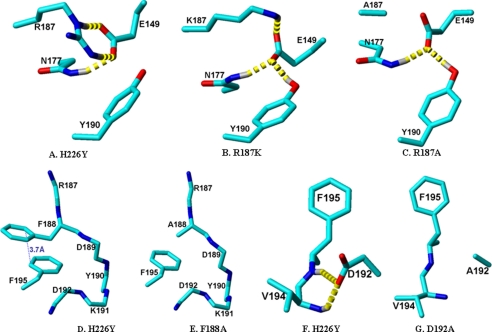
Molecular Modeling of the CSM Mutants Based on the CYP2B4-4-CPI Structure—To examine the role of the Arg187→Ala, Phe188→Ala, Tyr190→Ala, and Asp192→Ala substitutions in decreased protein stability, long range residue-residue interactions were analyzed in the energy-minimized CYP2B4-4-CPI structure using MolMol (54). We observed the residue-residue interaction sites Glu149-Asn177-Arg187-Tyr190, Asp192-Val194, and Phe188-Phe195. Furthermore, we generated models of the mutants by energy minimization using AMBER (55) to investigate the role of the substitutions in protein stability through altered hydrogen bonds, charge interactions, and π-π stacking (Fig. 4). Fig. 4A clearly shows that Arg187 forms two strong H-bonds with Glu149 (<1.9 Å) and an ionic interaction with Tyr190 (3.2 Å). Although the Arg187→ Ala substitution (Fig. 4C) completely abolished the H-bonds and ionic interaction, the Arg187→Lys substitution (Fig. 4B) retained one of the two H-bonds to Glu149. Tyr190 formed a relatively weak H-bond with Glu149 in R187K and R187A. In addition, Arg177 formed a H-bond with Glu149 in H226Y, R187K, and R187A. Phe188 and Phe195 were stabilized by very strong π-π stacking (Fig. 4D), which was disrupted by the Phe188→Ala substitution (Fig. 4E). Furthermore, Asp192 formed two strong hydrogen bonds to the backbone of Val194 (Fig. 4F), which were abolished by the Asp192→Ala substitution.
FIGURE 4.
Crystal structure of CYP2B4-4-CPI (1SUO) representing the hydrogen bond pattern (dashed yellow line) in residue-residue interactions. Panel A shows the interaction among Glu149-Asn177-Arg187-Tyr190, and panels B and C are models of R187K and R187A. Panel D shows the interaction between F188A-F195A and panel E is a model of F188A. Similarly, panel F shows the hydrogen bonds between Asp192 and Val194 in H226Y, and panel G shows the same region of the D192A mutant. Except for H226Y all the mutant models were energy-minimized using AMBER.
DISCUSSION
Data from numerous site-directed mutagenesis, directed evolution, and genetic polymorphism studies strongly suggest the role of non-active site regions/residues in P450 expression, stability, ligand binding, and/or catalytic activity (1-18). X-ray crystallography, solution thermodynamics, and site-directed and random mutagenesis have provided some clues about the role of these regions/residues in enzyme functions. As a complementary approach we utilized CSM analysis followed by site-directed mutagenesis and enzyme characterization. Analysis of available data suggested important roles of CSM in P450 structure-function relationships, and new experimental studies showed that the most conserved residues (Arg187, Phe188, Tyr190, and Asp192) in CSM 8 are involved in regulating enzyme catalysis, inhibition, and stability of CYP2B enzymes.
Conserved motifs are well known in several proteins and have been documented in ProSite (www.expasy.ch/prosite/). However, there is only one P450 ProSite motif, which describes the cysteine heme-iron ligand signature ([FW]-[SGNH]-X-[GD]-{F}-[RKHPT]-{P}-C-[LIVM-FAP]-[GAD]) (56). Additional known so-called, “conserved motifs” in P450s include: 1) a membrane-binding motif (57) and 2) Pro-rich motif (30PPGPTPFP37 in P450 2C2) (58). However, these motifs do not shed light on important P450 functions such as differential or overlapping substrate specificity, strict stereo- and regioselectivity, or protein stability. In this study we used a quantitative mathematical description of the naturally occurring 20 amino acids and performed statistical analysis of a multiple sequence alignment from 175 different CYP2 sequences, and identified 20 CSM. This approach may have been overlooked previously, because the enzymes share similar secondary or tertiary structural features irrespective of their diverse sequence identity (15-99%).
Although the mechanism by which the CSM 8 modulates enzyme function is not clear, it can be speculated that R187A, R187K, F188A, Y190A, and D192A exhibit altered dynamics, leading to opening of the active site either directly or by influencing the PR4 region (F/G helices; a proposed substrate access channel), which confers a preference for the larger ligand (BIF) over the smaller ligand (4-CPI). This conclusion is supported by the increase in differential substrate specificity (7-EFC/7-MFC) and kcat values, especially with R187A, R187K, and F188A. Furthermore, R187A, which showed the greatest alteration in substrate specificity (7-EFC/7-MFC, Tables 2 and 3), showed a >10-fold increased kcat for testosterone 16α-hydroxylation (6.6 min-1 versus 0.6 min-1) and a novel 16β-hydroxylation activity (1.5 min-1) (data not shown), suggesting that the Arg187→Ala substitution allows the enzyme to accommodate testosterone in both orientations. However, a reduced preference for the larger substrate 7-BFC over 7-MFC or 7-EFC in R187A, F188A, Y190A, and D192A is difficult to explain. In any event, the changes in substrate specificity, stereo- and regioselectivity, and inhibitor selectivity are in many ways just as or more striking than those of a recent study of active site substitutions in CYP2B4 (31). An altered interaction of P450 mutants with CPR or b5, leading to altered enzyme catalysis, can be ruled out, because most CSM 8 mutants showed similar changes in relative enzyme activity with 7-MFC, 7-EFC, and 7-BFC in CuOOH- to that of NADPH-supported reactions (data not shown). CuOOH is an alternate oxidant, which does not require redox partners for substrate metabolism.
The speculation that CSM 8 regulates a conformational transition is supported by our studies with a bacterial enzyme P450eryF, which suggest an important role for E helix residues 154-155 and E-F loop residues 160-164. The E-F loop is largely homologous to CSM 8 in CYP2 enzymes. Based on the effect of ionic strength, protein labeling by 7-(diethylamino)-3-(4′-maleimidylphenyl)-4-methyl coumarin at Cys154, and site-directed mutagenesis of Cys154 and Glu155 on the spin shifts and enzyme cooperativity, it has been proposed that Asp160-Arg164 and Glu161-Arg202 form salt links, which are involved in the conformational transition leading to altered hydration of the heme pocket and accessibility of substrates (59).
The basis of the decreased thermal stability and catalytic tolerance to temperature in the CSM mutants R187A and D192A can be explained by reduced charge pairs and hydrogen bonds, whereas a reduced protein stability in F188A can be explained by the absence of π-π stacking. An interaction between Glu149 and Tyr190 in CYP2B1 has already been postulated earlier based on the decreased thermal stability of Y190A, Y190F, E149Q, and E149A, presumably as the result of a disrupted H-bond (45). In all the cases, the interacting residues are far from the active site, and therefore, it is speculated that the reduced interactions lead to decreased thermal stability through unfolding of the protein on the surface rather than destabilizing the active site through increased heme dissociation. This hypothesis is also supported by the fact that the temperature inactivates the catalytic activity (T50) and heme protein (Tm) in a similar manner in most of the CSM mutants.
Comparison of the P450 expression and thermal stability of the CSM mutants reveals interesting similarities and differences. The findings suggest that the decreased thermal stability of R187A, Y190A, and D192A is associated with decreased P450 expression. In contrast, the Phe188→Ala substitution destabilizes the protein without altering P450 expression. P450 can be thermally inactivated by protein unfolding, inactivation of the heme moiety, and degradation into P420 (an inactive form of P450), whereas P450 expression can be influenced by the rate of incorporation of the heme in addition to the above mechanisms (11, 13). The results for 2B4dH are consistent with earlier observations for 2B6dH. Thus, although M198L and L390P showed enhanced expression, thermal stability was decreased, whereas L264F showed both enhanced P450 expression and stability (13).
Of the six residues tested in CSM 8, substitutions at four (Arg187, Phe188, Tyr190, and Asp192) lead to significantly altered enzyme functions, which correlate well with their rank orders of sequence conservation. Prior results from another laboratory on CYP2B1 suggest that Tyr190 is important for enzyme catalysis and protein stability (45). Using a negative control mutant K186A (juxtaposed to CSM 8), which does not show altered enzyme catalysis, inhibition, and stability, further suggests that the highly conserved CSM mutants are critical for P450 structure-function relationships. Further verification of the importance of specific residue positions is provided by the lack of functional alternations in D189A and K191A in contrast to the marked changes in R187A and D192A. In previous work, we successfully used the CSM approach followed by Ala substitutions to locate regions far from the active site that modulate substrate binding and processivity in apurinic/apyrimidinic endonuclease (APE1) and related nucleases (22-24). A comparative study with random Ala mutagenesis found fewer residues important for the function (27).
The CSM 8 mutants R187A and D192A, which showed decreased P450 expression and stability in CYP2B4dH, also exhibited similar effects in CYP2B1dH. In addition, the activity and inhibitor potency in R187A were increased for the larger compounds in both CYP2B4dH and CYP2B1dH. These results clearly suggest that the CSM 8 has an important and conserved functional role in CYP2B enzymes. For more than a decade the structural basis of substrate specificity and stereo- and regioselectivity of CYP2B enzymes has been studied by altering substrate recognition sites or substrate access channels (1, 20). However, growing evidence based on crystal structures and directed evolution of CYP2B enzymes and enzymes from other CYP subfamilies suggests that plastic regions and/or regions that regulate the ligand-induced enzyme plasticity also determine differential substrate specificity and stereoand regioselectivity (1, 29, 46, 48). The enzymes from different CYP subfamilies differ in the extent of plasticity, which could be associated with differential substrate specificity and stereo- and regioselectivity. For example, in contrast to the highly plastic CYP2B4 structures the CYP2A6 structure shows a compact, hydrophobic active site with one hydrogen bond donor, Asn297, which orients coumarin for regioselective oxidation. In addition, methoxsalen effectively fills the active site cavity without substantially perturbing the structure (48). Moreover, the CYP2C5 structures show an intermediate level of plasticity, with backbone motions on the order of 1-3 Å.4
Furthermore, to test the hypothesis that there are similarities and differences in CSM within the CYP2 family we analyzed the CYP2A, CYP2B, CYP2C, CYP2D, and CYP2J subfamilies separately (supplemental Table S3) and compared the CSM with the motifs for the whole CYP2 family. We identified motifs that are specific for each subfamily and those common to the whole CYP2 family. CSMs 1, 8, and 14 were found in all the subfamilies. Although, CYP2E was not analyzed because of the presence of only two sequences, they also contain CSM 8. The results suggest that CSM 8 is one of the most important motifs in all the subfamilies analyzed and is critical for structural stability and regulating enzyme functions. These analyses suggest that future functional studies are needed to investigate the role of CSM 1 and 14 and specific motifs of each subfamily in protein stability, differential substrate specificity, stereo- and regioselectivity, and enzyme plasticity.
In conclusion, we analyzed CSM in CYP2 enzymes. Detailed structural and prior functional analyses suggested that many of the 20 motifs identified are important for structure and function. Furthermore, study of motif 8 (187RFDYKD192) in CYP2B4 and CYP2B1 suggested that this is an important functional motif in helix E, perhaps mediating a structural change by an effect on the flexible helices F/G region, leading to an altered rate of substrate entry to the active site, ligand binding specificity, stereo- and regioselectivity, and/or dynamics of the protein. The study also tested an important hypothesis that the rank orders of the residue conservation within the motif are related to their functional importance. This first rational approach to analyze structure-function in P450 with regard to non-active site regions/residues is an important step forward for investigating the functional role of CSM in other P450 enzymes, such as families 1 and 3, which contain some of the most important human xenobiotic-metabolizing enzymes. In addition, the approach can be used to engineer other P450 enzymes with altered substrate specificity, stereo- and regioselectivity, or protein stability by site-directed mutagenesis.
Supplementary Material
Acknowledgments
We thank Dr. Yonghong Zhao for analyzing residue-residue interactions and Tori Strong for making the ligand structures.
This work was supported, in whole or in part, by National Institutes of Health Grant ES03619 and Center Grant ES06676 (to J. R. H.) and by NIH Grant AI 064913 (to W. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S3.
Footnotes
The abbreviations used are: CYP, cytochrome P450; 4-CPI, 4-(4-chlorophenyl)imidazole; 1-CPI, 1-(4-chlorophenyl)imdazole; BIF, bifonazole; SRS, substrate recognition sites; CSM, conserved sequence motif; APE1, apurinic/apyrimidinic endonuclease; 7-MFC, 7-methoxy-4-(trifluoromethyl)coumarin; 7-EFC, 7-ethoxy-4-(trifluoromethyl)coumarin; 7-BFC, 7-benzyloxy-4-(trifluoromethyl)coumarin; CuOOH, cumene hydroperoxide; PCP, physicochemical property; CPR, NADPH-cytochrome P450 reductase.
N. Oezguen, S. Kumar, A. Hindupur, W. Braun, B. K. Muralidhara, and J. R. Halpert, unpublished observations.
References
- 1.Zhao, Y. H., and Halpert, J. R. (2007) Biochim. Biophys. Acta 1770 402-412 [DOI] [PubMed] [Google Scholar]
- 2.Ekroos, M., and Sjögren, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13682-13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stout, C. D. (2004) Structure 12 1921-1922 [DOI] [PubMed] [Google Scholar]
- 4.Poulos, T. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13121-13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muralidhara, B. K., and Halpert, J. R. (2007) Drug Metab. Rev. 39 539-556 [DOI] [PubMed] [Google Scholar]
- 6.Scott, E. E., He, Y. A., Wester, M. R., White, M. A., Chin, C. C., Halpert, J. R., Johnson, E. F., and Stout, C. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13196-13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott, E. E., White, M. A., He, Y. A., Johnson, E. F., Stout, C. D., and Halpert, J. R. (2004) J. Biol. Chem. 279 27294-27301 [DOI] [PubMed] [Google Scholar]
- 8.Zhao, Y., White, M. A., Muralidhara, B. K., Sun, L., Halpert, J. R., and Stout, C. D. (2006) J. Biol. Chem. 281 5973-5981 [DOI] [PubMed] [Google Scholar]
- 9.Muralidhara, B. K., Negi, S., Chin, C. C., Braun, W., and Halpert, J. R. (2006) J. Biol. Chem. 281 8051-8061 [DOI] [PubMed] [Google Scholar]
- 10.Zhao, Y., Sun, L., Muralidhara, B. K., Kumar, S., White, M., Stout, C. D., and Halpert, J. R. (2007) Biochemistry 46 11559-11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., Sun, L., Liu, H., Muralidhara, B. K., and Halpert, J. R. (2006) Protein Eng. Des. Select. 19 547-554 [DOI] [PubMed] [Google Scholar]
- 12.Kumar, S., Chen, C. S., Waxman, D. J., and Halpert, J. R. (2005) J. Biol. Chem. 280 19569-19575 [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., Zhao, Y., Sun, L., Halpert, J. R., and Muralidhara, B. K. (2007) Mol. Pharmacol. 72 1191-1199 [DOI] [PubMed] [Google Scholar]
- 14.Kirchheiner, J., Klein, C., Meineke, I., Sasse, J., Zanger, U. M., Mürdter, T. E., Roots, I., and Brockmöller, J. (2003) Pharmacogenetics 13 619-626 [DOI] [PubMed] [Google Scholar]
- 15.Cho, J. Y., Lim, H. S., Chung, J. Y., Yu, K. S., Kim, J. R., Shin, S. G., and Jang, I. J. (2004) Drug Metab. Dispos. 32 1341-1344 [DOI] [PubMed] [Google Scholar]
- 16.Ward, B. A., Gorski, J. C., Jones, D. R., Hall, S. D., Flockhart, D. A., and Desta, Z. (2003) J. Pharm. Exp. Ther. 306 287-300 [DOI] [PubMed] [Google Scholar]
- 17.Lang, T., Klein, K., Richter, T., Zibat, A., Kerb, R., Eichelbaum, M., Schwab, M., and Zanger, U. M. (2004) J. Pharm. Exp. Ther. 311 34-43 [DOI] [PubMed] [Google Scholar]
- 18.Domanski, T. L., Schultz, K. M., Roussel, F., Stevens, J. C., and Halpert, J. R. (1999) J. Pharmacol. Exp. Ther. 290 1141-1147 [PubMed] [Google Scholar]
- 19.Gotoh, O. (1992) J. Biol. Chem. 267 83-90 [PubMed] [Google Scholar]
- 20.Domanski, T. L., and Halpert, J. R. (2001) Curr. Drug Metab. 2 117-137 [DOI] [PubMed] [Google Scholar]
- 21.Williams, P. A., Cosme, J., Sridhar, V., Johnson, E. F., and McRee, D. E. (2001) Mol. Cell 5 121-131 [DOI] [PubMed] [Google Scholar]
- 22.Schein, C. H., Zhou, B., and Braun, W. (2005) Virol. J. 2 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatarajan, S. M., Schein, C. H., and Braun, W. (2003) Bioinformatics 19 1381-1390 [DOI] [PubMed] [Google Scholar]
- 24.Schein, C. H.,Özgün, N., Izumi, T., and Braun, W. (2002) Bioinformatics 3 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schein, C. H., Zhou, B., Oezguen, N., Mathura, V. S., and Braun, W. (2005) Proteins 58 200-210 [DOI] [PubMed] [Google Scholar]
- 26.Negi, S. S., Kolokoltsov, A. A., Schein, C. H., Davey, R. A., and Braun, W. (2006) J. Mol. Model 12 921-929 [DOI] [PubMed] [Google Scholar]
- 27.Harlow, G. R., He, Y. A., and Halpert, J. R. (1997) Biochim. Biophys. Acta 1338 259-266 [DOI] [PubMed] [Google Scholar]
- 28.Venkatarajan, M. S., and Braun, W. (2001) J. Mol. Model 7 445-453 [Google Scholar]
- 29.Johnson, E. F., and Stout, C. D. (2005) Biochem. Biophys. Res. Commun. 338 331-336 [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez, C. E., Kumar, S., Liu, H., and Halpert, J. R. (2006) Arch. Biochem. Biophys. 455 61-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davydov, D. R., Deprez, E., Hoa, G. H., Knyushko, T. V., Kuznetsova, G. P., Koen, Y. M., and Archakov, A. I. (1995) Arch. Biochem. Biophys. 320 330-344 [DOI] [PubMed] [Google Scholar]
- 33.Scott, E. E., He, Y. Q., and Halpert, J. R. (2002) Chem. Res. Toxicol. 15 1407-1413 [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., Scott, E. E., Liu, H., and Halpert, J. R. (2003) J. Biol. Chem. 278 17178-17184 [DOI] [PubMed] [Google Scholar]
- 35.Hiratsuka, M., Hinai, Y., Konno, Y., Nozawa, H., Konno, S., and Mizugaki, M. (2004) Drug Metab. Pharmacokinet. 19 155-158 [DOI] [PubMed] [Google Scholar]
- 36.Klein, K., Lang, T., Saussele, T., Barbosa-Sicard, E., Schunck, W. H., Eichelbaum, M., Schwab, M., and Zanger, U. M. (2005) Pharmacogenet. Genomics 15 861-873 [DOI] [PubMed] [Google Scholar]
- 37.Lang, T., Klein, K., Fischer, J., Nüssler, A. K., Neuhaus, P., Hofmann, U., Eichelbaum, M., Schwab, M., and Zanger, U. M. (2001) Pharmacogenetics 11 399-415 [DOI] [PubMed] [Google Scholar]
- 38.Hesse, L. M., He, P., Krishnaswamy, S., Hao, Q., Hogan, K., von Moltke, L. L., Greenblatt, D. J., and Court, M. H. (2004) Pharmacogenetics 14 225-238 [DOI] [PubMed] [Google Scholar]
- 39.Honma, W., Li, W., Liu, H., Scott, E. E., and Halpert, J. R. (2005) Arch. Biochem. Biophys. 435 157-165 [DOI] [PubMed] [Google Scholar]
- 40.Lehnerer, M., Schulze, J., Achterhold, K., Lewis, D. F. V., and Hlavica, P. (2000) J. Biochem. 127 163-169 [DOI] [PubMed] [Google Scholar]
- 41.Bridges, A., Gruenke, L., Chang, Y. T., Vakser, I. A., Loew, G., and Waskell, L. (1998) J. Biol. Chem. 273 17036-17049 [DOI] [PubMed] [Google Scholar]
- 42.Blobaum, A. L., Harris, D. L., and Hollenberg, P. F. (2005) Biochemistry 44 3831-3844 [DOI] [PubMed] [Google Scholar]
- 43.Waller, S. C., He, Y. A., Harlow, G. R., He, Y. Q., Mash, E. A., and Halpert, J. R. (1999) Chem. Res. Toxicol. 12 690-699 [DOI] [PubMed] [Google Scholar]
- 44.Schenkman, J. B., and Jansson, I. (2003) Pharmacol. Ther. 97 139-152 [DOI] [PubMed] [Google Scholar]
- 45.Lin, H.-l., Zhang, H., Waskell, L., and Hollenberg, P. F. (2005) Chem. Res. Toxicol. 18 1203-1210 [DOI] [PubMed] [Google Scholar]
- 46.Kumar, S., and Halpert, J. R. (2005) Biochem. Biophys. Res. Commun. 338 456-465 [DOI] [PubMed] [Google Scholar]
- 47.Sun, L., Chen, C. C., Waxman, D. J., Liu, H., Halpert, J. R., and Kumar, S. (2007) Arch. Biochem. Biophys. 458 167-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yano, J. K., Hsu, M. H., Griffin, K. J., Stout, C. D., and Johnson, E. F. (2005) Nat. Struct. Mol. Biol. 12 822-823 [DOI] [PubMed] [Google Scholar]
- 49.Kim, D., and Guengerich, F. P. (2004) Arch. Biochem. Biophys. 432 102-108 [DOI] [PubMed] [Google Scholar]
- 50.Kim, D., and Guengerich, F. P. (2004) Biochemistry 43 981-988 [DOI] [PubMed] [Google Scholar]
- 51.Nakamura, K., Martin, M. V., and Guengerich, F. P. (2001) Arch. Biochem. Biophys. 395 25-31 [DOI] [PubMed] [Google Scholar]
- 52.Yun, C. H., Miller, G. P., and Guengerich, F. P. (2000) Biochemistry 39 11319-11329 [DOI] [PubMed] [Google Scholar]
- 53.Wu, Z. L., Podust, L. M., and Guengerich, F. P. (2005) J. Biol. Chem. 280 41090-41100 [DOI] [PubMed] [Google Scholar]
- 54.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graph. 14 51-55 [DOI] [PubMed] [Google Scholar]
- 55.Case, D. A., Darden, T. A., Cheatham, T. E., III, Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Merz, K. M., Pearlman, D. A., Crowley, M., Walker, R. C., Zhang, W., Wang, B., Hayik, S., Roitberg, A., Seabra, G., Wong, K. F., Paesani, F., Wu, X., Brozell, S., Tsui, V., Gohlke, H., Yang, L., Tan, C., Mongan, J., Hornak, V., Cui, G., Beroza, P., Mathews, D. H., Schafmeister, C., Ross, W. S., and Kollman, P. A. (2006) AMBER 9, University of California, San Francisco
- 56.Leys, L., Mowat, C. G., McLean, K. G., Richmond, A., Chapman, S. K., Walkinshaw, M. D., and Munro, A. W. (2003) J. Biol. Chem. 278 5141-5147 [DOI] [PubMed] [Google Scholar]
- 57.Braceya, M. H., Cravattabc, B. F., and Stevens, R. C. (2004) FEBS Lett. 567 159-165 [DOI] [PubMed] [Google Scholar]
- 58.Chen, C. D., and Kemper, B. (1996) J. Biol. Chem. 271 28607-28611 [DOI] [PubMed] [Google Scholar]
- 59.Davydov, D. R., Botchkareva, A. E., Kumar, S., He, Y. Q., and Halpert, J. R. (2004) Biochemistry 43 6475-6785 [DOI] [PubMed] [Google Scholar]
- 60.Kumar, S., Liu, H., and Halpert, J. R. (2006) Drug Metab. Dispos. 34 1958-1965 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.