Abstract
The Bcl-2 family proteins are important regulators of type I programmed cell death apoptosis; however, their role in autophagic cell death (AuCD) or type II programmed cell death is still largely unknown. Here we report the cloning and characterization of a novel Bcl-2 homology domain 3 (BH3)-only protein, apolipoprotein L1 (apoL1), that, when overexpressed and accumulated intracellularly, induces AuCD in cells as characterized by the increasing formation of autophagic vacuoles and activating the translocation of LC3-II from the cytosol to the autophagic vacuoles. Wortmannin and 3-methyladenine, inhibitors of class III phosphatidylinostol 3-kinase and, subsequently, autophagy, blocked apoL1-induced AuCD. In addition, apoL1 failed to induce AuCD in autophagy-deficient ATG5-/- and ATG7-/- mouse embryonic fibroblast cells, suggesting that apoL1-induced cell death is indeed autophagy-dependent. Furthermore, a BH3 domain deletion construct of apoL1 failed to induce AuCD, demonstrating that apoL1 is a bona fide BH3-only pro-death protein. Moreover, we showed that apoL1 is inducible by p53 in p53-induced cell death and is a lipid-binding protein with high affinity for phosphatidic acid (PA) and cardiolipin (CL). Previously, it has been shown that PA directly interacted with mammalian target of rapamycin and positively regulated the ability of mammalian target of rapamycin to activate downstream effectors. In addition, CL has been shown to activate mitochondria-mediated apoptosis. Sequestering of PA and CL with apoL1 may alter the homeostasis between survival and death leading to AuCD. To our knowledge, this is the first BH3-only protein with lipid binding activity that, when overproduced intracellularly, induces AuCD.
Bcl-2 family members play crucial roles in regulating programmed cell death (PCD).2 In stressed cells the live-or-die decision is largely determined by interplay between opposing members of the Bcl-2 protein family (1–4). Each member of the Bcl-2 family contains at least one of four conserved amino acid (aa) sequences, Bcl-2 homology (BH) domains 1–4. Based on their function in PCD, the Bcl-2 family members can be divided into three subgroups as follows: multi-BH domain anti-death, multi-BH domain pro-death, and BH3-only pro-death (1–3). The BH3 domain is conserved across all Bcl-2 family members and is the only domain retained by BH3-only proteins, further emphasizing the functional significance of this domain (4). In mammals, there are two major types of PCD, apoptosis and autophagic cell death (AuCD). Apoptosis (type 1) is characterized by nuclear condensation/DNA fragmentation and cell membrane alteration, without major ultrastructural changes of subcellular organelles. It usually involves the activation of caspases as well as the release of apoptogenic effectors, such as cytochrome c and apoptosis-inducing factor, from mitochondria that contribute to the acquisition of the apoptotic morphology (5–8). The molecular basis of AuCD (type 2), however, is not as well characterized in mammalian cells, even though many lines of evidence connect autophagy to human disease. Autophagy is generally defined as a lysosome-dependent mechanism of intracellular degradation that is used for the turnover of cytoplasmic constituents. There are three major classifications of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (9–12). Macroautophagy (hereafter referred to as autophagy) is a highly conserved eukaryotic catabolic process involving the sequestration of the cargo (i.e. fraction of cytosol or organelles) within double membranes, creating autophagosomes, which then fuse with the endosome and lysosome to create autolysosomes. In the lumen of autolysosomes, lysosomal enzymes operate at low pH to catabolize the autophagic material (12–14). Autophagosomes and autolysosomes, also called autophagic vacuoles (AV), are considered to be de novo subcellular organelles specific to the autophagic process. Autophagy allows cells to eliminate large portions of the cytoplasm and is directly involved to eliminate aberrant protein aggregates, remove damaged/aged organelles, defend against pathogen invasions, and cover an essential role in early neonatal development and differentiation (15–16). Upwards of 20 autophagy-related genes (ATG) have been identified, which regulate and/or directly participate in the autophagic pathway, particularly in AV formation (10, 13, 14). Deficiency of ATGs for example, ATG5 and ATG7, causes loss of AV formation and subsequently lack of autophagic response. These deficiencies correlate with various diseases such as cancer, cardiomyopathy, and neurodegenerative diseases (17–22). Paradoxically, recent evidence suggests that autophagy is, on the one hand, a survival pathway necessary to sustain mammalian viability during periods of starvation, and on the other hand, progressive autophagy can be a means to cell death (7, 11, 12). Significantly, Berry and Baehrecke (23) reported that autophagy is involved in physiological cell death during Drosophila development but is controlled by similar mechanisms as those that control autophagy function in cell survival. Furthermore, they provide in vivo evidence that both growth arrest and autophagy are required for physiological AuCD. However, the molecular determinants that dictate the control of autophagy for cell survival or death have not yet been revealed.
Aside from their role in apoptosis, the members of the Bcl-2 family also play an important role in autophagy. For example, when treating apoptosis-deficient cells, such as BAX and BAK double knock-out cells, with cytotoxic compounds (for example, etoposide), the primary response of those cells was AuCD, suggesting that homeostatic states of these knock-out Bcl-2 members play pivotal roles in regulating AuCD (24–27). In addition, it has been shown that the anti-death Bcl-2 and Bcl-xL can bind and inhibit Beclin1, a mammalian ortholog of yeast ATG6, and subsequently inhibit autophagy (28, 29). Overexpression of Beclin1 has been shown to induce formation of AV and autophagy in mammalian cells (28). Interestingly, Beclin1 has been found to be a haploinsufficient tumor suppressor frequently mutated/deleted in breast, prostate, and ovarian cancers (12, 30, 31). Heterozygous knock-out mice of Beclin1 (Beclin1+/-) are tumor-prone, suggesting that autophagy has a tumor suppression function (30). Importantly, structural and functional analyses showed that Beclin1 is a novel BH3-only protein, and this BH3 domain within Beclin1 mediates associations with Bcl-2 and Bcl-xL (32–34). In addition to Beclin1, recent studies showed other BH3-only members that also play regulatory roles in autophagy in physiological context. For example, Bid, a BH3-only protein, showed involvement in regulating both apoptosis and autophagy in breast cancer MCF-7 cells (35). Bad and BNIP3, two other BH3-only proteins, when overexpressed, can induce formation of AV in cancer cells (34, 36, 37). Interestingly, Bid, Bad, and BNIP3 are downstream targets of p53, a well characterized tumor suppressor and transactivating factor. It has been shown that p53 plays a critical role in the suppression of tumorigenesis, in part, by the induction of both type I and type II PCD (38–41).
To identify and characterize novel BH3-only proteins in AuCD and discover molecular regulators that dictate autophagy for death or survival, in this study, we utilized in silico data base-mining, functional expression, microscopy strategies, and lipid binding assay to examine a novel BH3-only protein, apolipoprotein L1 (apoL1).
EXPERIMENTAL PROCEDURES
Cell Lines, Chemicals, and Culture Media—DLD-1.TA14 (a p53-null, “Tet-Off” inducible colorectal cancer cell line) and Hec50co (a p53-null endometrial cancer cell line) and their corresponding growth medium and condition have been described previously (42–44). ATG5-/- and ATG7-/- mouse embryonic fibroblast (MEF) cells were gifts from Dr. Mizushima (15) and Dr. Komatsu (16), respectively. HEK293 and AD293 cell lines were purchased from ATCC and Clontech, respectively. All cell lines except DLD-1 were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% antimicrobial agents as described previously (44).
Identification and Sequence Analysis of Novel BH3-only Proteins by Functional Genomics and Biocomputing Approaches—The known BH3 domain consensus sequence and a data base-mining strategy were utilized to probe human genome and proteome data bases for the identification of novel BH3-only proteins as described previously (45). The annotated human open reading frames of candidate genes were downloaded from the most recent NCBI data base (build 36.2). In addition, candidate proteins were assessed for other putative domains/motifs, utilizing public sequence analysis programs and tools, such as the ExPASy proteomics server and ClustalW, as described previously (45). In addition, protein tertiary structure prediction was obtained from HMMSTR/ROSETTA (46, 47), and presentations of tertiary structure were made using Protein Data Bank files created with HMMSTR/ROSETTA and PyMOL.
Cloning of Human ApoL1 cDNA and Generation of Inducible ApoL1 Gene—Human apoL1 was identified as a putative BH3-only protein by in silico data base mining. PCR was used to amplified the 1,194-bp full-length apoL1 cDNA. Subsequently, the sequence-confirmed apoL1 and apoL1 with a C-terminal FLAG tag (apoL1.FLAG) were cloned into pBI-EGFP vector (Clontech). The primers used for amplifying apoL1 were as follows: reverse, 5′-CCGCTCCAGTCACAGTTCTTGGTC-3′ (bp +1,197 to +1,183), and forward, 5′-CGACGCGTATGGAGGGAGCTGCTTTGC-3′ (bp +1 to +19). The reverse primer used for amplifying apoL1.FLAG was 5′-CTAGCTAGCTCACTTATCGTCGTCATCCTTGTAATCCAGTTCTTGGTC-3′ (bp +1,197 to +1,183). The resulting plasmids were used to transfect a previously described “Tet-Off” inducible system in DLD-1.TA14 (44, 45). Stably transfected DLD-1 cells harboring pBI-apoL1 or pBI-apoL1.FLAG, namely DLD-1.apoL1 and DLD-1.apoL1.FLAG, respectively, were selected in noninduction medium (D.20) with 20 ng/ml doxycycline (Dox) as described previously (44, 45). We assayed inducible expression of apoL1 transcripts by semi-quantitative RT-PCR and immunoblot analyses.
Site-specific Deletion Mutagenesis—To generate the BH3 domain deletion allele of apoL1, that is to delete the 27 bp that encode the 9-amino acid BH3 domain (codons 158–166; base pairs +474 to +498), a PCR-based site-directed mutagenesis kit (ExSite, Stratagene) and the deletion-specific primers were used. Primer sequences were as follows: forward, 5′-AAGGTCCACAAAGGCACCACC, and reverse, 5′-(p)CCTTCTTATGTTATCCTACAGC. The deletion construct pBI-apoL1.dBH3 was confirmed by sequencing and was used to construct pAD-apoL1.dBH3 plasmid as described above (44).
ApoL1 Adenovirus (AD-ApoL1)—Construction, amplification, and assaying for titers of adenoviruses (AD) harboring apoL1 and apoL1.dBH3 alleles, and viral infection of mammalian cells were performed as described previously (44). A variety of cells were infected with control AD, AD-apoL1, and AD-apoL1.dBH3 with a range of concentrations from 0.01 to 5 multiplicity of infection (or plaque-forming units/cell, which was estimated to be 0.2–100 virus particles/cell) for 3 days. Cell numbers, cell death, and survival were measured over time by counting the attached cells, and by microscopy analysis in cell morphology (roundup) and increase of intracellular vacuole formation.
Semi-quantitative RT-PCR—Total RNA was isolated from cells using a Purescript kit (Gentra Systems). RT was conducted using random hexamers, and PCR was conducted using gene-specific primers and a standard protocol as described (36, 37). The primers used for PCR were as follows: reverse, 5′-CCGCTCCAGTCACAGTTCTTGGTC-3′ (bp +1,197 to +1,183), and forward, 5′-CAGCAGTACCATGGACTACGG3′ (bp +672 to +692).
Generation of Anti-peptide Antibodies against Human ApoL1—To raise anti-peptide antisera specifically against human apoL1, a peptide corresponding to the codons 384–398 of apoL1 (ILNNNYKILQADQEL) was used to immunize rabbits (Anaspec). Antibody was affinity-purified by chromatography on peptide (antigen)-linked Sepharose and designated as anti-apoL1.
Immunoblot Analysis—Total cellular extracts were isolated and used for immunoblot analysis from indicated cells as described previously (42, 43). Proteins were separated using 10% SDS-PAGE (20 μg of protein/lane) and transferred to nitrocellulose membranes, which were then incubated with anti-apoL1, anti-activated caspase-9 (Cell Signaling Technology), anti-caspase 8 (Cell Signaling), anti-caspase 3, anti-poly-(ADP-ribosyl) polymerase (PARP) (Santa Cruz Biotechnology), anti-LC3 (MBL), and anti-β-actin (Oncogene Research Products). Subsequently, blots were incubated with goat anti-rabbit or anti-mouse horseradish peroxidase-labeled secondary antibodies (Bio-Rad).
Analysis of Dose-dependent, Time Course-dependent Cell Death—Taking advantage of the inducibility of the Tet-Off system, dose-dependent and time course-dependent cell death studies were conducted. Briefly, stably transfected DLD-1.apoL1 or DLD-1.apoL1.FLAG cells were grown in D.20. For induction studies, adherent cells were rinsed in phosphate-buffered saline, refed with the induction medium containing gradually decreasing concentrations of Dox (5 ng/ml, D.5; 1 ng/ml, D.1; 0 ng/ml, D.0), and harvested at the indicated time points as described previously (31, 38).
Assay of Protein Localization by Immunofluorescence Microscopy—Cells were grown on glass coverslips coated with collagen (Roche Applied Science). After induction of apoL1 for various times, cells were rinsed and then fixed with 4% (v/v) paraformaldehyde solution at room temperature. Coverslips were rinsed twice with HBSS and blocked with 3% (w/v) BSA in TBST for 30 min at room temperature. After incubation overnight at 4 °C in 3% (w/v) BSA containing the anti-LC3 antibody (P0036, MBL), coverslips were rinsed with HBSS four times and incubated in 3% (w/v) BSA in TBST containing Texas Red-labeled secondary antibody (T2767, Invitrogen) for 1 h at room temperature. Finally, coverslips were rinsed with HBSS, mounted onto slides, and subjected to fluorescent microscope analysis.
Transmission Electron Microscopy—Cells on coverslips were fixed in 3% formaldehyde, 2% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.4) for 45 min, washed, and finally fixed for 1 h in 1% osmium tetroxide, 0.5% potassium ferrocyanide in cacodylate buffer. The samples were then washed again, dehydrated with graded alcohol, and embedded in Epon-Araldite resin (Canemco). The resin was cured at 65 °C for 2 days, after which the glass coverslips were removed from the resin blocks and the samples were sectioned. Ultrathin sections were collected on copper grids and examined in an Hitachi H-7500 electron microscope. Images were captured and processed with an AMT XR60 B camera and Image Capture software version 5.42.
Effect of 3-Methyladenine (3-MA), Wortmannin, and ATGs on ApoL1-induced Cell Death—To investigate whether apoL1-induced cell death was through autophagy, two different experiments were conducted. First, 3-MA (5 mm) and wortmannin (1 μg/ml), two inhibitors of PI3Ks and autophagy, were added to DLD-1.apoL1 cells 8 h prior to the induction of apoL1. Cell death was monitored by light microscopy analysis 24 h after apoL1 induction. Second, wild type (WT), ATG5-/-, and ATG7-/- MEF cells were infected with AD-apoL1. Induction of cell death, overexpression of apoL1, and activation of LC3-II were assays 36 h after viral infection.
Induction, Immunoprecipitation, and Elution of the ApoL1.FLAG Fusion Protein—DLD-1.apoL1.FLAG cells were grown in the induction medium for 24 h, rinsed twice with cold phosphate-buffered saline, pelleted, and lysed in lysis buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100). Total soluble protein extract was incubated with Anti-FLAG-agarose affinity gel (Sigma, F2426) at 4 °C overnight with gentle agitation. The agarose beads were then washed two times with ice-cold TBS (50 mm Tris-HCl and 150 mm NaCl, pH 7.4) and incubated with 400 μl (150 ng/μl) of 3×FLAG peptide (Sigma, F4799) in TBS at 4 °C overnight for eluting apoL1-FLAG fusion protein.
In Vitro Lipid Binding Assay—Similar to an immunoblot analysis, phosphoinositide (PIP), and membrane lipid strips, nitrocellulose membranes pre-spotted with various indicated lipid species (P-6001, P-6002, Echelon, Salt Lake City, UT) were blocked in 3% fat-free BSA for 60 min at room temperature, probed with the purified apoL1.FLAG protein (∼100 ng/ml) in 3% fat-free BSA in TBST for 1 h at room temperature, followed by primary anti-apoL1 antibody, secondary goat anti-rabbit antibody conjugated with horseradish peroxidase, and ECL detection and imaging.
RESULTS
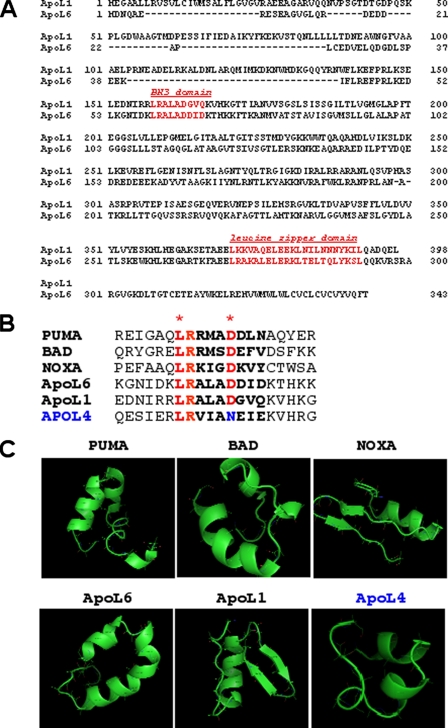
ApoL1 Is a BH3 Domain Containing Protein—We utilized a 9-residue consensus sequence of the BH3 domain constructed from known human Bcl-2 proteins to probe the human proteome data base that identified a BH3 domain embedded in the apoL1 (GenBank™ accession number NM_003661). We then retrieved full-length cDNA and protein sequences of apoL1 from the National Center for Biotechnology Information (NCBI) (ncbi.nih.gov) nucleotide and protein sequence data bases. Previously, using the same methodology, we reported the cloning and characterization of a bona fide BH3-only pro-death gene apolipoprotein L6 (apoL6) and showed that overexpression of apoL6 induces mitochondria- and caspase 8-mediated apoptosis (45). Both apoL1 and apoL6 belong to a newly identified apolipoprotein L family that share significant sequence identity within the predicted amphipathic α-helix. Besides apoL1 and L6, apoL family has four other members, L2–5. It has been suggested that these proteins play important roles in lipid binding and transport (48–50). Sequence analysis showed that apoL1 possesses a putative BH3 domain (aa 158–166) with 67% sequence identity with that of apoL6 (Fig. 1A). Over all, apoL1 and apoL6 possess 27% sequence identity and 42% similarity at the polypeptide level. In addition to the putative BH3 domain, apoL1 also possesses a leucine zipper domain (aa 365–392) (Fig. 1A). Sequence alignment of the 20-aa putative BH3 domain with a number of known BH3-only proteins (PUMA, BAD, NOXA, and apoL6) and apoL4, further showed that the 9-aa BH3 domain of apoL1 harbors the two important amino acids, Leu and Asp, conserved in all known BH3 domains (2, 4). By contrast, apoL4 does not possess the conserved Asp residue, which is substituted by an Asn instead (Fig. 1B). Tertiary structure prediction, of the 20-aa putative BH3 domain in the indicated proteins, showed that the 9-aa BH3 domain of apoL1 possessed amphipathic α-helical structure, similar to other BH3-only proteins of PUMA, BAD, NOXA, and apoL6 (Fig. 1C). In contrast, apoL4 does not possess a canonical α-helical structure in that region (Fig. 1C). Finally, sequence analysis suggests that apoL1 does not possess BH1, BH2, or BH4 domains. Taken together, these results suggest that apoL1 is a putative BH3-only protein.
FIGURE 1.
Sequence and domain analysis of human apoL1. A, polypeptide sequence alignment of human apoL1 and apoL6. Over all, apoL1 and apoL6 shows 27% identity and 42% similarity at the aa level. Putative domains in apoL1 include a BH3 domain (aa 158–166) and a leucine zipper domain (aa 365–392). B, sequence alignment of the 20-aa region containing putative BH3 domain in known BH3-only proteins (PUMA, BAD, NOXA, and apoL6) and apoL1 and apoL4. The 9-aa BH3 domain are in boldface and the two amino acids, L and D, which are conserved in all known BH3 domains, are labeled in red with asterisks. ApoL4 does not possess the conserved Asp residue, instead, it is substituted by an Asn (in blue). C, predicted tertiary structure of the 20-aa region containing putative BH3 domain in the indicated proteins. As expected, the 9-aa BH3 domains of known BH3-only proteins, PUMA, BAD, NOXA, and apoL6, show amphipathic α-helical structures. Importantly, the putative BH3 domain of apoL1 also possesses an α-helical domain. In contrast, apoL4 does not possess a complete, canonical α-helical structure in that region.
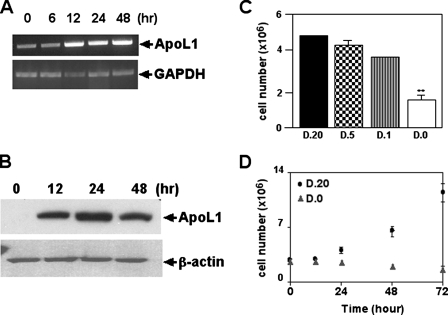
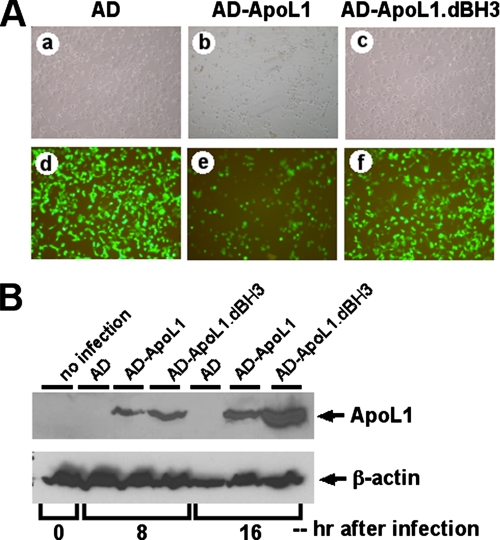
ApoL1 Induces Cell Death in a Variety of Cell Types—To investigate whether apoL1 induced cell death, we constructed a Tet-Off apoL1-inducible system in DLD-1 cells (DLD-1.apoL1) and adenovirus harboring wild type apoL1 (AD-apoL1) or a BH3 domain deletion construct of apoL1 (AD-apoL1.dBH3). We first examined whether apoL1 expression was inducible in a time-dependent manner in stably transfected DLD-1.apoL1 cells. Using semi-quantitative RT-PCR and total RNA isolated from DLD-1.apoL1 cells induced for 0, 6, 12, 24, and 48 h, we showed that there was time-dependent induction of apoL1 in induced DLD-1.apoL1 cells at the RNA level (Fig. 2A). We observed that expression of apoL1 was increased ∼4-fold 12 h after the removal of Dox (Fig. 2A). ApoL1 transcripts were also detectable in the DLD-1.TA14 control cells, thereby indicating that DLD-1 cells have some endogenous apoL1 expression (Fig. 2A). To confirm the induction of apoL1 expression at the protein level, we conducted immunoblot analysis on induced DLD-1.apoL1 cells using anti-apoL1 antibody. As shown in Fig. 2B, the amount of apoL1 protein was greatly increased (>6-fold) 12 h after the induction. We then investigated whether overexpression of apoL1 could promote cell death. As shown in Fig. 2C, induction of apoL1 in DLD-1.apoL1 cells had a negative effect on cell proliferation significantly reducing the growth rate at 48 h when grown in the absence of Dox (D.0; Fig. 2, C, 4th column, and D). Dying/dead cells showed a change in cell morphology, increasing intracellular vacuoles, and detachment from culture plates. In contrast, DLD-1.apoL1 cells grown in D.20 exhibited normal growth (Fig. 2, C, 1st column, and D). Similar results were observed in DLD-1.apoL1.FLAG cells (data not shown). Because apoL1 expression was Tet-Off-regulated by the concentration of Dox in the culture medium, a cell growth study was conducted at various concentrations of Dox. In the presence of gradually increasing Dox concentrations, such as 1 ng/ml (D.1) and 5 ng/ml (D.5), cell death was decreased in DLD-1.apoL1 cells, implying that the cell death rate was related to the level of apoL1 expression. Furthermore, to investigate the importance of the BH3 domain of apoL1 in the induction of cell death, we constructed adenovirus harboring WT apoL1 (AD-apoL1) or a BH3 domain deletion mutant of apoL1 (AD-apoL1.dBH3) and used those to infect a variety of cell types, including HEK293 cells. As shown in Fig. 3A, cells overexpressing green fluorescence protein also overexpressed apoL1 protein. It is evident that greater than 95% of cells infected with AD-apoL1 were dead, same as those inducing DLD-1.apoL1 cells, 72 h after infection (Fig. 3A, panels b and e). In contrast, AD only or AD-apoL1.dBH3 failed to induce cell death (Fig. 3A, panels a and d, and c and f, respectively). Immunoblot analysis showed the protein products of WT apoL1 and apoL1.dBH3 were made in a time-dependent manner and were stable (Fig. 3B, 3rd and 6th lanes for WT apoL1 and 4th and 7th lanes for apoL1.BH3). In addition, endogenous expression of apoL1 was very low in HEK293 cells with or without AD infection (Fig. 3B, 1st, 2nd, and 5th lanes). Identical results were observed for the same set of experiments in four different cell lines, DLD-1, HepG2 (liver cancer), MCF-7 (breast cancer), and LNCaP, PC3, and DU145 (prostate cancer) (data not shown). Thus, AD-apoL1 is a potent inducer of AuCD in a variety of cells and may be a general autophagy mediator.
FIGURE 2.
Time- and dose-dependent induction of apoL1 and cell death in DLD-1.ApoL1 cells. Induction of apoL1 expression is time-dependent as indicated by semi-quantitative RT-PCR using total RNA (A) or immunoblot analysis using total soluble proteins isolated from cells grown in induction medium (D.0) for the indicated time (B). C, apoL1 induces dose-dependent cell death. DLD-1.apoL1 cells were cultured in media containing Dox (D; ng/ml) as indicated. Attached cells were harvested and counted after 48 h. D, apoL1 induces time-dependent cell death. DLD-1.apoL1 cells were cultured in D.20 (Dox, 20 ng/ml) medium (squares) or D.0 (Dox, 0 ng/ml) medium (triangles), harvested, and counted at the times indicated. Each experiment was repeated at least three times. *, p < 0.05, indicates significant difference compared with control by Student's t test.
FIGURE 3.
BH3 domain deletion construct of apoL1 failed to induce cell death in DLD-1 cells. A, adenoviruses harboring WT apoL1 (AD-apoL1) and BH3 domain deletion mutant of apoL1 (AD-apoL1.dBH3) were constructed and used to infect DLD-1 cells. Cells overexpressing green fluorescence protein also overexpressed apoL1 protein. AD-apoL1 induced cell death 72 h after infection (panels b (bright field) and e). In contrast, AD only or AD-apoL1.dBH3 failed to induce cell death (panels a (bright field) and d, and c (bright field) and f, respectively). B, time-dependent protein production of WT apoL1 and apoL1.dBH3. Hours after infection were as indicated. Note that endogenous expression of apoL1 was very low, almost undetectable, in DLD-1 cells (1st, 2nd, and 5th lanes). Both proteins were stably produced in time-dependent manner (3rd and 6th and 4th and 7th lanes for WT apoL1 and apoL1.dBH3, respectively). β-Actin was used as a loading control.
ApoL1-induced Cell Death Is Not through Caspase-dependent Apoptosis—Because apoL1 and apoL6 have high sequence and structure similarity and apoL6 induces caspase-dependent apoptosis, we examined whether apoL1-induced cell death was mediated through apoptosis. Fig. 4 showed that caspases 9 and 3 were not activated, and PARP, a well known substrate of caspases, was not cleaved during apoL1-induced cell death in DLD-1.apoL1 cells (Fig. 4A). By contrast, apoptotic DLD-1.apoL6 cells showed activation of caspase 9 8 h after apoL6 induction, which is consistent with previous findings (45).
FIGURE 4.
ApoL1-induced cell death is through autophagy, not by apoptosis. A, caspases 9 and 3 were not activated, and PARP was not cleaved during apoL1-induced cell death. By contrast, apoL6 induces cell death through apoptosis by activation of caspases 9 as described previously (45). B, apoL1 induced activation andtranslocationofLC3.ImmunoblotanalysisofLC3inautophagicDLD-1.apoL1 cells. Total soluble proteins were isolated at indicated time points from DLD-1.apoL1 cells cultured in induction medium. Times after induction of apoL1 are as indicated. Accumulation of LC3-II in induced DLD-1.apoL1 cells can be seen 12 h after induction. The ratios between LC3-II and LC3-I were 0.1, 2, and 20, 0, 12, and 24 h after induction, respectively. C, immunofluorescence staining of LC3 (red) in DLD-1.apoL1 cells. Panel a, DLD-1.apoL1 cells cultured in normal medium, and panel b, DLD-1.apoL1 cells cultured in induction medium for 24 h. At least 300 cells were analyzed for each experiment.
ApoL1 Induces Activation and Translocation of LC3-II—To investigate whether apoL1-induced cell death acted through autophagy, we analyzed apoL1-induced activation and translocation of LC3-II to AV, a critical step in autophagic pathway. The amount of LC3-II has been shown previously to correlate with the number of AV (13, 14, 51). Immunoblot analysis of LC3 indicated a time-dependent accumulation of LC3-II and increase in the ratios of LC3-II to LC3-I in induced and autophagic DLD-1.apoL1 cells (Fig. 4B). After induction of apoL1, the ratios of LC3-II and LC3-I were 1:10, 2:1, and 20:1 0, 12, and 24 h, respectively. In addition, immunofluorescence microscopy analysis showed a pattern of punctate staining of LC3-II in induced DLD-1.apoL1 cells (Fig. 4C, panel b), indicating the translocation of LC3-II from cytosol to AV. In contrast, control DLD-1.apoL1 cells showed a cytosolic staining pattern of LC3 (Fig. 4C, panel a), which indicates physiological localization of LC3-I in normal cells.
ApoL1 Induces Increased Formation of AV and Autophagy Which Can Be Inhibited by 3-MA and Wortmannin—Transmission electron microscopy (TEM) is one of the most sensitive and definitive methods to detect autophagic compartments in mammalian cells (52, 53). To further confirm that apoL1-induced cell death was indeed through autophagy, we conducted both light microscopy and TEM analyses. Normal DLD-1. apoL1 cells under light microscopy and TEM were displayed in Fig. 5, A, panel a, and B, panel a, respectively. Induced DLD-1.apoL1 cells with increased expression of apoL1 showed significant formation of AV under the TEM (Fig. 5B, panels b and c). The magnified images showed an enlarged empty vacuole (Fig. 5B, panel d), a double-membraned AV (Fig. 5B, panel e), and an autophagosome engulfing an electron-dense body (Fig. 5B, panel f) in an induced DLD-1.apoL1 cell (Fig. 5B, panel c). Induced DLD-1.apoL1 cells were deformed and slightly enlarged under the light microscopy (Fig. 5A, panel b). In addition, we examined the effect of 3-MA and wortmannin, two inhibitors of PI3Ks and autophagy (13, 14, 54), on apoL1-induced cell death. Fig. 5A, panels c and d, showed that 3-MA and wortmannin partially blunt apoL1-induced cell death in DLD-1.apoL1 cells cultured in induction medium. These data suggest that apoL1-induced cell death can be blocked by inhibitors of autophagy.
FIGURE 5.
ApoL1 induces accumulation of AV and autophagic cell death, which can be inhibited by 3-MA and wortmannin. A, effect of 3-MA (5 mm) and wortmannin (1 μg/ml) on apoL1-induced cell death was observed by light microscopy analysis. 3-MA and wortmannin were added to noninduction medium 8 h prior to the induction of apoL1. Panel a, DLD-1.apoL1 cells in noninduction (regular) medium; panel b, in induction medium; panel c, in induction medium + 3-MA; and panel d, in induction medium + wortmannin. Obviously, 3-MA and wortmannin partially blunt apoL1-induced cell death in DLD-1.apoL1 cells by 55 and 40%, respectively. All the images were taken by the same inverted microscope (Olympus, CK40) with a MacroFire digital camera (OPTRONIC, model S99831). B, electron microscopy analysis of autophagic DLD-1.apoL1 cells. Panel a, control DLD-1.apoL1 cells; panels b and c, autophagic DLD-1.apoL1 cells under two different magnifications; panels d–f, magnified views of vacuoles in the cell shown in panel c. Panel d, an enlarged empty vacuole; panel e, a double-membraned structure presumably an autophagosome; panel f, an AV, presumably an autolysosome, engulfing an electron-dense body. Scale bars are as indicated. Data are representative of four experiments. Each experiment was repeated at least three times.
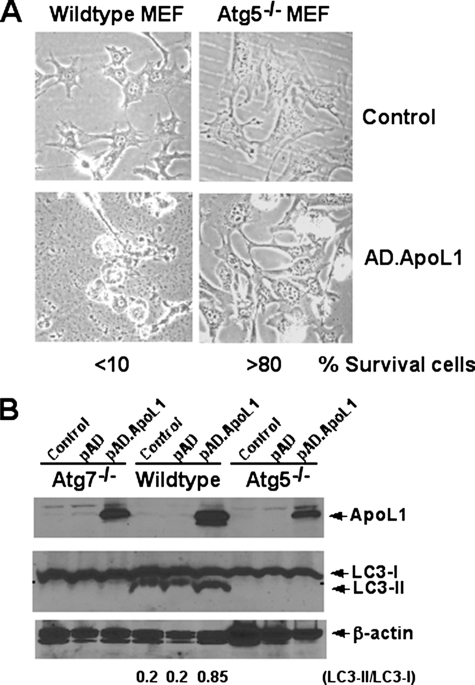
ApoL1 Fails to Induce Cell Death in Autophagy-deficient MEFs—To further confirm apoL1 induces AuCD, two well studied, autophagy-deficient MEFs, ATG5-/- and ATG7-/-, were used. It has been well documented that ATG5 and ATG7 are essential for the initiation of autophagosome formation. ATG5 is an acceptor molecule for the ubiquitin-like molecule ATG12, and the proper conjugation of ATG5 with ATG12 is required for the elongation of the autophagic membrane (15). ATG7, an E1-like enzyme, together with ATG10, an E2-like enzyme, catalyze the covalently conjugated complex of ATG5-ATG12. In addition, ATG7 is required for the lipidation/activation of LC3-II and subsequently translocation of LC3-II to the autophagic membrane (10, 16). Thus, cells lacking either ATG5 or ATG7 are defective in LC3-II activation and autophagosome formation. MEF cells were infected with AD-apoL1 for 36 h at which point induction of apoL1, cell death, and LC3 activation were assayed. Fig. 6A showed that greater than 90% of WT MEF cells infected with AD-apoL1 had died, whereas less than 20% of the ATG5-/- cells were dead. Fig. 6B showed overexpression of apoL1 protein was achieved at 36 h after infection of AD-apoL1 in ATG5-/-, ATG7-/-, and WT MEFs. Importantly, apoL1 again induced activation of LC3 II and increased the ratio of LC3-II to LC3-I in WT MEF cells (Fig. 6B, 6th lane). By contrast, apoL1 failed to induce activation of LC3 and autophagy, as expected, in both the ATG5-/- and ATG7-/- cells. These results further indicate that apoL1-induced cell death is autophagy-dependent.
FIGURE 6.
ApoL1 fails to induce cell death in autophagy-deficient ATG5-/- and ATG7-/- MEFs. Wild type (WT), ATG5-/-, and ATG7-/- MEF cells were grown in Dulbecco's modified Eagle's medium to 30% confluence, infected with control AD or AD-apoL1 viruses for 36 h, and followed by light microscopy analysis (A). Greater than 90% of WT MEF cells infected with AD-apoL1 had died, whereas less than 20% of the ATG5-/- or ATG7-/- (not shown) cells were dead. B, immunoblot analysis showed overexpression of apoL1 protein was achieved at 36 h after infection of AD-apoL1 in ATG7-/-, WT, and ATG5-/- MEFs. Importantly, apoL1 again induced activation of LC3 II and increased the ratio of LC3-II to LC3-I (6th lane). By contrast, apoL1 failed to induce activation of LC3 in the ATG5-/- and ATG7-/- cells. Of note, it has been reported that MEFs have high endogenous LC3-II (15, 16).
Expression of ApoL1 Is Up-regulated by P53 in P53-induced Cell Death—Previous studies showed that p53, one of the master regulators in PCD, induces both apoptosis and autophagy (39, 40). To investigate whether the expression of apoL1 was regulated by p53, we took advantage of two of our previously used cancer cell models, DLD-1.p53 and Hec50co, in p53-induced apoptosis. Overexpression of p53 was achieved in DLD-1.p53 cells by Tet-Off induction and in Hec50co cells by AD-p53 infection as described previously (44). Immunoblot analysis showed time-dependent expression of apoL1 under the influence of p53 in both cell lines. As shown in Fig. 7, induction of apoL1 was detected 8 and 12 h after p53 overexpression in DLD-1.p53 cells and Hec50co cells, respectively, and 16 and 12 h prior to p53-induced cell death in DLD-1.p53 cells and Hec50co cells, respectively. This indicates that apoL1 is inducible by p53 during p53-induced cell death.
FIGURE 7.
Expression of apoL1 is up-regulated by p53 in p53-induced cell death. Immunoblot analysis of time-dependent expression of apoL1 under the influence of p53 was assayed. Overexpression of p53 was achieved in DLD-1.p53 cells and in Hec50co cells as described previously (45). Hours after p53 overexpression are as indicated. Induction of apoL1 was detected 8 and 12 h after p53 overexpression in DLD-1.p53 cells and Hec50co cells, respectively. β-Actin was used as loading control.
ApoL1 Binds Strongly with PA and CL and Less with Some Phosphoinositides—To explore the binding of apoL1 to lipids, apoL1.FLAG fusion protein was first immunoprecipitated by anti-FLAG antibody and subsequently eluted by 3× FLAG peptide. The purified apoL1.FLAG protein was confirmed by immunoblot analysis using anti-FLAG or anti-apoL1 antibodies (Fig. 8A) and was subsequently used for protein lipid overlay assay. Two membranes pre-spotted with a variety of lipid species were probed with the apoL1.FLAG fusion protein. As expected, Fig. 8B showed that the apoL1.FLAG, but not 3× FLAG itself (negative control), bound to a variety of lipid species immobilized on the nitrocellulose membranes in vitro. The apoL1.FLAG showed strong binding affinity for PA and CL but less affinity with different species of PIPs and other phospholipids in the following order: phosphatidylinositol 3,5-bisphosphate (PI-3,5-P2) > PI4P > PI5P > PI3P > PI-4,5-P2 > PI-3,4,5-P3 > PI-3,4-P2 > 3-sulfogalactosylceramide > phosphoserine > phosphoglycerol (PG) > phosphoinositol (PI) (Fig. 8B). Interestingly, apoL1 did not bind cholesterol, phosphoethanolamine (PE), phosphocholine (PC), sphingomyelin (SM), triglyceride (TG), diacylglycerol, sphingosine 1-phosphate, lysophosphatidic acid, and lysophosphocholine.
FIGURE 8.
Interacting lipid species of apoL1.FLAG protein. Affinity-purified apoL1.FLAG fusion protein was used to conduct lipid overlay assay (see “Experimental Procedures” for details). 3× FLAG peptide solution was used as a negative control. A, purified apoL1.FLAG was confirmed by immunoblot analysis using both anti-apoL1 and anti-FLAG antibodies. B, apoL1.FLAG bound strongly with CL and PA, followed by PI-3,5-P2, PI4P, PI5P, PI3P, PI-4,5-P2, PI-3,4,5-P3, PI-3,4-P2, 3-sulfogalactosylceramide (3-SGCer), phosphoserine, PG, and PI (very weak). ApoL1.FLAG did not bind cholesterol (CHO), TG, PE, PC, SM, diacylglycerol (DAG), sphingosine 1-phosphate, lysophosphatidic acid (LPA), and lysophosphocholine (LPC). Lipid abbreviations are as follows: TG (triglyceride), PI (phosphatidylinositol), DAG (diacylglycerol), PI4P (PtdIns(4)P), PA (phosphatidic acid), PI4,5P2 (PtdIns(4,5)P2), PS (phosphatidylserine), PI3,4,5P3 (PtdIns(3,4,5)P3), PE (phosphatidylethanolamine), CHO (cholesterol), PC (phosphatidylcholine), SM (sphingomyelin), PG (phosphatidylglycerol), 3-SGCer (3-sulfogalactosylceramide (Sulfatide)), CL (cardiolipin), S1P (sphingosine 1-phosphate), LPA (lysophosphatidic acid), LPC (lysophosphocholine), PI3,4P3 (PtdIns(3,4)P2), PI3,5P2 (PtdIns(3,5)P2), PI3P (PtdIns(3)P), PI4P (PtdIns(4)P), PI5P (PtdIns(5)P), and Solvent blank (Blank).
DISCUSSION
The role of the Bcl-2 family in regulating apoptosis is well studied; however, its role in mediating AuCD is still elusive. In an effort to identify and characterize novel BH3-only proteins in AuCD, we identified a novel BH3-only pro-death protein apolipoprotein L1 (apoL1) in silico. Subsequently we showed that WT apoL1 induces AuCD in a variety of cells, whereas the BH3 deletion construct of apoL1 failed to induce cell death.
Interestingly, apoL1 protein has been found both inside the cells and circulating in the blood. When it is in the serum, apoL1 binds apolipoprotein A-I and is one of the components of high density lipoprotein (HDL) particles, suggesting apoL1 has a role in lipid metabolism (48, 49). In addition, protein secondary structure analysis identified four lipid-binding amphipathic α-helices in apoL1, which may function as a catalyst for intra- and extracellular lipid transfer (48–50). Importantly, we previously reported that apoL6, a closely related protein of apoL1 and a BH3-only pro-death protein, induces mitochondria- and caspase-mediated apoptosis (45). By contrast we showed apoL4, another member of the apoL family not possessing BH3 domain, failed to induce cell death. Sequence analysis showed that six of the nine residues of the BH3 domain are identical between apoL1 and apoL6 (Fig. 1A). In addition, the BH3 domain of apoL1 showed an amphipathic α-helical structure similar to other known BH3 domains, including that of apoL6. Taken together, our results indicated that both apoL1 and apoL6 are bona fide BH3-only pro-death proteins. ApoL1 and apoL6 possess conserved primary sequence and tertiary structure; nevertheless, they function differently and nonredundantly in the induction of PCD. ApoL6 induces apoptosis (type I), whereas apoL1 induces autophagy (type II). It is not unreasonable to postulate that their interacting partners, protein and/or lipid species, play important role in mediating different death-signaling pathways. As a matter of fact, we showed that apoL1 has very strong affinity for PA and CL. CL is a dimer of PG and PA and includes four acyl chains, two phosphate groups, and three glycerols. As apoL1 binds strongly with PA, it is logical to speculate that apoL1 binds CL through its PA moiety. Interestingly, Fang et al. (55) previously showed that PA is a critical component of mammalian target of rapamycin (mTOR) signaling. They showed mitogenic stimulation of mammalian cells led to a phospholipase D-dependent accumulation of cellular PA, which was required for activation of mTOR downstream effectors. PA directly interacted with the domain in mTOR that is targeted by rapamycin, and this interaction was positively correlated with the ability of mTOR to activate downstream effectors. It has also been shown that mTOR is a negative regulator of autophagy (40, 56). Thus, binding of PA by apoL1 might inhibit mTOR activation and subsequently induce autophagy. In addition, accumulating evidence suggests that CL has active roles in mitochondria-mediated apoptosis (57). Binding of CL with apoL1 may prevent apoptosis in favor of induction of AuCD.
PIPs play critical roles in regulating cellular signaling pathways. The homeostatic equilibrium of PIPs determines cellular proliferation, differentiation, trafficking, and PCD (58, 59). It has been shown that PI4P is the most abundant member of PIPs. Furthermore, PI-3,4,5-P3 and PI-3,4-P2 can induce cell growth through the Akt pathway, but PI3P has been shown to induce autophagy and endosomal trafficking. Our results showed that binding of apoL1 to various species of PIPs may alter the homeostasis of PIPs leading to the AuCD. Interestingly, we found that apoL1 did not bind cholesterol, TG, PC, PE, or SM, the major lipid components in HDL (Fig. 8B). It has been shown that apoL1, a minor protein component, can directly interact with apoAI, the major protein component in HDL (48). It is logical to postulate that apoL1, which does not interact with HDL lipid species, binds and modulates the activities of apoAI. To our best knowledge, apoL1 is the first BH3-only, lipid-binding apolipoprotein that induces AuCD.
Interestingly, the genes encoding apoLs are localized to chromosome 22q12.3-13.1, a high susceptibility locus for schizophrenia. Importantly, expression of apoL1 has been shown to be significantly up-regulated (>1.41-fold) in the brains of schizophrenics (60). It will be interesting to see whether overexpression of apoL1 induces AuCD in central nervous system cells, and if so what role of apoL1 plays in the etiology of schizophrenia.
ApoL1 was recently found to be the trypanosome lytic factor in human serum. The parasite Trypanosoma brucei rhodesiense causes human sleeping sickness in Africa. The molecular basis of this disease is the product of resistance-associated protein, an inhibitory protein that binds specifically to apoL1 to prevent cell lysis. Unsequestered apoL1 promotes forming anion channels on lysosomal membranes of trypanosome and subsequently kills the parasite (61, 62). Thus, in addition to the aforementioned functions, apoL1 can also play an important role in innate immunity.
It is well documented that LC3 is required for complete autophagosome formation during autophagic pathway in mammalian cells. The combination of alteration of subcellular localization of LC3-II with increased ratio of LC3-II to LC3-I is one of the hallmarks of autophagy (13, 14). This study demonstrates that apoL1 is a potent and specific inducer of AuCD that stimulates time-dependent accumulation and translocation of LC3-II, formation of the AV (confirmed by TEM analysis), and subsequently cell death in apoL1-overexpressing cells. Importantly, no apoptotic hallmarks, such as release of apoptogenic factors from mitochondria or activation of caspases, were detected in apoL1-induced cell death. We also showed that expression of apoL1 is inducible by p53, a well known tumor suppressor and transactivating factor frequently mutated in cancers, which regulates both types of PCD (38–41). Regarding the role of p53 in autophagy, it has been shown that direct activation of p53 by nutrient starvation and serum deprivation induces autophagy in cancer cells (40). In addition, damage-regulated autophagy modulator, a p53 target gene encoding a lysosomal protein, has been shown to be involved in the induction of autophagy (63). Furthermore, Bad and Bid, two BH3-only proteins and direct downstream effectors of p53, not only play important roles in apoptosis but also in autophagy (34, 36, 37). Our results suggest that apoL1 is a novel BH3 only protein and is a p53 downstream effector functioning as a molecular determinant that dictates autophagy for death (Fig. 9).
FIGURE 9.
Hypothetical model of apoL1-induced autophagic death in cancer cells. Expression of apoL1 is inducible by p53, a tumor suppressor gene. Intracellular accumulation of apoL1 alters lipid homeostasis and signaling, induces activation and translocation of LC3, and generation of autophagic vacuoles leading to cell death. Inhibitors of autophagy, such as 3-MA or wortmannin, blunt apoL1-induced autophagic cell death.
Regarding other BH3-only proteins in PCD, Beclin1 interacts with anti-apoptotic Bcl-2 by virtue of its BH3 domain, an amphipathic α-helix, that binds to the hydrophobic cleft of Bcl-2. Interestingly, the association of Beclin1 with Bcl-2 is of low affinity (1–2 μm) as compared with other BH3-only proteins that bind to Bcl-2 (in low nanomolar range) (32–34). In fact, it has been shown that Bad, as well as BH3-mimetic compounds such as ABT737, competitively disrupt the inhibitory interaction between Beclin1 and Bcl-2 (34). Thus, it is logical to speculate that when the other BH3-only proteins, such as apoL1 and apoL6, are induced by various stimuli/stresses, they can displace Beclin1, rendering the cell susceptible to autophagy as well as apoptosis, respectively. It will be of interest to investigate whether apoL1 and apoL6 interact with anti-death Bcl-2 family members and the functional consequences of the interactions in PCD. Finally, apoL1 provides an excellent model to further study the molecular basis of gene-induced AuCD.
Acknowledgments
We thank Dr. Steven Jett and Tamara Howard for their excellent EM service at the EM Core Facility, University of New Mexico School of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grant 5RO1CA106644-01A1 from NCI and New Mexico-IdeA Network of Biomedical Research Excellence (NM-INBRE) Program P20RR016480-04 (to C.-A. A. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PCD, programmed cell death; AuCD, autophagic cell death; AV, autophagic vacuoles; BH, Bcl-2 homology domain; CL, cardiolipin; LC3, microtubule associated proteins light chain 3; PA, phosphatidic acid; PI, phosphatidylinositol; PI3K, phosphatidylinositol 3-phosphate kinase; PIP, phosphoinositide; 3-MA, 3-methyladenine; PARP, poly-(ADP-ribosyl) polymerase; TEM, transmission electron microscopy; aa, amino acid; mTOR, mammalian target of rapamycin; MEF, mouse embryonic fibroblast; RT, reverse transcription; BSA, bovine serum albumin; HBSS, Hanks' balanced salt solution; PE, phosphoethanolamine; PG, phosphoglycerol; PI, phosphatidylinositol; SM, sphingomyelin; TG, triglyceride; AD, adenovirus; Dox, doxycycline; HDL, high density lipoprotein; PI-3,4,5-P3, phosphatidylinositol 3,4-trisphosphate; PI-3,4-P2, phosphatidylinositol 3,4-bisphosphate; PI3P, phosphatidylinositol 3-phosphate; PI4P, phosphatidylinositol 4-phosphate; Ptd, phosphatidylinositol.
References
- 1.Tsujimoto, Y. (2003) J. Cell. Physiol. 195 158-167 [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. M., and Cory, S. (2007) Curr. Opin. Immunol. 19 488-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scorrano, L., and Korsmeyer, S. J. (2003) Biochem. Biophys. Res. Commun. 304 437-444 [DOI] [PubMed] [Google Scholar]
- 4.Huang, D. C., and Strasser, A. (2000) Cell 103 839-842 [DOI] [PubMed] [Google Scholar]
- 5.Chipuk, J. E., Bouchier-Hayes, L., and Green, D. R. (2006) Cell Death Differ. 13 1396-1402 [DOI] [PubMed] [Google Scholar]
- 6.Danial, N. N., and Korsmeyer, S. J. (2004) Cell 116 205-219 [DOI] [PubMed] [Google Scholar]
- 7.Maiuri, M. C., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007) Nat. Rev. Mol. Cell Biol. 8 741-752 [DOI] [PubMed] [Google Scholar]
- 8.Tsujimoto, Y., and Shimizu, S. (1995) Cell Death Differ. 12 1528-1534 [DOI] [PubMed] [Google Scholar]
- 9.Shintani, T., and Klionsky, D. J. (2004) Science 306 990-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima, N. (2007) Genes Dev. 21 2861-2873 [DOI] [PubMed] [Google Scholar]
- 11.Cuervo, A. M. (2004) Trends Cell Biol. 14 70-77 [DOI] [PubMed] [Google Scholar]
- 12.Levine, B., and Klionsky, D. J. (2004) Dev. Cell 6 463-477 [DOI] [PubMed] [Google Scholar]
- 13.Mizushima, N., Ohsumi, Y., and Yoshimori, T. (2002) Cell Struct. Funct. 27 421-429 [DOI] [PubMed] [Google Scholar]
- 14.Xie, Z., and Klionsky, D. J. (2007) Nat. Cell Biol. 9 1102-1109 [DOI] [PubMed] [Google Scholar]
- 15.Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., Tokuhisa, T., and Mizushima, N. (2004) Nature 432 1032-1036 [DOI] [PubMed] [Google Scholar]
- 16.Komatsu, M., Waguri, S., Ueno, T., Iwata, J., Murata, S., Tanida, I., Ezaki, J., Mizushima, N., Ohsumi, Y., Uchiyama, Y., Kominami, E., Tanaka, K., and Chiba, T. (2005) J. Cell Biol. 169 425-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gozuacik, D., and Kimchi, A. (2004) Oncogene 23 2891-2906 [DOI] [PubMed] [Google Scholar]
- 18.Kondo, Y., Kanzawa, T., Sawaya, R., and Kondo, S. (2005) Nat. Rev. Cancer 5 726-734 [DOI] [PubMed] [Google Scholar]
- 19.Levine, B., and Deretic, V. (2007) Nat. Rev. Immunol. 7 767-777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paludan, C., Schmid, D., Landthaler, M., Vockerodt, M., Kube, D., Tuschl, T., and Munz, C. (2005) Science 307 593-596 [DOI] [PubMed] [Google Scholar]
- 21.Anglade, P., Vyas, S., Javoy-Agid, F., Herrero, M. T., Michel, P. P., Marquez, J., Mouatt-Prigent, A., Ruberg, M., Hirsch, E. C., and Agid, Y. (1997) Histol. Histopathol. 12 25-31 [PubMed] [Google Scholar]
- 22.Kaneda, D., Sugie, K., Yamamoto, A., Matsumoto, H., Kato, T., Nonaka, I., and Nishino, I. (2003) Neurology 61 128-131 [DOI] [PubMed] [Google Scholar]
- 23.Berry, D. L., and Baehrecke, E. H. (2007) Cell 131 1137-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, S., Kanaseki, T., Mizushima, N., Mizuta, K., Arakawa-Kobayashi, S., Thompson, C. B., and Tsujimoto, Y. (2004) Nat. Cell Biol. 6 1221-1228 [DOI] [PubMed] [Google Scholar]
- 25.Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., Packer, M., Schneider, M. D., and Levine, B. (2005) Cell 122 927-939 [DOI] [PubMed] [Google Scholar]
- 26.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B., and Korsmeyer, S. J. (2001) Science 292 727-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis, S. N., Fletcher, J. I., Kaufmann, T., van Delft, M. F., Chen, L., Czabotar, P. E., Ierino, H., Lee, E. F., Fairlie, W. D., Bouillet, P., Strasser, A., Kluck, R. M., Adams, J. M., and Huang, D. C. (2007) Science 315 856-859 [DOI] [PubMed] [Google Scholar]
- 28.Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., and Levine, B. (1999) Nature 402 672-676 [DOI] [PubMed] [Google Scholar]
- 29.Pattingre, S., and Levine, B. (2006) Cancer Res. 66 2885-2888 [DOI] [PubMed] [Google Scholar]
- 30.Yue, Z., Jin, S., Yang, C., Levine, A. J., and Heintz, N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15077-15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., Rosen, J., Eskelinen, E. L., Mizushima, N., Ohsumi, Y., Cattoretti, G., and Levine, B. (2003) J. Clin. Investig. 112 1809-18020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberstein, A., Jeffrey, P. D., and Shi, Y. (2007) J. Biol. Chem. 282 13123-13132 [DOI] [PubMed] [Google Scholar]
- 33.Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) EMBO J. 26 2527-2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiuri, M. C., Criollo, A., Tasdemir, E., Vicencio, J. M., Tajeddine, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) Autophagy 3 374-376 [DOI] [PubMed] [Google Scholar]
- 35.Lamparska-Przybysz, M., Gajkowska, B., and Motyl, T. (2005) J. Physiol. Pharmacol. 56 Suppl. 3, 159-179 [PubMed] [Google Scholar]
- 36.Fei, P., Wang, W., Kim, S. H., Wang, S., Burns, T. F., Sax, J. K., Buzzai, M., Dicker, D. T., McKenna, W. G., Bernhard, E. J., and El-Deiry, W. S. (2004) Cancer Cell 6 597-609 [DOI] [PubMed] [Google Scholar]
- 37.Hamacher-Brady, A., Brady, N. R., Logue, S. E., Sayen, M. R., Jinno, M., Kirshenbaum, L. A., Gottlieb, R. A., and Gustafsson, A. B. (2007) Cell Death Differ. 14 146-157 [DOI] [PubMed] [Google Scholar]
- 38.Chipuk, J. E., and Green, D. R. (2006) Cell Death Differ. 13 994-1002 [DOI] [PubMed] [Google Scholar]
- 39.Lum, J. J., Bauer, D. E., Kong, M., Harris, M. H., Li, C., Lindsten, T., and Thompson, C. B. (2005) Cell 120 237-248 [DOI] [PubMed] [Google Scholar]
- 40.Levine, A. J., Feng, Z., Mak, T. W., You, H., and Jin, S. (2006) Genes Dev. 20 267-275 [DOI] [PubMed] [Google Scholar]
- 41.Gu, S., Liu, Z., Pan, S., Jiang, Z., Lu, H., Amit, O., Bradbury, E. M., Hu, C. A., and Chen, X. (2004) Mol. Cell. Proteomics 3 998-1008 [DOI] [PubMed] [Google Scholar]
- 42.Liu, Z., Lu, H., Shi, H., Du, Y., Yu, J., Gu, S., Chen, X., Liu, K. J., and Hu, C. A. (2005) Cancer Res. 65 1647-1654 [DOI] [PubMed] [Google Scholar]
- 43.Hu, C. A., Donald, S. P., Yu, J., Lin, W. W., Liu, Z., Steel, G., Obie, C., Valle, D., and Phang, J. M. (2007) Mol. Cell. Biochem. 295 85-92 [DOI] [PubMed] [Google Scholar]
- 44.Liu, Z., Wan, G., Heaphy, C., Bisoffi, M., Griffith, J. K., and Hu, C. A. (2007) Mol. Cell. Biochem. 297 179-187 [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z., Lu, H., Jiang, Z., Pastuszyn, A., and Hu, C. A. (2005) Mol. Cancer Res. 3 21-31 [PubMed] [Google Scholar]
- 46.Bystroff, C., and Shao, Y. (2002) Bioinformatics (Oxf.) 18 S54-S61 [DOI] [PubMed] [Google Scholar]
- 47.Bystroff, C., Thorsson, V., and Baker, D. (2000) J. Mol. Biol. 301 173-190 [DOI] [PubMed] [Google Scholar]
- 48.Duchateau, P. N., Pullinger, C. R., Orellana, R. E., Kunitake, S. T., Naya-Vigne, J., O'Connor, P. M., Malloy, M. J., and Kane, J. P. (1997) J. Biol. Chem. 272 25576-25582 [DOI] [PubMed] [Google Scholar]
- 49.Duchateau, P. N., Pullinger, C. R., Cho, M. H., Eng, C., and Kane, J. P. (2001) J. Lipid Res. 42 620-630 [PubMed] [Google Scholar]
- 50.Page, N. M., Butlin, D. J., Lomthaisong, K., and Lowry, P. J. (2001) Genomics 74 71-78 [DOI] [PubMed] [Google Scholar]
- 51.Kabeya, Y. (2000) EMBO J. 19 5720-5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klionsky, D. J., Cuervo, A. M., and Seglen, P. O. (2007) Autophagy 3 181-206 [DOI] [PubMed] [Google Scholar]
- 53.Eskelinen, E.-L. (2005) Autophagy 1 1-10 [DOI] [PubMed] [Google Scholar]
- 54.Seglen, P. O., and Gordon, P. B. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 1889-1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A., and Chen, J. (2001) Science 294 1942-1945 [DOI] [PubMed] [Google Scholar]
- 56.Backer, J. M. (2008) Biochem. J. 410 1-17 [DOI] [PubMed] [Google Scholar]
- 57.Gonsalvez, F., and Gottlieb, E. (2007) Apoptosis 12 877-885 [DOI] [PubMed] [Google Scholar]
- 58.Sasaki, T., Sasaki, J., Sakai, T., Takasuga, S., and Suzuki, A. (2007) Biol. Pharm. Bull. 30 1599-1604 [DOI] [PubMed] [Google Scholar]
- 59.Wymann, M. P., and Schneiter, R. (2008) Nat. Rev. Mol. Cell Biol. 9 162-176 [DOI] [PubMed] [Google Scholar]
- 60.Mimmack, M. L., Ryan, M., Baba, H., Navarro-Ruiz, J., Iritani, S., Faull, R. L., McKenna, P. J., Jones, P. B., Arai, H., Starkey, M., Emson, P. C., and Bahn, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4680-4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanhamme, L., Paturiaux-Hanocq, F., Poelvoorde, P., Nolan, D. P., Lins, L., Van Den Abbeele, J., Pays, A., Tebabi, P., Van Xong, H., Jacquet, A., Moguilevsky, N., Dieu, M., Kane, J. P., De Baetselier, P., Brasseur, R., and Pays, E. (2003) Nature 422 83-87 [DOI] [PubMed] [Google Scholar]
- 62.Perez-Morga, D., Vanhollebeke, B., Paturiaux-Hanocq, F., Nolan, D. P., Lins, L., Homble, F., Vanhamme, L., Tebabi, P., Pays, A., Poelvoorde, P., Jacquet, A., Brasseur, R., and Pays, E. (2005) Science 309 469-472 [DOI] [PubMed] [Google Scholar]
- 63.Crighton, D., Wilkinson, S., O'Prey, J., Syed, N., Smith, P., Harrison, P. R., Gasco, M., Garrone, O., Crook, T., and Ryan, K. M. (2006) Cell 126 121-134 [DOI] [PubMed] [Google Scholar]