Abstract
The small leucine-rich proteoglycan (SLRP) family has significantly expanded in the past decade to now encompass five discrete classes, grouped by common structural and functional properties. Some of these gene products are not classical proteoglycans, whereas others have new and unique features. In addition to being structural proteins, SLRPs constitute a network of signal regulation: being mostly extracellular, they are upstream of multiple signaling cascades. They affect intracellular phosphorylation, a major conduit of information for cellular responses, and modulate distinct pathways, including those driven by bone morphogenetic protein/transforming growth factor β superfamily members, receptor tyrosine kinases such as ErbB family members and the insulin-like growth factor I receptor, and Toll-like receptors. The wealth of mechanistic insights into the molecular and cellular functions of SLRPs has revealed both the sophistication of this family of regulatory proteins and the challenges that remain in uncovering the totality of their functions. This review is focused on novel biological functions of SLRPs with special emphasis on their protein cores, newly described genetic diseases, and signaling events in which SLRPs play key functions.
General Structural Features and New Small Leucine-rich Proteoglycan Families
The SLRP2 gene family (1–3) has expanded in the past decade to encompass 17 genes. Although some of these gene products are not true proteoglycans, classically defined as harboring at least one glycosaminoglycan side chain, we have listed those as members of the SLRP family based primarily on functional commonality. We now classify the SLRPs into five distinct families based on several parameters, including conservation and homology at the protein and genomic levels, the presence of characteristic N-terminal Cys-rich clusters with defined spacing, and chromosomal organization (Fig. 1). The first three canonical classes of SLRPs have been amply covered previously (1–7).
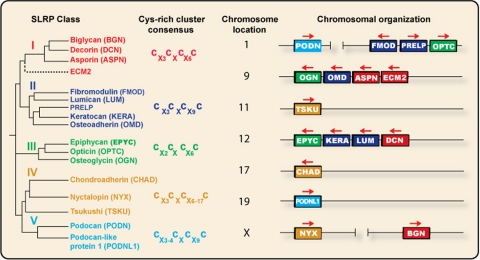
FIGURE 1.
Phylogenetic analysis and chromosomal organization of various human SLRP classes. The color-coded dendrogram (left) shows the presence of five distinct families of SLRP and related LRR proteins. The consensus for the N-terminal Cys-rich cluster is also shown. The chromosomal arrangement of the various SLRP genes is shown in a telomeric orientation (right). Transcriptional direction is shown by the arrows above the color-coded boxes. The horizontal distance between genes is not to scale. This figure was modified after Henry et al. (7). Sequences were retrieved from Swiss-Prot and inserted into Jalview 2.3, and the rooted dendrogram was generated with the ClustalW2 algorithm and plotted with Biology Workbench 3.2 DRAWGRAM.
In addition to the well described LRRs (1), typical SLRP features include the presence of N-terminal Cys clusters, usually four cysteine residues with finite intervening amino acid sequences that define the various classes. Another typical feature of SLRPs is the presence of the recently described “ear repeat” (5). In the case of decorin, LRR11 contains one of the two C-terminal Cys residues that form a disulfide bond with the other Cys in LRR12. Classes I–III and ECM2 contain the ear repeat, whereas the other two classes do not. On this basis, it has been proposed that the ear repeat is the hallmark of the true SLRP family (5). Although this might be true on a structural basis, it is apparently different on a functional level. For instance, tsukushi (class IV) is a potent modulator of BMP (8), a role shared by biglycan (class I) (9, 10). Podocan (class V) binds collagen I and inhibits cell growth (11), two biological activities shared by class I–III SLRPs.
Class I—In this class, we include decorin, biglycan, and asporin. The N termini have a typical cluster of Cys residues that form two disulfide bonds (Fig. 1). Although decorin and biglycan can be substituted with either one or two chondroitin/dermatan sulfate side chains, asporin lacks the typical Ser-Gly dipeptide and flanking amino acids required for glycanation. Thus, asporin is likely not a classical proteoglycan. However, asporin contains a stretch of Asp residues, an acidic domain also found in class II (osteoadherin), class III (epiphycan), and class V (podocan) members, located in either the N- or C-terminal region. All class I SLRPs have a similar exonic organization (eight exons), with highly conserved intron/exon junctions. We have tentatively included ECM2 in this class. Although ECM2 is much larger and structurally different from conventional SLRPs, its LRRs are 35% identical to the corresponding domains of decorin, and the ECM2 gene is physically linked to asporin on chromosome 9.
Class II—This class can be subdivided into three subgroups based on protein homology (Fig. 1). Notably, class II SLRPs contain clusters of Tyr sulfate residues at their N termini that could contribute to the polyanionic nature of SLRPs. Class II members contain primarily keratan sulfate and polylactosamine, an unsulfated form of keratan sulfate, and their respective genes have a similar exonic organization (three exons), with a large central exon encoding most of LRRs.
Class III—This class contains three members characterized by a relatively low number of LRRs (seven LRRs) and a genomic organization comprising seven exons. Again, albeit most of these SLRPs have a consensus sequence for glycanation, some of them exist as glycoproteins in tissues.
Class IV—We propose a new non-canonical class of SLRPs, class IV, composed of related chondroadherin and nyctalopin (12, 13) and of a new member called tsukushi because its expression pattern is similar to the shape of the Japanese horsetail plant tsukushi (8). Nyctalopin is very interesting, being the first described glycosylphosphatidylinositol-anchored member (see below) and the second linked to the X chromosome. Both tsukushi and nyctalopin have 11 homologous LRRs flanked by an N-terminal Cys-rich region. Tsukushi shares functional properties with class I SLRPs (9, 10) insofar as it is a BMP inhibitor that forms a ternary complex with BMP and chordin (8, 14).
Class V—This is a new non-canonical class of SLRPs and contains two genes, podocan located on chromosome 1 (15) and a highly homologous podocan-like protein 1 (NCBI accession number 079101) located on chromosome 19. Podocan was originally cloned from a library derived from human immunodeficiency virus transgenic podocytes and hence its eponym (15). Although these proteins have a different C-terminal Cysrich cluster, they have 20 LRRs with homology to class I and II molecules. Moreover, podocan binds collagen I and inhibits cell growth via induction of p21 (11), both functional properties shared by other SLRP members.
Chromosomal Clusters—Chromosomal clustering of SLRP genes suggests that they arose by duplication of chromosomal segments. For instance, chromosomes 9 and 12 contain four SLRP genes, with class I genes being centromeric to class II and III genes (Fig. 1). Likewise class III members always lie telomeric to class II members. Classes IV and V appear not to cluster with other SLRPs with the exception of podocan and nyctalopin, which are on chromosomes 1 and X, respectively, where other SLRP members are situated. The significance of the SLRP clusters is unclear. Because several of the SLRP genes have been retained in the clusters during evolution, it is likely that a degree of functional redundancy has also been maintained (7).
Dimerization: Dimer-Monomer Transition—Some SLRPs have been shown to dimerize with high affinity (5, 16). Although this may be true in vitro, in vivo they likely function as monomers (17). The sequence in decorin that binds collagen I is located in the concave face of LRR6 (18), a position that would be less accessible to triple-helical collagen. Given the overall dimensions of the decorin protein core (16), a dimeric decorin would not fit in the EGFR groove where the EGF binds. Moreover, the reported interaction between decorin and the EGFR in the yeast two-hybrid system (19) is likely a 1:1 interaction. Finally, truncated forms of decorin protein core lacking the first five LRRs are still capable of functionally interacting with the EGFR (19), whereas truncated mutant forms of decorin harboring the first five LRRs bind and activate the IGF-IR (20): both mutants likely will not be able to form dimers. Thus, it is difficult to envisage the binding and blocking of TGFβ or BMP by dimeric decorin or biglycan. A plausible scenario is that SLRPs undergo a dimer-monomer transition that would expose key sites involved in specific bindings. Thus, their functional activity in vivo would be regulated by the intrinsic affinity of each SLRP for its cognate receptor. We favor this possibility insofar as it would contribute to specialization and functional differentiation (5).
Diseases Genetically Linked to SLRP Mutations
In the past decade, several SLRP-linked genetic diseases have been reported, and curiously, all the inherited disorders cause ocular abnormalities (Table 1). Truncated forms of decorin lacking the C-terminal 33 amino acids comprising the ear repeat cause congenital stromal dystrophy, an autosomal dominant disorder characterized by opacities in the corneal stroma (21). Interestingly, heterozygotes containing both a normal and a truncated decorin have corneal clouding. Thus, the truncated form of decorin may act in a dominant-negative fashion by altering the orthogonal arrangement of corneal collagen fibrils required for transparency. Regulation of collagen fibrillogenesis is an important function shared by several SLRPs, and null mutations of lumican and fibromodulin also lead to abnormal collagen architecture (22, 23). However, a critical concept is the compensation of one SLRP function over another. For example, in the absence of fibromodulin, lumican accumulates (22), whereas in the absence of biglycan, decorin is up-regulated in repairing muscle, diseased kidney, and bone cells (24). Considering tissue context, the same SLRP could have distinct roles in different organs or even species.
TABLE 1.
Human ocular diseases linked to mutations in SLRP-encoding genes
SNPs, single-nucleotide polymorphisms.
| Gene | Mutation | Inheritance | Chromosome | Phenotype |
|---|---|---|---|---|
| Decorin | Frameshift mutation generating a C-terminally truncated decorin protein core | Autosomal dominant | 12 | Congenital stromal dystrophy of the cornea: corneal opacities caused by deposition of white fluffy material in the corneal stroma (21) |
| Lumican, fibromodulin, PRELP, and opticin | Intronic variations, non-synonymous and synonymous changes, SNPs in promoter | Autosomal dominant | 1 and 12 | High myopia: a common cause of blindness secondary to corneal detachment and choroidal neovascularization (25, 26) |
| Keratocan | Missense and frameshift mutations generating a single amino acid substitution or a C-terminally truncated keratocan | Autosomal recessive | 12 | Cornea plana (CNA2): corneal radius of curvature larger than normal, producing high hypermetropia with astigmatism and poor acuity (30) |
| Nyctalopin | Intragenic deletions, missense mutations, nonsense mutations, and in-frame insertions | X-linked | X | Congenital stationary night blindness with associated myopia, hyperopia, nystagmus, and reduced visual acuity (12, 13) |
High myopia is caused by loss-of-function mutations involving several SLRPs, including lumican, fibromodulin, PRELP, and opticin (25, 26). Notably, gene targeting studies on lumican-null (27) and lumican-fibromodulin-double-null mice (28) have shown analogous ocular abnormalities that can be partially rescued by re-expression of lumican in the cornea (29). Missense and frameshift mutations generating a C-terminal truncated keratocan cause cornea plana (30), an autosomal recessive disease in which the corneal radius of curvature is larger than normal, producing high hypermetropia with astigmatism and poor acuity.
A number of mutations in the nyctalopin gene cause X-linked congenital stationary night blindness, a group of stable retinal disorders that are characterized by abnormal nocturnal vision (12, 13). Often these disorders are associated with myopia, hyperopia, nystagmus, and reduced visual acuity. The proposed pathogenetic mechanism of action is that the disruption of the glycosylphosphatidylinositol-anchored nyctalopin causes a concurrent alteration of developing retinal interconnections involving the ON bipolar cells, leading to loss of nocturnal vision.
Gene targeting in mice has revealed widespread involvement of SLRP genes in various pathogenetic mechanisms causing skin fragility, osteoporosis, and cardiovascular disease (31–33). Decorin deficiency (34) enhances renal fibrosis (35) and progressive nephropathy in diabetic mice with increased mesangial matrix accumulation and fibrin deposits (36), partly regulated by the ability of decorin to bind fibrinogen and sterically modulate fibrin assembly (37).
Multiple Signaling Pathways Evoked by SLRPs
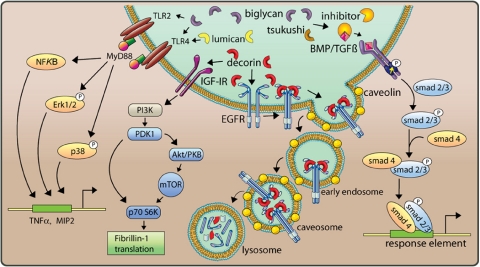
Receptor Tyrosine Kinase: EGFR and IGF-IR—SLRPs are involved in the initial triggering of multiple cellular responses. In tumor cells, there is a host of evidence that decorin LRR7 binds to the EGFR and ErbB4 and leads to activation of the MAPK pathway, Ca2+ influx, induction of the cyclin-dependent kinase inhibitor p21, and subsequently down-regulation of the receptor (19, 38–43). The decorin-bound EGFR is internalized via caveolin-mediated pathways (43) and reaches caveosomes and then lysosomes, where the receptor is degraded (Fig. 2). These studies have implications for the potential protein-based therapy for solid tumors: local or systemic delivery of decorin can retard the growth of primary as well as metastatic carcinomas and reduces EGFR levels (44–46).
FIGURE 2.
Multiple signaling pathways evoked by SLRPs. Several distinct pathways are affected by various SLRPs as indicated. The diagram is obviously incomplete insofar as it does not include cross-talks among the various signal transduction pathways. For additional information, see text. PI3K, phosphatidylinositol 3-kinase; PKB, protein kinase B.
In normal cells such as endothelial and renal cells, decorin affects different pathways (20, 35, 47, 48). The N-terminal region of decorin protein core binds the IGF-IR, resulting in its phosphorylation and activation, followed by receptor down-regulation (20, 48). Decorin-mediated regulation of IGF-IR signaling leads to enhanced phosphorylation of protein kinase B (Akt) with subsequent induction of p21 by a MAPK-independent pathway, resulting in the inhibition of apoptosis in endothelial cells (47). Thus, decorin stimulates the IGF-IR without inhibiting signaling, as has been shown for its interaction with receptors of the ErbB family. The affinities of decorin and IGF-I for the receptor under similar binding conditions in epithelial cells and renal fibroblasts differ only by 10–20-fold (20, 48). Therefore, when decorin is abundantly expressed or when the amount of IGF available for binding to the IGF-IR is limited by the IGF-binding proteins, decorin may effectively compete with IGF-I for binding to the IGF-IR and preempt IGF signaling. In addition to p21, the related cyclin-dependent kinase inhibitor p27 is also induced, although through an Akt- and MAPK-independent mechanism (47). After unilateral ureteral obstruction, decorin-null mice reveal enhanced apoptosis of tubular epithelial cells, resulting in accelerated renal fibrosis (35). Notably, the IGF-IR is up-regulated, presumably to compensate for the lack of decorin, suggesting that in vivo decorin and the IGF-IR may functionally cooperate. Furthermore, interaction of decorin with the IGF-IR/mTOR/p70 S6 kinase signaling pathway leads to enhanced translation of fibrillin-1 (Fig. 2) and its deposition in the extracellular environment (48, 50). Thus, decorin acts in normal cells as a signaling molecule through the canonical IGF signaling cascade and directly regulates cell death and synthesis of other matrix constituents, potentially influencing the pathophysiology of several diseases.
Toll-like Receptors—A striking concurrence of biglycan over-expression and enhanced numbers of infiltrating cells has been observed in animal models of renal inflammation (35), suggesting the involvement of biglycan in regulation of the inflammatory response reaction. In fact, biglycan acts as an endogenous ligand of the innate immunity receptors TLR4 and TLR2 in macrophages. Because of a MyD88-dependent induction of extracellular signal-regulated kinase (Erk), p38, and NF-κB, biglycan stimulates expression of the inflammatory mediators TNFα and MIP2 (Fig. 2), the murine interleukin-8 analog (51). Activation of both TLRs requires intact and soluble biglycan, suggesting that both the protein core and glycosaminoglycan side chains are required and that proteolytic release from the extracellular matrix is necessary to initiate the pro-inflammatory function of biglycan. Notably, activated macrophages synthesize and secrete biglycan. Such mechanisms likely play a role in vivo insofar as in TLR4- and TLR2-dependent models of sepsis, biglycan-null mice do survive for longer periods of time due to lower circulating TNFα and less pulmonary mononuclear cell infiltration (51). The fact that biglycan-induced signaling in macrophages is mediated by two receptors important in the recognition of both Gram-negative and Gram-positive pathogens emphasizes the biological relevance of this proteoglycan within the innate immune system as a TLR ligand analogous to the PAMPs. Notably, lumican has also been shown to interact with the TLR4 pathway by presenting lipopolysaccharide to CD14, a cell-surface lipopolysaccharide-binding protein required for TLR4 activation (52). Thus, akin to gene products involved in pathogen recognition, SLRPs may either react as PAMP analogs or present PAMPs to the receptor complex, thereby influencing TLR signaling.
BMP/TGFβ Receptors—Many members of the SLRP gene family are able to bind to and modulate BMP/TGFβ pathways. Decorin, biglycan, and asporin (class I) and fibromodulin (class II) bind to TGFβ (53). Moreover, decorin modulates the TGFβ pathway through interaction with LRP1 (54) and regulates matrix organization and mechanical characteristics of three-dimensional collagen matrices (55) and skeletal muscle differentiation (56). The lessons from biglycan-deficient mice, which develop age-dependent osteopenia (24) due to a decreased ability to make new bone because of a reduced response of bone marrow stromal cells to TGFβ (9), presaged the functional relationship between biglycan and BMPs in controlling skeletal cell differentiation. In fact, osteoblasts lacking biglycan displayed a defect in differentiation due to reduced BMP4 binding, followed by lower BMP4 sensitivity, resulting in less BMP4 signal transduction and decreased expression of core binding factor α1, an essential transcription factor for osteoblast differentiation (9). However, in studies addressing the regulation of BMP4 signaling pathways during embryonic development, biglycan has been shown to bind BMP4 and accelerate the inhibitory effects of chordin and Tsg (Twisted gastrulation) on BMP4 activity by increasing the binding of BMP4 to chordin and improving the efficiency of chordin-Tsg complexes to inactivate BMP4. The biological relevance of these biochemical findings was further confirmed in Xenopus embryos, where microinjection of biglycan mRNA inhibited BMP4 activity and influenced embryonic development in a chordin-dependent manner (10). In the absence of biglycan and fibromodulin, tendon progenitor cells are more sensitive to BMP2 (57), known to inhibit tendon formation during development. Thus, biglycan might play a crucial role in the network of secreted proteins regulating BMP signaling. The effects of biglycan on BMP signaling, observed in bone and tendon of biglycan-null mice, could indicate a tissue-specific function of biglycan potentially influenced by different players in BMP signaling, e.g. competitive binding of BMP2 and BMP4 to biglycan (10) and other SLRPs (9). In this context, tsukushi, which functions as a BMP4 antagonist by binding to both BMP and chordin (14), might be of particular relevance (Fig. 2). Tsukushi regulates BMP4 transcription indirectly via binding to X-delta-1 and by modulating Notch signaling, thereby controlling ectodermal patterning and neural crest specification (58). Two tsukushi isoforms modulate VG1 signaling, a crucial pathway in the development of the chick embryo (14). Recently, fibroblast growth factor and Xnr2 (Xenopus nodal-related protein-2) were added to the complex extracellular network of tsukushi-regulated signaling (49).
Conclusions
The signaling network of SLRPs provides an additional layer of control during tissue morphogenesis, cancer growth, and native immunity, among other functions. Abundance of certain SLRPs at sites of remodeling may switch one pathway, whereas their absence is permissive for other pathways. Future research should focus on translating some of this information generated in the past decade into the clinics by, for instance, utilizing protein-based therapy for fibrosis, cancer, and inflammatory disorders.
Supplementary Material
Acknowledgments
We thank A. Nyström and A. McQuillan for help with the phylogenetic tree and graphics, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA39481, RO1 CA47282, and RO1 CA120975 (to R. V. I.). This work was also supported by Deutsche Forschungsgemeinschaft Grant SFB 492 (to L. S.). This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: SLRP, small leucine-rich proteoglycan; LRR, leucine-rich repeat; BMP, bone morphogenetic protein; EGFR, epidermal growth factor receptor; IGF-IR, insulin-like growth factor I receptor; TGFβ, transforming growth factor β; MAPK, mitogen-activated protein kinase; TLR, Toll-like receptor; PAMP, pathogen-associated molecular pattern.
References
- 1.Iozzo, R. V. (1997) Crit. Rev. Biochem. Mol. Biol. 32 141–174 [DOI] [PubMed] [Google Scholar]
- 2.Hocking, A. M., Shinomura, T., and McQuillan, D. J. (1998) Matrix Biol. 17 1–19 [DOI] [PubMed] [Google Scholar]
- 3.Iozzo, R. V. (1999) J. Biol. Chem. 274 18843–18846 [DOI] [PubMed] [Google Scholar]
- 4.Iozzo, R. V. (1998) Annu. Rev. Biochem. 67 609–652 [DOI] [PubMed] [Google Scholar]
- 5.McEwan, P. A., Scott, P. G., Bishop, P. N., and Bella, J. (2006) J. Struct. Biol. 155 294–305 [DOI] [PubMed] [Google Scholar]
- 6.Huxley-Jones, J., Robertson, D. L., and Boot-Handford, R. P. (2007) Matrix Biol. 26 2–11 [DOI] [PubMed] [Google Scholar]
- 7.Henry, S. P., Takanosu, M., Boyd, T. C., Mayne, P. M., Eberspaecher, H., Zhou, W., Crombrugghe, B., Höök, M., and Mayne, R. (2001) J. Biol. Chem. 276 12212–12221 [DOI] [PubMed] [Google Scholar]
- 8.Ohta, K., Lupo, G., Kuriyama, S., Keynes, R., Holt, C. E., Harris, W. A., Tanaka, H., and Ohnuma, S.-I. (2004) Dev. Cell 7 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, X.-D., Fisher, L. W., Robey, P. G., and Young, M. F. (2004) FASEB J. 18 948–958 [DOI] [PubMed] [Google Scholar]
- 10.Moreno, M., Muñoz, R., Aroca, F., Labarca, M., Brandan, E., and Larraín, J. (2005) EMBO J. 24 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu-Hirota, R., Sasamura, H., Kuroda, M., Kobayashi, E., and Saruta, T. (2004) FEBS Lett. 563 69–74 [DOI] [PubMed] [Google Scholar]
- 12.Bech-Hansen, N. T., Naylor, M. J., Maybaum, T. A., Sparkes, R. L., Koop, B., Birch, D. G., Bergen, A. A. B., Prinsen, C. F. M., Polomeno, R. C., Gal, A., Drack, A. V., Musarella, M. A., Jacobson, S. G., Young, R. S. L., and Weleber, R. G. (2000) Nat. Genet. 26 319–323 [DOI] [PubMed] [Google Scholar]
- 13.Pusch, C. M., Zeitz, C., Brandau, O., Pesch, K., Achatz, H., Feil, S., Scharfe, C., Maurer, J., Jacobi, F. K., Pinckers, A., Andreasson, S., Hardcastle, A., Wissinger, B., Berger, W., and Meindl, A. (2000) Nat. Genet. 26 324–327 [DOI] [PubMed] [Google Scholar]
- 14.Ohta, K., Kuriyama, S., Okafuji, T., Gejima, R., Ohnuma, S.-I., and Tanaka, H. (2006) Development (Camb.) 133 3777–3786 [DOI] [PubMed] [Google Scholar]
- 15.Ross, M. D., Bruggeman, L. A., Hanss, B., Sunamoto, M., Marras, D., Klotman, M. E., and Klotman, P. E. (2003) J. Biol. Chem. 278 33248–33255 [DOI] [PubMed] [Google Scholar]
- 16.Scott, P. G., McEwan, P. A., Dodd, C. M., Bergmann, E. M., Bishop, P. N., and Bella, J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15633–15638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldoni, S., Owens, R. T., McQuillan, D. J., Shriver, Z., Sasisekharan, R., Birk, D. E., Campbell, S., and Iozzo, R. V. (2004) J. Biol. Chem. 279 6606–6612 [DOI] [PubMed] [Google Scholar]
- 18.Kalamajski, S., Aspberg, A., and Oldberg, Å. (2007) J. Biol. Chem. 282 16062–16067 [DOI] [PubMed] [Google Scholar]
- 19.Santra, M., Reed, C. C., and Iozzo, R. V. (2002) J. Biol. Chem. 277 35671–35681 [DOI] [PubMed] [Google Scholar]
- 20.Schönherr, E., Sunderkötter, C., Iozzo, R. V., and Schaefer, L. (2005) J. Biol. Chem. 280 15767–15772 [DOI] [PubMed] [Google Scholar]
- 21.Bredrup, C., Knappskog, P. M., Majewski, J., Rodahl, E., and Boman, H. (2005) Investig. Ophthalmol. Vis. Sci. 46 420–426 [DOI] [PubMed] [Google Scholar]
- 22.Svensson, L., Aszódi, A., Reinholt, F. P., Fässler, R., HeinegÅrd, D., and Oldberg, Å. (1999) J. Biol. Chem. 274 9636–9647 [DOI] [PubMed] [Google Scholar]
- 23.Jepsen, K. E., Wu, F., Peragallo, J. H., Paul, J., Roberts, L., Ezura, Y., Oldberg, Á., Birk, D. E., and Chakravarti, S. (2002) J. Biol. Chem. 277 35532–35540 [DOI] [PubMed] [Google Scholar]
- 24.Ameye, L., and Young, M. F. (2002) Glycobiology 12 107R–116R [DOI] [PubMed] [Google Scholar]
- 25.Majava, M., Bishop, P. N., Hägg, P., Scott, P. G., Rice, A., Inglehearn, C., Hammond, C. J., Spector, T. D., Ala-Kokko, L., and Männikkö, M. (2007) Hum. Mutat. 28 336–344 [DOI] [PubMed] [Google Scholar]
- 26.Wang, I.-J., Chiang, T.-H., Shih, Y.-F., Hsiao, C. K., Lu, S.-C., Hou, Y.-C., and Lin, L. L. K. (2006) Mol. Vision 12 852–857 [PubMed] [Google Scholar]
- 27.Chakravarti, S., Magnuson, T., Lass, J. H., Jepsen, K. J., LaMantia, C., and Carroll, H. (1998) J. Cell Biol. 141 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarti, S., Paul, J., Roberts, L., Chervoneva, I., Oldberg, A., and Birk, D. E. (2003) Investig. Ophthalmol. Vis. Sci. 44 2422–2432 [DOI] [PubMed] [Google Scholar]
- 29.Meij, J. T. A., Carlson, E. C., Wang, L., Liu, C.-Y., Jester, J. V., Birk, D. E., and Kao, W. W. Y. (2007) Mol. Vis. 13 2012–2018 [PubMed] [Google Scholar]
- 30.Pellegata, N. S., Dieguez-Lucena, J. L., Joensuu, T., Lau, S., Montgomery, K. T., Krahe, R., Kivela, T., Kucherlapati, R., Forsius, H., and de la Chapelle, A. (2000) Nat. Genet. 25 91–95 [DOI] [PubMed] [Google Scholar]
- 31.Reed, C. C., and Iozzo, R. V. (2003) Glycoconj. J. 19 249–255 [DOI] [PubMed] [Google Scholar]
- 32.Corsi, A., Xu, T., Chen, X.-D., Boyde, A., Liang, J., Mankani, M., Sommer, B., Iozzo, R. V., Eichstetter, I., Robey, P. G., Bianco, P., and Young, M. F. (2002) J. Bone Miner. Res. 17 1180–1189 [DOI] [PubMed] [Google Scholar]
- 33.Heegaard, A.-M., Corsi, A., Danielsen, C. C., Nielsen, K. L., Jorgensen, H. L., Riminucci, M., Young, M. F., and Bianco, P. (2007) Circulation 115 2731–2738 [DOI] [PubMed] [Google Scholar]
- 34.Danielson, K. G., Baribault, H., Holmes, D. F., Graham, H., Kadler, K. E., and Iozzo, R. V. (1997) J. Cell Biol. 136 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer, L., Macakova, K., Raslik, I., Micegova, M., Gröne, H.-J., Schönherr, E., Robenek, H., Echtermeyer, F. G., Grässel, S., Bruckner, P., Schaefer, R. M., Iozzo, R. V., and Kresse, H. (2002) Am. J. Pathol. 160 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, K. J., Qiu, G., Usui, H. K., Dunn, S. R., McCue, P., Bottinger, E., Iozzo, R. V., and Sharma, K. (2007) Am. J. Pathol. 171 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dugan, T. A., Yang, V. W. C., McQuillan, D. J., and Höök, M. (2006) J. Biol. Chem. 281 38208–38216 [DOI] [PubMed] [Google Scholar]
- 38.De Luca, A., Santra, M., Baldi, A., Giordano, A., and Iozzo, R. V. (1996) J. Biol. Chem. 271 18961–18965 [DOI] [PubMed] [Google Scholar]
- 39.Moscatello, D. K., Santra, M., Mann, D. M., McQuillan, D. J., Wong, A. J., and Iozzo, R. V. (1998) J. Clin. Investig. 101 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iozzo, R. V., Moscatello, D., McQuillan, D. J., and Eichstetter, I. (1999) J. Biol. Chem. 274 4489–4492 [DOI] [PubMed] [Google Scholar]
- 41.Santra, M., Mann, D. M., Mercer, E. W., Skorski, T., Calabretta, B., and Iozzo, R. V. (1997) J. Clin. Investig. 100 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CsordÁs, G., Santra, M., Reed, C. C., Eichstetter, I., McQuillan, D. J., Gross, D., Nugent, M. A., Hajnóczky, G., and Iozzo, R. V. (2000) J. Biol. Chem. 275 32879–32887 [DOI] [PubMed] [Google Scholar]
- 43.Zhu, J.-X., Goldoni, S., Bix, G., Owens, R. A., McQuillan, D., Reed, C. C., and Iozzo, R. V. (2005) J. Biol. Chem. 280 32468–32479 [DOI] [PubMed] [Google Scholar]
- 44.Reed, C. C., Gauldie, J., and Iozzo, R. V. (2002) Oncogene 21 3688–3695 [DOI] [PubMed] [Google Scholar]
- 45.Reed, C. C., Waterhouse, A., Kirby, S., Kay, P., Owens, R. A., McQuillan, D. J., and Iozzo, R. V. (2005) Oncogene 24 1104–1110 [DOI] [PubMed] [Google Scholar]
- 46.Seidler, D. G., Goldoni, S., Agnew, C., Cardi, C., Thakur, M. L., Owens, R. A., McQuillan, D. J., and Iozzo, R. V. (2006) J. Biol. Chem. 281 26408–26418 [DOI] [PubMed] [Google Scholar]
- 47.Schönherr, E., Levkau, B., Schaefer, L., Kresse, H., and Walsh, K. (2001) J. Biol. Chem. 276 40687–40692 [DOI] [PubMed] [Google Scholar]
- 48.Schaefer, L., Tsalastra, W., Babelova, A., Baliova, M., Minnerup, J., Sorokin, L., Gröne, H.-J., Reinhardt, D. P., Pfeilschifter, J., Iozzo, R. V., and Schaefer, R. M. (2007) Am. J. Pathol. 170 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris, S. A., Almeida, A. D., Tanaka, H., Ohta, K., and Ohnuma, S.-I. (2007) PLoS ONE e1004. [DOI] [PMC free article] [PubMed]
- 50.Schaefer, L., Mihalik, D., Babelova, A., Krzyzankova, M., Grone, H. J., Iozzo, R. V., Young, M. F., Seidler, D. G., Lin, G., Reinhardt, D., and Schaefer, R. M. (2004) Am. J. Pathol. 165 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaefer, L., Babelova, A., Kiss, E., Hausser, H.-J., Baliova, M., Krzyzankova, M., Marsche, G., Young, M. F., Mihalik, D., Götte, M., Malle, E., Schaefer, R. M., and Gröne, H.-J. (2005) J. Clin. Investig. 115 2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, F., Vij, N., Roberts, L., Lopez-Briones, S., Joyce, S., and Chakravarti, S. (2007) J. Biol. Chem. 282 26409–26417 [DOI] [PubMed] [Google Scholar]
- 53.Hildebrand, A., Romaris, M., Rasmussen, L. M., HeinegÅrd, D., Twardzik, D. R., Border, W. A., and Ruoslahti, E. (1994) Biochem. J. 302 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabello-Verrugio, C., and Brandan, E. (2007) J. Biol. Chem. 282 18842–18850 [DOI] [PubMed] [Google Scholar]
- 55.Ferdous, Z., Wei, V. M., Iozzo, R. V., Höök, M., and Grande-Allen, K. J. (2007) J. Biol. Chem. 282 35887–35898 [DOI] [PubMed] [Google Scholar]
- 56.Droguett, R., Cabello-Verrugio, C., Riquelme, C., and Brandan, E. (2006) Matrix Biol. 25 332–341 [DOI] [PubMed] [Google Scholar]
- 57.Bi, Y., Ehirchiou, D., Kilts, T. M., Inkson, C. A., Embree, M. C., Sonoyama, W., Li, L., Leet, A. I., Seo, B.-M., Zhang, L., Shi, S., and Young, M. F. (2007) Nat. Med. 13 1219–1227 [DOI] [PubMed] [Google Scholar]
- 58.Kuriyama, S., Lupo, G., Ohta, K., Ohnuma, S.-I., Harris, W. A., and Tanaka, H. (2005) Development (Camb.) 133 75–88 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.