Abstract
The discovery of β-arrestin-related ∼46-kDa polypeptide in transfected cells and mouse hearts led us to examine angiotensin II type 1 receptor (AT1R)-dependent proteolytic cleavage of β-arrestin(s). Receptor-ligand induced proteolysis of β-arrestin(s) is novel, especially in the endocrine system, since proteolytic and/or splice variants of nonvisual arrestins are unknown. We used a strategy to retrieve AT1R-engaged isoforms of β-arrestin 1 to confirm direct interaction of fragments with this G protein-coupled receptor and determine cleavage sites. Here we show that the angiotensin II-AT1R complex is associated with full-length and ∼46-kDa β-arrestin forms. Mass spectrometric analysis of the AT1R-associated short form suggested a scissile site located within the Arg363–Arg393 region in the bovine β-arrestin 1. Edman degradation analysis of a β-arrestin 1 C-terminal fragment fused to enhanced green fluorescent protein confirmed the major cleavage to be after Phe388 and a minor cleavage after Asn375. Rather unexpectedly, the inverse agonist EXP3174-bound AT1R generated different fragmentation of bovine β-arrestin 1, at Pro276. The angiotensin II-induced cleavage is independent of inositol 1,4,5-trisphosphate- and Ca2+-mediated signaling pathways. The proteolysis of β-arrestin 2 occurs, but the pattern is more complex. Our findings suggest that β-arrestin cleavage upon AT1R stimulation is a part of the unraveling β-arrestin-mediated G protein-coupled receptor signaling diversity.
Signaling mechanism(s) involving the G protein-coupled receptors (GPCRs)4 and β-arrestins is gaining considerable attention. GPCRs are the largest family of cell surface receptors that share structural and regulatory features, including sequence similarity, a seven-transmembrane helical bundle, the ability to recruit and activate G proteins, and desensitization by phosphorylation and the binding of β-arrestins (1). Physiologically, GPCRs carry out one of the most fundamental cellular processes, transmission of specific signals across the plasma membrane in response to an extracellular stimulus. The β-arrestins turn off G protein-mediated signals and independently serve as multifunctional adaptors and scaffold proteins that recruit a broad spectrum of signaling molecules to the specific conformational isomers of a GPCR (2). Quite surprisingly, this novel means of β-arrestin-mediated signal transduction by GPCRs seems to be universal and versatile, and its involvement in physiology and pathological processes may be more important than previously considered. Both β-arrestin 1 and 2 are involved in signal transduction, and they share many common structural and functional features. Both are ubiquitously expressed and interact with GPCRs to initiate clathrin-dependent internalization and cellular signaling pathways by serving as multifunctional scaffold/adapter proteins (2). The recruitment and activation of numerous kinases (c-Src family and mitogen-activated protein kinases, tyrosine kinases, Akt, and phosphoinositide 3-kinase), phosphatases, and NFκB have been reported (2, 3).
Because of the diversity among GPCRs, it is perhaps not surprising that arrestins mediate different functions of distinct GPCRs. β-Arrestin signaling in the context of angiotensin II type I receptor (AT1R) is very important. The AT1R, which is a GPCR, plays an important role in blood pressure regulation and water-electrolyte balance (3). It mediates various Ang II-dependent physiological responses, such as cardiac contraction and growth, smooth muscle cell constriction, proliferation, and motility, and aldosterone secretion (3). Upon Ang II binding, the AT1R activates G protein-dependent formation of IP3 and diacylglycerol, leading to release of stored Ca2+ from the endoplasmic reticulum and Ca2+-dependent vasoconstriction. The AT1R is rapidly phosphorylated by GPCR kinases, and β-arrestin is recruited to turn off G protein activation (4).
Using specifically designed analogs of Ang II and AT1R mutants, stimulation of AT1R has been shown to lead to slow but persistent β-arrestin-mediated activation of ERK1/2 in the cytosol in the absence of Gq activation (10). The existence of β-arrestin-dependent signaling strongly suggests novel physiological end points linked to AT1R, but the cellular processes that regulate β-arrestin-mediated physiological events are not completely described. Activated receptor has been shown to induce conformational changes in β-arrestin leading to interaction with other molecules or post-translational modifications, such as phosphorylation and ubiquitination (5, 6). These β-arrestin-regulatory mechanisms play a crucial role in both internalization and signaling of GPCRs (2).
In the visual system, splice variants of the visual arrestin that lack the C-terminal tail have been reported (7, 8). A splice variant of visual arrestin, termed p44, is identical to full-length arrestin except that the last exon encoding the C-terminal 35 amino acids is replaced by a single alanine residue and binds to both nonphosphorylated and phosphorylated rhodopsin, whereas the full-length visual arrestin binds to only phosphorylated rhodopsin (7). In purified bovine rod outer segments, visual arrestin was shown to be proteolyzed in light conditions, and the proteolysis occurred only when the visual arrestin was bound to rhodopsin (9). Splice variants of bovine β-arrestin 1 and 2 in some tissues have been reported (10). Although the short form of visual arrestin involved in rhodopsin desensitization has been studied, the proteolysis of β-arrestin in the endocrine system has not been reported. Here we show that β-arrestin is cleaved following interaction with the AT1R, and proteolysis occurs at different cleavage sites, depending on whether agonist or inverse agonist is bound to AT1R. We conclude that proteolysis of β-arrestin is an important previously unknown aspect of AT1R signaling. Our finding has important implications with regard to the expanding signaling repertoires of GPCR scaffolding proteins and their ligands.
EXPERIMENTAL PROCEDURES
Materials—The cDNA clone for bovine β-arrestin 1 was kindly provided by Jeffrey Benovic (Thomas Jefferson University, Philadelphia, PA). The cDNA for human β-arrestin 2 was obtained from Origene TrueClone (Rockville, MD). [Sar1]Ang II was purchased from Bachem (Torrance, CA). [Sar1, Ile4,Ile8]Ang II was synthesized at the Cleveland Clinic Foundation Molecular Biotechnology Core (Cleveland, OH). Losartan and EXP3174 were gifts from DuPont Merck Co. (Wilmington, DE). Candesartan was a gift from AstraZeneca (Wilmington, DE). BAPTA acetoxymethyl ester and Lipofectamine 2000 were purchased from Invitrogen. Monoclonal antibody to HA high affinity (3F10; rat) and horseradish peroxidase-conjugated HA (12CA5; mouse) were purchased from Roche Applied Science. Monoclonal antibody to β-arrestin 2 (H-9) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal antibody to phospho-β-arrestin 1 (Ser412) was purchased from Cell Signaling Technology (Danvers, MA). Monoclonal antibody to green fluorescent protein (GFP) was purchased from Clontech (Mountain View, CA). Monoclonal antibody to Myc was obtained from the Cleveland Clinic Foundation Hybridoma Core Facility (Cleveland, OH). Sheep anti-mouse IgG secondary antibody was purchased from GE Healthcare. COS-1 cells and rat aortic smooth muscle cells (A7r5) were from the American Type Culture Collection (Manassas, VA). Western blot stripping buffer was purchased from Pierce. The HisTrap column and Protein G-Sepharose were purchased from GE Healthcare. All other reagents unless stated otherwise were from Sigma.
Cell Culture and Expression of AT1R—The synthetic rat AT1R gene, cloned in the shuttle expression vector pMT3, was used for expression. Creation of the constitutively active AT1R mutant AT1N111G by site-directed mutagenesis was described in a previous study (11). To express the AT1R protein, 60–65% confluent COS-1 cells grown in a 10-cm Petri dish cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum were transfected with 10 μg of purified plasmid DNA using Lipofectamine 2000 according to the manufacturer's instructions.
Immunoprecipitations and Western Blotting—The β-arrestin bands shown in Figs. 1, 2, 3 are HA-AT1R-associated fraction isolated using the following protocol. Transfected cells were cultured for 48 h, treated with 1 μm [Sar1]Ang II for 1 h, and then washed with cold PBS. Cells were scraped and suspended in lysis buffer (25 mm HEPES, pH 7.2, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 130 μm bestatin, 14 μm E-64, 1 μm leupeptin, 0.3 μm aprotinin, 10 mm sodium pyrophosphate, 10 mm sodium fluoride, 1 mm sodium orthovanadate, 10% glycerol) for 30 min. The protease inhibitors in lysis buffer were from Sigma protease inhibitor mixture (P2714). Cell lysates were centrifuged at 13,000 rpm for 10 min at 4 °C to remove cell debris. 10 μg of Anti-HA high affinity antibody was added to the supernatant with 20 μl of Protein G-Sepharose to immunoprecipitate the HA-AT1Rs. After overnight incubation at 4 °C on a rocker platform, the immunoprecipitates were collected by centrifugation at 10,000 rpm for 1 min at 4 °C. Pellets were dissolved in Laemmli's sample buffer, boiled for 5 min at 95 °C, and separated by SDS-polyacrylamide gel electrophoresis with a 10% separation gel. Following electrophoresis, the proteins were transferred to nitrocellulose membranes and then blocked for 1 h at room temperature in 5% nonfat dry milk and 0.1% Tween 20 in PBS (pH 7.4). Incubation with primary antibodies was carried out overnight at 4 °C. Following washes with PBS, incubation with horseradish peroxidase-conjugated secondary antibody was carried out for 1 h at room temperature. The detection was made with enhanced chemiluminescence (GE Healthcare), and the films were scanned for densitometry analysis. All immunoprecipitation experiments were repeated 3–5 times, and representative examples are shown under “Results.”
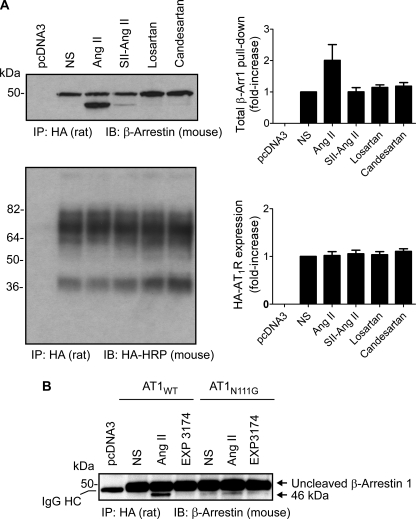
FIGURE 1.
Site-specific proteolysis of AT1R-bound β-arrestin 1 upon Ang II treatment. A, HA-tagged AT1R and β-arrestin 1 were co-expressed in COS-1 cells. Sepharose-immobilized rat high affinity monoclonal antibody to HA (3F10) was used for immunoprecipitation, and mouse monoclonal antibody to β-arrestin (H-9) was used for Western blotting. Ang II treatment increases total β-arrestin 1 coupled to the AT1R 2-fold (2.03 ± 0.48) and generates the cleavage of β-arrestin 1 in the C-terminal region. The amount of HA-AT1R expressed in each sample is similar. -Fold increases of total β-arrestin 1 pull-down were calculated compared with the value of β-arrestin 1 in nonstimulated cells. Data correspond to the mean ± S.D. from five independent experiments. A representative blot is shown. B, β-arrestin proteolysis requires stable interaction with Ang II-activated AT1R. β-Arrestin proteolysis is absent or substantially reduced in phosphorylation-deficient AT1N111G-expressing cells. There is cross-reactivity of sheep anti-mouse IgG secondary antibody with rat anti-HA high affinity antibody; thus, IgG heavy and light chains bands are seen in varying degrees, depending upon x-ray film exposure time in all samples. The IgG heavy chain band (IgG HC) is marked. Experiments were repeated three times. A representative blot is shown. NS, nonstimulated. SII-Ang II, [Sar1,Ile4,Ile8]Ang II. IP, immunoprecipitation. IB, immunoblot.
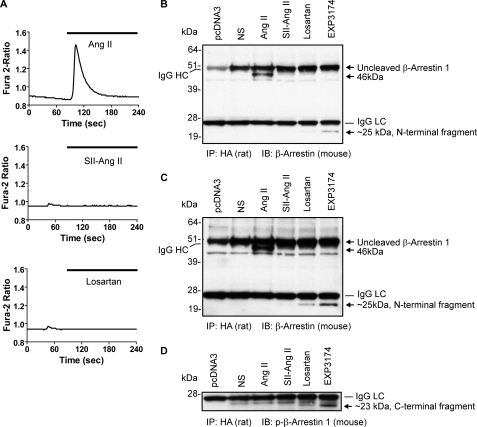
FIGURE 2.
Ang II-induced β-arrestin 1 proteolysis is distinct from losartan and EXP3174-induced proteolysis. A, Ca2+ mobilization in rat aortic smooth muscle (A7r5) cells upon Ang II, [Sar1,Ile4,Ile8]Ang II, and losartan (EXP3174; not shown) treatment was measured, respectively. Values are averages of 6–8 cells in each experiment, and the experiments were repeated three times. B, β-arrestin 1-coupling to the AT1R following different ligand treatment was determined with anti-β-arrestin antibody (H-9) after immunoprecipitation with HA-AT1R with rat anti-HA high affinity antibody (3F10). The IgG heavy (IgG HC) and light chains (IgG LC) are marked. The uncleaved β-arrestin 1 and the 46- and 25-kDa N-terminal fragments of β-arrestin 1 are marked with an arrow, respectively. The β-arrestin 1 band is over and above the IgG heavy chain band in this 1-min exposure of the film. Experiments were repeated three times, and a representative blot is shown. C, a 5-min exposure of the x-ray film shown in B to demonstrate the N-terminal fragment of β-arrestin 1 induced by losartan and EXP3174. D, β-arrestin proteolysis upon losartan and EXP3174 treatment was probed with anti-phospho-β-arrestin 1 (Ser412) antibody. The ∼23-kDa β-arrestin 1 C-terminal fragment is marked with an arrow. NS, nonstimulated. IP, immunoprecipitation. IB, immunoblot.
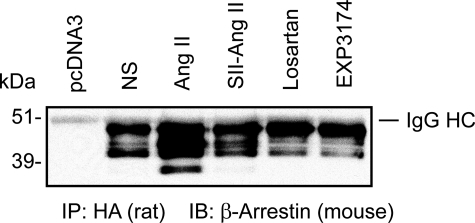
FIGURE 3.
The proteolysis of β-arrestin 2 occurs differently from β-arrestin 1. The proteolysis of transfected β-arrestin 2 in COS-1 cells was probed with mouse anti-β-arrestin antibody (H-9) after immunoprecipitation of HA-AT1R with rat anti-HA high affinity antibody (3F10). Experiments were repeated four times. A representative blot is shown. NS, nonstimulated. SII-Ang II, [Sar1,Ile4,Ile8]Ang II. IP, immunoprecipitation. IB, immunoblot.
Intracellular Ca2+ Measurement—Changes in cytoplasmic Ca2+ were measured using the fluorescent calcium indicator dye Fura-2 (excitation maximum ∼340 and 380 nm; emission maximum 510 nm) (Invitrogen). A7r5 cells growing on coverglasses in a 6-well culture plate were loaded with 2 μm Fura-2/AM in a Ca2+-free balanced salt solution (BSS; 140 mm NaCl, 2.8 mm KCl, 2 mm MgCl2, 0.5 mm EGTA, and 10 mm HEPES, pH 7.2) containing 0.1% Pluronic 127 and 2 mm Ca2+ at 37 °C for 30 min, followed by washing with BSS to remove the extracellular dye. Calcium measurements were done in single cells using an inverted microscope (Zeiss Axiovert 135) connected to a CCD camera (Photon Technology International). The data were collected at 1.5-s intervals and analyzed using Image Master software (Photon Technology International). The release of intracellular Ca2+ in individual cells was measured after exposure to 1 μm [Sar1]Ang II, 1 μm [Sar1,Ile4,Ile8]Ang II, or 1 μm losartan, respectively, in BSS by rapid solution exchange. Results are presented as average changes in the ratio of Fura-2 fluorescence upon excitation at 340 nm and 380 nm. Data from 6–8 cells were collected and plotted using GraphPad Prism software.
Construction of β-Arrestin 1-GFP-Myc-His6 and β-Arrestin 1-Myc-His6 Fusion Plasmids—To retrieve the cleaved C-terminal β-arrestin 1 fragment and determine the cleavage site by mass spectrometry and Edman degradation, β-Arrestin 1-GFP-Myc-His6 was generated by PCR in two steps. First, GFP cDNA was amplified from pEGFP vector (Clontech) and subcloned into pcDNA3.1(-)/Myc-His vector (Invitrogen) using the following primers with Pfu DNA polymerase. The forward primer was 5′-GATATCTGCAGAATTCCAATGGTGAGCAAGGGCGAGGAG-3′ (EcoRI site underlined and the N-terminal part of GFP in boldface type), and the reverse primer was 5′-GTTCGGGCCCAAGCTTCTTGTACAGCTCGTCCATG-3′ (HindIII site underlined and the C-terminal part of GFP in boldface type). Next, bovine β-arrestin 1 cDNA amplified by the following primers was subcloned into pcDNA3.1(-)/GFP-Myc-His vector. The forward primer was 5′-CGGGCCCTCTAGACATGGGC GACAAAGGGACG-3′ (XbaI site underlined and the N-terminal part of β-arrestin 1 in boldface type), and the reverse primer was 5′-CAGCACAGTGGCGGCCGCTCTGTCGTTGAGCCGCGGAG-3′ (NotI site underlined and the C-terminal part of β-arrestin 1 in boldface type). For the construction of β-arrestin 1-Myc-His6 plasmid, bovine β-arrestin 1 cDNA amplified to generate β-arrestin 1-GFP-Myc-His6 was subcloned into XbaI and NotI sites of the pcDNA3.1(-)/Myc-His vector. Accuracy of the fusion constructs in the expression vector was confirmed by DNA sequence analysis.
Mass Spectrometry—COS-1 cells growing in six 10-cm plates were prepared for transfection. Each 5 μg of plasmid DNA for the AT1WT receptor and β-arrestin 1-GFP-Myc-His6 fusion construct were transfected into COS-1 cells with 60 μl of Lipofectamine 2000. 48 h post-transfection, cells were treated with 1 μm [Sar1]Ang II for 1 h. Ang II-treated cells were washed with cold PBS and scraped, and cell lysates were prepared as described above. Total proteins were resolved by preparative SDS-PAGE. The cleaved (∼46 kDa) β-arrestin 1 band was identified by immunoblotting with anti-β-arrestin antibody (H-9). The gel region was marked and provided to the mass spectrometry core. For the protein digestion, the bands were cut from the gel as closely as possible to minimize excess polyacrylamide; washed/destained in 50% ethanol, 5% acetic acid; and reduced and alkylated with DTT and iodoacetamide. The gel pieces were then dehydrated in acetonitrile and dried in a SpeedVac. In-gel proteolytic digestion using trypsin was accomplished by adding 5 μl of trypsin (20 ng/μl) in 50 mm ammonium bicarbonate. The sample was digested overnight at room temperature. The peptides that were formed were extracted from the polyacrylamide in two aliquots of 30 μl of 50% acetonitrile-5% formic acid. These extracts were combined and evaporated to <10 μl in a SpeedVac and then resuspended in 1% acetic acid to make up a final volume of ∼30 μl for liquid chromatographymass spectrometry analysis (Finnigan LTQ linear ion trap mass spectrometer system) equipped with a microelectrospray ion source. The samples were also analyzed on a Finnigan LCQ-Deca ion trap mass spectrometer system with a Protana microelectrospray ion source. The data were analyzed by using all CID spectra collected in the experiment to search the NCBI nonredundant data base with the search program Mascot. All matching spectra were verified in addition by manual interpretation. The interpretation process was also aided by additional searches using the Sequest program. Blast searches were conducted as needed. The data shown under “Results” are a summary of analyses of five independent preparations of the 46 kDa band.
N-terminal Sequencing of Cleaved β-Arrestin 1—COS-1 cells growing in six 10-cm plates were prepared for transfection. Each 5 μg of plasmid DNA for the AT1WT receptor and β-arrestin 1-GFP-Myc-His6 fusion construct were transfected into COS-1 cells with 60 μl of Lipofectamine 2000. 48 h post-transfection, cells were treated with 1 μm [Sar1]Ang II for 1 h and then washed with cold PBS. Cells were scraped and suspended in lysis buffer for 30 min. Cell lysates were centrifuged at 13,000 rpm for 10 min at 4 °C. Cleaved β-arrestin 1 C-terminal fragment fused to GFP-Myc-His6 protein was purified by affinity chromatography using a 1-ml HisTrap column. The column was washed with 10 ml of 20 mm Tris, 500 mm NaCl, 15 mm imidazole, pH 8.0, and was eluted in 5 ml of the same buffer containing 250 mm imidazole. Purified protein was dialyzed at 4 °C against PBS and concentrated using a Vivascience concentrator (10,000 molecular weight cut-off). Laemmli's sample buffer was added to the sample, boiled for 5 min at 95 °C, and separated by SDS-PAGE with a 10% separation gel. A small amount of samples were used for Western blotting, probed with anti-β-arrestin antibody (1:1000) and anti-GFP antibody (1:3000) to determine the band for the cleaved construct. The cleaved β-arrestin band was identified, cut, and analyzed using Edman degradation.
RESULTS
Interaction of β-Arrestin 1 Fragment with Ang II-activated AT1R—To test β-arrestin coupling to the AT1R, the N-terminal HA-tagged AT1R and β-arrestin 1 were co-expressed in COS-1 cells. Upon treatment with Ang II, [Sar1,Ile4,Ile8]Ang II, losartan, and candesartan, respectively, the HA-AT1R was immunoprecipitated from solubilized lysate with rat anti-HA high affinity antibody (3F10) and protein G-Sepharose. The immunoprecipitates were resolved by SDS-PAGE followed by Western blotting with mouse anti-β-arrestin antibody (H-9). A significant amount of full-length β-arrestin 1 was co-immunoprecipitated with HA-AT1R in all samples, and Ang II treatment increased the total β-arrestin pulled down. In the Ang II-treated sample, the recruitment of the 46-kDa β-arrestin 1 fragment in addition to the full-length β-arrestin 1 to the AT1R was evident (Fig. 1A). The amount of total β-arrestin in Ang II-AT1R sample was 2-fold (2.03 ± 0.48-fold) compared with β-arrestin 1 bound to the AT1R in nonstimulated cells. Because the epitope of anti-β-arrestin antibody is localized in the N-terminal region, the cleavage site of β-arrestin 1 is considered to be in the C-terminal region.
A pull-down experiment from sample treated with the biased AT1R agonist, [Sar1,Ile4,Ile8]Ang II, which recruits β-arrestin upon binding to AT1R and activates ERK1/2 via a Gq-independent pathway (12), indicated small amount of 46-kDa fragment recruitment, whereas the antagonist-bound HA-AT1R did not appear to engage the 46 kDa band in COS-1 cells. To ensure that the increased β-arrestin 1 recruitment upon Ang II treatment is not because of a higher AT1R expression from variable transfection, nitrocellulose membrane was stripped and probed again with horseradish peroxidase-conjugated anti-HA antibody (12CA5). The result shows similar levels of unglycosylated and glycosylated AT1R in each lane (Fig. 1A, lower panel), as shown by Thomas et al. (13).
In the next experiment, we co-expressed the constitutively active AT1R mutant AT1N111G with β-arrestin 1 in COS-1 cells. The AT1N111G receptor was shown to be defective in coupling to β-arrestin and instead preferentially couples to G protein, Gq (14). The AT1N111G receptor was reported to increase IP3 level (∼42%) in the absence of Ang II and induce the same maximal IP3 production as the wild-type receptor upon Ang II treatment (11). A similar level of IP3 induction from the AT1N111G receptor in the absence of Ang II stimulation was measured in the present experiments (data not shown). Under these conditions, we found that the 46-kDa β-arrestin 1 band is absent (Fig. 1B). These data suggest that the ligand-induced proteolysis of β-arrestin 1 requires stable interaction with the receptor-ligand complex in a specific conformation. In addition, this experiment also suggests that Gq-phospholipase C-mediated intracellular Ca2+ flux is not the trigger for the proteolysis. To further assess the role of Ca2+ release from the endoplasmic reticulum in β-arrestin 1 proteolysis, we incubated COS-1 cells with the Ca2+-chelator BAPTA for 90 min before Ang II treatment. BAPTA did not inhibit Ang II-induced β-arrestin 1 proteolysis, supporting the above observation (data not shown).
AT1R Agonist and Inverse Agonist Induce Cleavage of β-Arrestin 1 at Distinct Sites—In order to evaluate the role of AT1R in β-arrestin 1 cleavage in a different cell line, β-arrestin 1 was co-expressed with the HA-AT1R in rat aortic vascular smooth muscle (A7r5) cells. Following treatment of transfected A7r5 smooth muscle cells with different AT1R-selective ligands, receptor activity was measured by Ca2+ flux, and the state of β-arrestin was examined (Fig. 2). Increasing the level of AT1R expression by itself had no effect, but the activation of receptor by the full agonist Ang II induced generation of the 46 kDa band. The biased agonist [Sar1,Ile4,Ile8]Ang II did not induce intracellular Ca2+ release and also did not generate the 46 kDa band. To our surprise, exposure to AT1R-selective inhibitor EXP3174 generated a ∼25-kDa N-terminal β-arrestin 1 (Fig. 2B). Longer exposure (5 min) of the x-ray film showed that the ∼25-kDa fragment N-terminal β-arrestin 1 was present in a losartan-treated sample (Fig. 2C). Note that the short exposure (1 min) of the film distinguished the IgG heavy chain band from the uncleaved β-arrestin 1 band in Fig. 2B. Thus the different intensity of the ∼25 kDa band seen may be reflecting a difference in the extent of cleavage induced by losartan and EXP3174. It is known that losartan is a weaker inverse agonist than EXP3174. The β-arrestin antibody clone (H-9) used in our experiments showed experiment-to-experiment variation in detecting losartan and EXP3174-induced β-arrestin 1 fragment. We speculated that proteolysis altered the affinity of H-9 antibody for the N-terminal fragment, or its stability in cells varied. To independently confirm the generation of the β-arrestin 1 C-terminal fragment upon losartan and EXP3174 treatment, immunoprecipitates resolved by SDS-PAGE were transferred onto nitrocellulose membrane and probed with anti-phospho-β-arrestin 1 antibody directed against Ser412 in the C-terminal region of β-arrestin 1. This antibody recognized the cleaved ∼23-kDa C-terminal fragment of β-arrestin 1 only in losartan and EXP3174-treated samples (Fig. 2D). There is no increase of β-arrestin 1 recruitment by losartan and EXP3174 as shown in Fig. 1. Our data suggest that these drugs bind to β-arrestin-engaged AT1Rs under basal conditions. Because losartan and EXP3174 are inverse agonists, upon binding they induce a conformational change (different from Ang II-induced conformation) in the receptor to inhibit basal activity. We speculate that the conformational change in the AT1R is transmitted in turn to the β-arrestin molecule bound to the receptor, resulting in exposure of a protease site in the middle of β-arrestin 1 distinct from the Ang II-induced site. Losartan and its active metabolite EXP3174 are commonly used antihypertensive drugs and reduce blood pressure by inhibiting AT1R-mediated IP3 and Ca2+ signaling responsible for vasoconstriction in smooth muscle cells (15).
β-Arrestin 2 Cleavage Is Different from β-Arrestin 1—Next, we asked the question whether β-arrestin 2 is cleaved upon AT1R engagement. Human β-arrestin 2 was co-expressed with the HA-AT1R. The β-arrestin 2 cleavage appeared to be more complex. In addition to the predominant 46 kDa band, a 36 kDa molecular mass band was observed after Ang II treatment (Fig. 3). Other ligands, including [Sar1,Ile4, Ile8]Ang II, losartan, and EXP3174, did not affect β-arrestin 2. This finding supports the idea that β-arrestin 1 and 2 are regulated differently and may play distinct roles in Ang II-AT1R signal transduction.
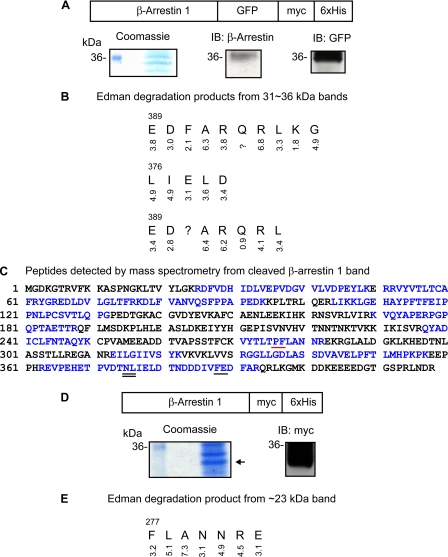
β-Arrestin 1 Is Cleaved after Phe388 by Ang II Treatment—The cleavage site is anticipated to be in the C-terminal region of β-arrestin 1, because the epitope of anti-β-arrestin antibody is localized in the N-terminal region. In multiple attempts, we were unable to recover the C-terminal 4-kDa fragment in cell extract to determine Ang II-induced β-arrestin 1 cleavage site. To overcome this problem, we generated a C-terminal fusion construct (see “Experimental Procedures”) of β-arrestin 1 and GFP. Myc and His6 tag were introduced next to the C-terminal end of GFP for detection of the fusion construct by anti-Myc antibody and affinity column purification (see Fig. 4A). The fusion construct was co-expressed in COS-1 cells with the HA-AT1R. Following AT1R stimulation by Ang II, the cleaved C-terminal fragment of β-arrestin 1 fusion protein was purified from cell lysate using a 1-ml HisTrap column and resolved on a preparative SDS-polyacrylamide gel. Proteins were transferred onto polyvinylidene difluoride membrane. In parallel experiment, Western blotting with anti-β-arrestin antibody (H-9) and anti-GFP antibody confirmed the presence of the desired band in the selected 31–36 kDa region on preparative SDS gel (see Coomassie-stained gel in Fig. 4A). The 4-mm region of the polyvinylidene difluoride membrane was cut into 1-mm strips and subjected to Edman sequencing, and the sequence was deduced based on the yield of PTH-derivatives recovered at each step. The Edman sequencing results are shown in Fig. 4B. Two unrelated sequences obtained (data not shown) matched perfectly the N-terminal sequences of ribosomal protein S4 and ATP/ADP translocator. Three β-arrestin 1 peptide sequences were obtained, all starting within the region spanning from residue 376 to 388. Since the PTH-derivative yield of products at each step of Edman degradation was high, the cleavage site was assigned for Phe388 (underlined in Fig. 4C) and Asn375 (double underlined in Fig. 4C), respectively. To independently confirm the Edman sequencing observations, we isolated the N-terminal 46-kDa β-arrestin 1 from preparative gel, subjected the band to digestion by trypsin, and analyzed the tryptic fragments by mass spectrometry (see “Experimental Procedures”). Tryptic peptides recovered from the 46-kDa fragment of β-arrestin 1 are shown in blue in Fig. 4C. The 363–393 tryptic fragment was recovered in one of five experiments, and none of the peptides beyond the Phe388 region were found in all other experiments. This observation supports the cleavage site determined by N-terminal sequencing. Thus, Phe388 is most likely the preferred cleavage site of β-arrestin 1 upon Ang II treatment. Finding this cleavage site is consistent with the work of Azarian et al. (9) on the in vitro proteolysis of visual arrestin by calpain, where the visual arrestin was cleaved after Phe located in the corresponding position. To elucidate the role of calpain in the cleavage of β-arrestin 1, we incubated the cells with a cell-permeable calpain inhibitor, calpeptin, for 30 min before AT1R stimulation, but Ang II-activated β-arrestin 1 cleavage in COS-1 cells was not prevented (data not shown).
FIGURE 4.
Determination of Ang II and EXP3174-induced β-arrestin 1 cleavage site. A, schematic representation of the β-arrestin 1-GFP-Myc-His6 construct designed for retrieval of the cleaved C-terminal peptide. A Coomassie-stained gel shows ∼31–36 kDa bands obtained after 1-ml HisTrap column purification (left). A small amount of samples were transferred onto nitrocellulose membrane after SDS-PAGE to identify the cleaved β-arrestin 1-GFP-Myc-His6 band. The membrane was probed with anti-β-arrestin antibody (H-9) (middle) and anti-GFP antibody after stripping (right). B, Edman degradation sequence analysis results obtained from 1-mm strips from the 31–36 kDa region. The PTH-derivative yield from each degradation step is shown below the residue assigned. C, mass spectrometric analysis of trypsin-digested 46-kDa N-terminal fragment of β-arrestin 1. The regions indicted in blue represent the β-arrestin 1 sequence recovered in tryptic digestion. Note that tryptic peptides beyond Phe388 were not found. The data shown under “Results” are a summary of analyses of five independent preparations of the 46 kDa band. The major cleavage site Phe388-Gln is underlined. The minor cleavage site Asn375-Leu is double underlined. D, schematic representation of β-arrestin-Myc-His6 constructs to determine the β-arrestin 1 cleavage site by EXP3174. A Coomassie-stained gel shows the band obtained after HisTrap column purification. Samples transferred onto nitrocellulose membrane were probed with anti-Myc antibody. E, Edman degradation sequence results obtained from 1-mm strips from the ∼23 kDa band. The PTH-derivative yield from each degradation step is shown below the residue assigned. The cleavage site Pro276-Phe is underlined in red. IB, immunoblot.
β-Arrestin 1 Is Cleaved after Pro276 by EXP3174 Treatment—To determine the EXP3174-induced β-arrestin 1 cleavage site, another fusion construct that has Myc-His6 next to the C terminus of β-arrestin 1 was generated (Fig. 4D) and co-expressed with the HA-AT1R in COS-1 cells. After EXP3174 treatment, cell lysates were purified through a HisTrap column and resolved by SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride membrane, and the band in the ∼23 kDa region was identified by Western blotting with anti-Myc antibody (Fig. 4D) and subjected to Edman degradation. The yields of PTH-derivatives are shown in Fig. 4E. The N-terminal sequencing result shows that β-arrestin 1 is cleaved after Pro276 upon EXP3174 treatment.
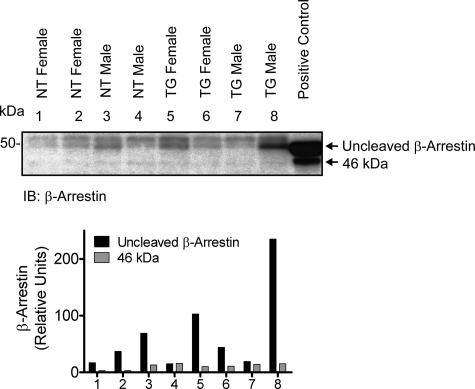
Steady-state Accumulation of β-Arrestin Fragment in Vivo—Studies on various AT1R ligand-induced β-arrestin 1 cleavages carried out in the present study were from cultured cell lines, such as COS-1 and A7r5 cells. Previous studies on the AT1R signaling have indicated that β-arrestin mediates ligand-specific and G protein-independent signals (2), and these concepts also have mainly come from studies carried out in cultured cell models. We examined β-arrestin steady state in the mouse model overexpressing AT1R in the cardiomyocytes (16). Detergent extracts from heart tissue obtained from transgenic mice and nontransgenic littermates were blotted with anti-β-arrestin antibody. The β-arrestin appeared to be cleaved in both nontransgenic and transgenic mouse heart tissue samples (Fig. 5). The cleaved β-arrestin did not increase in proportion to the 100-fold increase of AT1R in the transgenic heart tissue. This is probably because the cleavage is proportional to the circulating concentration of Ang II both in the transgenic mice and nontransgenic littermates, which is not altered, whereas the increased AT1Rs in the transgenic mice perhaps contributed to the pool of spare receptors (17). The anti-β-arrestin antibody used in this experiment recognized both β-arrestin 1 and 2; which subtype of β-arrestin 1 is actually cleaved is not certain. A similar pattern of cleavage of β-arrestin was seen in H295R adrenocarcinoma cells upon Ang II activation and was blocked by the AT1R antagonist, losartan (data not shown). Next, we chronically treated mice with losartan for 14 days to block the formation of the potential 46 kDa band and demonstrate the formation of the 25 kDa band in vivo. Contrary to our expectations, the full-length β-arrestin band was almost absent, and both 46 kDa and 25 kDa bands were not detected in the drug-treated mice (data not shown). The role of β-arrestin proteolysis upon Ang II and AT1R blockers in cellular physiology in vivo needs to be further elucidated.
FIGURE 5.
Proteolysis of β-arrestin in vivo from mouse heart tissue. Equal amounts of total protein (100 μg) extracted from four nontransgenic (NT) and four AT1R-transgenic mouse (TG) heart samples were separated by SDS-PAGE. Immunoprecipitated sample from co-expression of the wild-type HA-AT1R and β-arrestin 1 was used as a positive control. The membrane was probed with anti-β-arrestin antibody. The arrow (46 kDa) indicates cleaved β-arrestin. IB, immunoblot.
DISCUSSION
The most important finding in this study is that a fraction of β-arrestin recruited to AT1R is cleaved, and the site-specificity of the cleavage depends on the particular ligand-receptor-β-arrestin complex. Thus, β-arrestin 1 and β-arrestin 2 are cleaved with distinct patterns when in complex with Ang II-AT1R, and even more interestingly, the β-arrestin 1 cleavage differed when in complex with EXP3174-AT1R. While analyzing β-arrestin steady-state in mouse hearts, cultured smooth muscle, and adrenal cell lines, we first found the ∼46-kDa β-arrestin isoform that could result from either proteolysis or alternative splicing. The in vivo proteolysis of β-arrestin data from mouse heart tissue suggests that normal circulating concentrations of Ang II are enough to induce cleavage of β-arrestin, and overexpression of receptor alone did not enhance it. Co-expression of AT1R and β-arrestin 1 established that the appearance of this band requires acute activation of AT1R by Ang II, which suggests a post-translational mechanism and rules out alternative mRNA splicing. We speculated that proteolysis generated the 46-kDa isoform, and the location of the epitope for the β-arrestin 1 antibody used in our study suggested that cleavage occurs in the C-terminal region of β-arrestin 1. A combination of mass spectrometry and Edman sequencing determined two sites of cleavage, the major site after residue Phe388 and second after Asn375 in bovine β-arrestin 1. Azarian et al. (9) found that a short form (p46) of visual arrestin generated by light exposure of bovine rod outer segments is identical to that generated by in vitro proteolysis of visual arrestin by calpain. The short form of visual arrestin (p44) resulting from alternative mRNA splicing (7, 8) differs in biochemical characteristics compared with the full-length visual arrestin. For instance, the full-length visual arrestin binds only to phosphorylated rhodopsin, but p44 binds to both nonphosphorylated and phosphorylated rhodopsin with similar affinity (7). Surprisingly, the major site of β-arrestin 1 cleavage we found corresponds to the site of calpain cleavage of visual arrestin at Phe377; however, the proteolysis of β-arrestin 1 in our experiment was not prevented by the calpain inhibitor, calpeptin, and other routinely used protease inhibitors. We conclude that the short form of β-arrestin 1 described here is the product of unknown protease action rather than alternative mRNA splicing.
Much evidence suggested that the ligand-induced conformation of the AT1R regulates the cleavage process involved in the formation of the 46-kDa β-arrestin 1 isoform. The most potent inducer of cleavage was the native hormone Ang II, and peptide analogs of Ang II and AT1R-selective antagonists were ineffective in producing this isoform. To our surprise, the [Sar1,Ile4,Ile8]Ang II, which induces AT1R phosphorylation (13, 18) and recruitment of β-arrestin to AT1R, leading to ERK1/2 activation (12), did not promote β-arrestin cleavage. This suggests that recruitment of β-arrestin to ligand-activated receptor is not sufficient for cleavage. Since this analog does not induce Ca2+ release, we tested the constitutively active mutant AT1R, which induces Ca2+ release in basal and Ang II-activated conditions. Lack of β-arrestin cleavage in this mutant suggests that G protein-dependent Ca2+ release is not obligatory for cleavage. We have shown previously that AT1N111G mutant receptor is defective in forming a stable complex with β-arrestin (14). Our co-immunoprecipitation data are consistent with initial binding of full-length β-arrestin with ligand-activated receptor and subsequent generation of the cleaved short form in the complex. Our co-immunoprecipitation data are consistent with constitutive binding of full-length β-arrestin 1 to the AT1R. Previously, Qian et al. (19) showed that FLAG-tagged β-arrestin 1 is associated with the HA-AT1R in an Ang II-dependent manner by cross-linking studies in Chinese hamster ovary cells. We found that the total β-arrestin 1 pull-down was increased 2-fold, including the 46-kDa form, upon agonist binding. The level of β-arrestin 1 expression may account for constitutive association of the receptor with β-arrestin 1 in our experiments. Most likely, the constitutive association of β-arrestins with the unphosphorylated receptor in the high expression system facilitated detection of the site-specific proteolysis event, which still could occur at endogenous levels. The conformation of β-arrestin required for Ang II-induced proteolysis needs to be further elucidated. The short form remains associated with the receptor, and the lack of the 46-kDa β-arrestin 1 isoform in the pull-down assays from samples treated with different AT1R ligands suggests that formation of this isoform in the cytoplasm is less likely. The signaling pathways leading to this proteolysis are unknown, but our data on the constitutively active AT1N111G mutant receptor and Ca2+-chelator BAPTA indicate that the proteolysis is independent of the IP3- and Ca2+-mediated mechanism. Together, this analysis leads us to propose that a particular conformation among the ensemble of Ang II-activated conformations is necessary for inducing the cleavage of AT1R-engaged β-arrestin. This particular conformation is not attained when [Sar1,Ile4,Ile8]Ang II binds to AT1R or when Ang II binds to the AT1N111G receptor.
Induced conformational change in the receptor-bound β-arrestin was described previously (20). The activated receptor induces the β-arrestin C-terminal region to interact with clathrin (residues 375–380) and AP2 (residues 393–395) (20). The β-arrestin 1 C-terminal tail is constitutively phosphorylated at Ser412 and, once recruited to the plasma membrane, it is dephosphorylated by protein phosphatase 2A for interaction with clathrin (5, 21) and AP2 for internalization of GPCRs (22, 23). The C-terminal region is normally buried inside the β-arrestin structure, and recruitment by the activated GPCR induces an open conformation enabling multiple interactions (20). The location of the cleavage sites determined in this study in the region critical for interaction with protein phosphatase 2A, clathrin, and AP2 and the observation that the C-terminal fragment of β-arrestin 1-GFP generated is still phosphorylated at Ser412 (data not shown) imply that a specific conformation of ligand-receptor-β arrestin complex recruits a protease, leading to cleavage of the C-terminal regulatory region of β-arrestin 1, rendering the complex unsuitable for clathrin-mediated endocytosis. The β-arrestin-GFP fusion, usually attached to the C terminus of β-arrestin, has been widely used to monitor the translocation of β-arrestin from the cytoplasm to the activated GPCRs upon agonist stimulation (24, 25) and used as a tool to screen for ligands for orphan GPCRs and other potential drugs that regulate GPCRs (26). Using the β-arrestin-GFP construct might only show the translocation, endocytosis, and redistribution of uncleaved β-arrestin-GFP proteins and fate of cleaved β-arrestin-GFP fusion protein we report here, although a small fraction is not known. A more sophisticated tool that can monitor the trafficking of the cleaved 46-kDa β-arrestin fragment needs to be developed.
Therefore, once β-arrestin is cleaved, the β-arrestin-mediated internalization of AT1R may be attenuated. However, a number of signaling molecules, such as mitogen-activated protein kinases, JNK3 (c-Jun N-terminal kinase 3), Src, guanidine nucleotide exchange factors, and small G proteins (27), could still interact with the receptor-bound β-arrestin N-terminal domain through a conformation-driven process. β-Arrestins interact with these molecules only in the receptor-bound state (28, 29) while on the plasma membrane, much earlier than what was previously thought to occur at endosomes (30). This may account for both the regulation of late onset AT1R signals through β-arrestins and the slower internalization process. Alternatively, proteolytic cleavage may be a mechanism to channel the complex toward clathrin-independent (4) and other unconventional internalization pathways (31). The fate of cleaved β-arrestin C-terminal fragment needs to be elucidated for a possible role in signal transduction. The cellular effects of β-arrestin cleavages are not known but will require development of some critical tools, which is beyond the scope of current study.
Cleavage of β-arrestin 1 at Pro276 in the presence of inverse agonists, losartan and its active metabolite EXP3174, is intriguing and may be indicative of a different conformational change in β-arrestin 1 and/or recruitment of a protease with different specificity. In conventional signaling studies, inverse agonists inhibit basal and agonist-dependent AT1R signals, but in this study, both drugs induced proteolysis of β-arrestin 1 at a different site. Whether the net effect of this cleavage is the inhibition of β-arrestin N-terminal domain signaling is not clear at this time. In recent years, evidence for distinct signaling states of GPCRs generated by agonist, antagonist, or inverse agonist that could yield different functional profiles has been described (32). In principle, certain ligands characterized as agonist or antagonist could exert the opposite effect on another signaling pathway. For example, β2-adrenergic receptor inverse agonists for Gs-coupled adenylyl cyclase activity, such as ICI118551 and propranolol, were found to provoke partial agonism for ERK1/2 activation through the β-arrestin-mediated pathway (33). Another β2-adrenergic receptor inverse agonist, carvedilol, although uncoupled from Gs, upon its binding to the β2-adrenergic receptor stimulated activation of ERK1/2 through β-arrestin (34). Thus, the AT1R-antagonist induction of β-arrestin 1 cleavage could be a signal in itself, but the functional consequences are unknown at this time. Both of these AT1R blockers are used in cardioprotection, antihypertensive, and anti-diabetes therapies, but their molecular mechanism is largely unknown (35, 36). Our finding of losartan-induced β-arrestin proteolysis provides a novel insight into the molecular mechanism of losartan action through the AT1R that could be physiologically important.
The proteolysis of β-arrestin following the AT1R activation presents a new paradigm of GPCR signal transduction. The role of β-arrestins in terminating G protein coupling and receptor endocytosis following agonist stimulation of GPCRs is well characterized. However, subcellular events that enable receptor-engaged β-arrestins to guide the ligand-receptor complexes toward specific and diverse functional paths are poorly characterized. Discovery of β-arrestin cleavage reported here suggests the existence of an exciting hitherto unknown mode of regulation of Ang II-AT1R complex in vivo and perhaps other related ligand-GPCR systems. Our finding of antagonist-induced proteolysis provides a novel insight into the molecular mechanism of action of GPCR antagonists, which are highly successful therapeutic agents used in clinical practice today.
Acknowledgments
We thank members of the Karnik laboratory for helpful comments. We thank Michael Kinter for mass spectrometric analysis of samples.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HL57470 (to S. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GPCR, G protein-coupled receptor; Ang II, angiotensin II; BAPTA, 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; ERK, extracellular signal-regulated kinase; IP3, inositol 1,4,5-trisphosphate; GFP, green fluorescent protein; HA, hemagglutinin; PBS, phosphate-buffered saline; PTH, phenylthiohydantoin.
References
- 1.Ji, T. H., Grossmann, M., and Ji, I. (1998) J. Biol. Chem. 273 17299-17302 [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz, R. J., and Shenoy, S. K. (2005) Science 308 512-517 [DOI] [PubMed] [Google Scholar]
- 3.Hunyady, L., and Catt, K. J. (2006) Mol. Endocrinol. 20 953-970 [DOI] [PubMed] [Google Scholar]
- 4.Zhang, J., Ferguson, S. S., Barak, L. S., Menard, L., and Caron, M. G. (1996) J. Biol. Chem. 271 18302-18305 [DOI] [PubMed] [Google Scholar]
- 5.Lin, F. T., Krueger, K. M., Kendall, H. E., Daaka, Y., Fredericks, Z. L., Pitcher, J. A., and Lefkowitz, R. J. (1997) J. Biol. Chem. 272 31051-31057 [DOI] [PubMed] [Google Scholar]
- 6.Shenoy, S. K., McDonald, P. H., Kohout, T. A., and Lefkowitz, R. J. (2001) Science 294 1307-1313 [DOI] [PubMed] [Google Scholar]
- 7.Palczewski, K., Buczylko, J., Ohguro, H., Annan, R. S., Carr, S. A., Crabb, J. W., Kaplan, M. W., Johnson, R. S., and Walsh, K. A. (1994) Protein Sci. 3 314-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith, W. C., Milam, A. H., Dugger, D., Arendt, A., Hargrave, P. A., and Palczewski, K. (1994) J. Biol. Chem. 269 15407-15410 [PubMed] [Google Scholar]
- 9.Azarian, S. M., King, A. J., Hallett, M. A., and Williams, D. S. (1995) J. Biol. Chem. 270 24375-24384 [DOI] [PubMed] [Google Scholar]
- 10.Sterne-Marr, R., Gurevich, V. V., Goldsmith, P., Bodine, R. C., Sanders, C., Donoso, L. A., and Benovic, J. L. (1993) J. Biol. Chem. 268 15640-15648 [PubMed] [Google Scholar]
- 11.Noda, K., Feng, Y. H., Liu, X. P., Saad, Y., Husain, A., and Karnik, S. S. (1996) Biochemistry 35 16435-16442 [DOI] [PubMed] [Google Scholar]
- 12.Wei, H., Ahn, S., Shenoy, S. K., Karnik, S. S., Hunyady, L., Luttrell, L. M., and Lefkowitz, R. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10782-10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas, W. G., Qian, H., Chang, C. S., and Karnik, S. (2000) J. Biol. Chem. 275 2893-2900 [DOI] [PubMed] [Google Scholar]
- 14.Lee, C., Hwang, S. A., Jang, S. H., Chung, H. S., Bhat, M. B., and Karnik, S. S. (2007) FEBS Lett. 581 2517-2522 [DOI] [PubMed] [Google Scholar]
- 15.Timmermans, P. B., Carini, D. J., Chiu, A. T., Duncia, J. V., Price, W. A., Jr., Wells, G. J., Wong, P. C., Wexler, R. R., and Johnson, A. L. (1991) Hypertension 18 136-142 [DOI] [PubMed] [Google Scholar]
- 16.Paradis, P., Dali-Youcef, N., Paradis, F. W., Thibault, G., and Nemer, M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 931-936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendelson, C., Dufau, M., and Catt, K. (1975) J. Biol. Chem. 250 8818-8823 [PubMed] [Google Scholar]
- 18.Holloway, A. C., Qian, H., Pipolo, L., Ziogas, J., Miura, S., Karnik, S., Southwell, B. R., Lew, M. J., and Thomas, W. G. (2002) Mol. Pharmacol. 61 768-777 [DOI] [PubMed] [Google Scholar]
- 19.Qian, H., Pipolo, L., and Thomas, W. G. (2001) Mol. Endocrinol. 15 1706-1719 [DOI] [PubMed] [Google Scholar]
- 20.Han, M., Gurevich, V. V., Vishnivetskiy, S. A., Sigler, P. B., and Schubert, C. (2001) Structure (Camb.) 9 869-880 [DOI] [PubMed] [Google Scholar]
- 21.Hupfeld, C. J., Resnik, J. L., Ugi, S., and Olefsky, J. M. (2005) J. Biol. Chem. 280 1016-1023 [DOI] [PubMed] [Google Scholar]
- 22.Goodman, O. B., Jr., Krupnick, J. G., Gurevich, V. V., Benovic, J. L., and Keen, J. H. (1997) J. Biol. Chem. 272 15017-15022 [DOI] [PubMed] [Google Scholar]
- 23.Laporte, S. A., Oakley, R. H., Holt, J. A., Barak, L. S., and Caron, M. G. (2000) J. Biol. Chem. 275 23120-23126 [DOI] [PubMed] [Google Scholar]
- 24.Barak, L. S., Ferguson, S. S., Zhang, J., and Caron, M. G. (1997) J. Biol. Chem. 272 27497-27500 [DOI] [PubMed] [Google Scholar]
- 25.Groarke, D. A., Wilson, S., Krasel, C., and Milligan, G. (1999) J. Biol. Chem. 274 23263-23269 [DOI] [PubMed] [Google Scholar]
- 26.Hudson, C. C., Oakley, R. H., Sjaastad, M. D., and Loomis, C. R. (2006) Methods Enzymol. 414 63-78 [DOI] [PubMed] [Google Scholar]
- 27.Ahn, S., Wei, H., Garrison, T. R., and Lefkowitz, R. J. (2004) J. Biol. Chem. 279 7807-7811 [DOI] [PubMed] [Google Scholar]
- 28.McDonald, P. H., Chow, C. W., Miller, W. E., Laporte, S. A., Field, M. E., Lin, F. T., Davis, R. J., and Lefkowitz, R. J. (2000) Science 290 1574-1577 [DOI] [PubMed] [Google Scholar]
- 29.Luttrell, L. M., Ferguson, S. S., Daaka, Y., Miller, W. E., Maudsley, S., Della Rocca, G. J., Lin, F., Kawakatsu, H., Owada, K., Luttrell, D. K., Caron, M. G., and Lefkowitz, R. J. (1999) Science 283 655-661 [DOI] [PubMed] [Google Scholar]
- 30.Luttrell, L. M., Roudabush, F. L., Choy, E. W., Miller, W. E., Field, M. E., Pierce, K. L., and Lefkowitz, R. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2449-2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng, Y. H., Ding, Y., Ren, S., Zhou, L., Xu, C., and Karnik, S. S. (2005) Hypertension 46 419-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez, D. M., and Karnik, S. S. (2005) Pharmacol. Rev. 57 147-161 [DOI] [PubMed] [Google Scholar]
- 33.Azzi, M., Charest, P. G., Angers, S., Rousseau, G., Kohout, T., Bouvier, M., and Pineyro, G. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11406-11411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wisler, J. W., DeWire, S. M., Whalen, E. J., Violin, J. D., Drake, M. T., Ahn, S., Shenoy, S. K., and Lefkowitz, R. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 16657-16662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diez, J. (2006) Clin. Ther. 28 832-848 [DOI] [PubMed] [Google Scholar]
- 36.Reid, J. L. (2005) J. Renin Angiotensin Aldosterone Syst. 6 15-24 [DOI] [PubMed] [Google Scholar]