Abstract
The hyaluronic acid (HA) receptor for endocytosis (HARE) is the primary scavenger receptor for HA and chondroitin sulfates in mammals. The two human isoforms of HARE (full-length 315-kDa and a 190-kDa proteolytic cleavage product), which are type I single-pass membrane proteins, are highly expressed in sinusoidal endothelial cells of lymph nodes, liver, and spleen. Their identical HARE cytoplasmic domains contain four candidate AP-2/clathrin-mediated endocytic signaling motifs as follows: YSYFRI2485, FQHF2495, NPLY2519, and DPF2534 (315-HARE numbering). Stably transfected cells expressing 190-HARE(ΔYSYFRI), 190-HARE(ΔFQHF), or 190-HARE(ΔNPLY) (lacking Motifs 1, 2, or 3) had decreased 125I-HA endocytosis rates of ∼49, ∼39, and ∼56%, respectively (relative to wild type). In contrast, 190-HARE(ΔDPF) cells (lacking Motif 4) showed no change in HA endocytic rate. Deletions of motifs 1 and 2 or of 1, 2, and 4 decreased the rate of HA endocytosis by only ∼41%. Endocytosis was ∼95% decreased in mutants lacking all four motifs. Cells expressing a 190-HARE(Y2519A) mutant of the NPLY motif retained 85–90% of wild type endocytosis, whereas this mutation in the triple motif deletant decreased endocytosis to ∼7% of wild type. Tyr in NPLY2519 is thus important for endocytosis. All HARE mutants showed similar HA binding and degradation of the internalized HA, indicating that altering endocytic motifs did not affect ectodomain binding of HA or targeting of internalized HA to lysosomes. We conclude that, although NPLY may be the most important motif, it functions together with two other endocytic motifs; thus three signal sequences (YSYFRI, FQHF, and NPLY) provide redundancy to mediate coated pit targeting and endocytosis of HARE.
The GAGs,2 which are primarily located at the cell surface or in the ECM, constitute a polysaccharide family that includes the chondroitin sulfates, heparin, heparan sulfate, keratan sulfate, dermatan sulfate, and HA. HA is found in essentially all vertebrate tissues, is often a major ECM constituent (1), and is abundant in young skin, synovial fluid, vitreous humor of the eye, and other tissues (e.g. umbilical cord). The size range of HA (up to ∼107 Da) is much greater than other GAGs (<105 Da). Unlike other cell surface receptors for HA, such as CD44 and CD168, HARE mediates the rapid endocytosis of HA via the clathrin-coated pit pathway (2–4) in an endocytic process similar to that for transferrin, asialoglycoprotein, mannose 6-phosphate, and low density lipoprotein receptors (5, 6). Although named for its first known function (3), HARE is a general receptor for GAG clearance, mediating the endocytosis of CS-A, CS-C, CS-D, CS-E, heparin, and dermatan sulfate (CS-B) (7, 8). Heparan sulfate and keratan sulfate are the only GAGs that are not ligands for HARE (63).
Despite its simple linear structure (i.e. a polymer of 2-deoxy,2-acetamido-d-glucopyranosyl-β(1,4)-d-glucuronopyranosyl-β(1,3) disaccharide units), HA is a very dynamic molecule, whose synthesis and degradation appear tightly regulated. Mammals degrade and resynthesize about one-third (∼5 g) of their total body HA daily. The metabolic half-life of HA varies from ∼1.5 day in skin (9) to 2–3 weeks in cartilage (10). Similarly, circulating HA has a half-life of 2–5 min, because of its rapid clearance by liver sinusoidal endothelial cells (10, 11). In mammals, large native HA (∼107 Da) in ECMs throughout the body is partially degraded to fragments of ∼106 Da that are released into lymphatic fluid and flow to lymph nodes where sinusoidal endothelial cells remove and degrade ∼85% of the HA. The remaining HA (∼15%) passes through the lymph nodes, enters the blood, and is removed by the liver via receptor-mediated endocytosis. Although HA turnover is high, HARE in sinusoidal endothelial cells of lymph nodes and liver keeps the steady-state HA concentration in blood low (10–100 ng/ml). A high level of HA in the blood circulation could elevate blood viscosity and impair the microcirculation in capillaries.
Full-length hHARE is a 2551-aa type I single transmembrane glycoprotein (∼315 kDa in SDS-PAGE) encoded by the 180.2-kb STAB-2 gene on chromosome 12q23.3; the gene has 69 exons. HARE proteins were first purified from rat liver (12) and human spleen (13). Two isoforms of HARE are present in tissues (e.g. 190- and 315-kDa hHARE). The smaller HARE isoform is derived by proteolysis from the full-length protein (7, 14). Both HARE isoforms are functional membrane receptors that bind and internalize ligands via the clathrin-coated pit pathway, targeting them to lysosomes for degradation (2, 4, 7, 8). The 190-HARE (Fig. 1A) has a large extracellular domain (1323 aa), a transmembrane domain (21 aa), and a small (72 aa) COOH-terminal CD (13, 15). The CD (Tyr2480–Leu2551) contains 17 aa that could be phosphorylated: four Tyr, seven Ser, one His, and five Thr. Only Ser2497, Ser2537, Thr2523, Tyr2519, and Tyr2531 are predicted to be phosphorylated (by NetPhos 2.0). The CD does not contain PEST sequences for rapid protein degradation and turnover or for O-glycosylation with GlcNAc.
FIGURE 1.
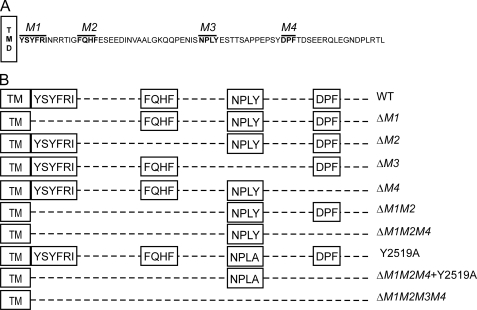
The 190- and 315-HARE CD and mutants used in this study. A, four putative endocytic motifs (indicated in boldface and overlined) are present in the 72-aa CD of recombinant 190-HARE (1416 aa long) or 315-HARE (2551 aa long). The four sequence motifs (YSYFRI2485, FQHF2495, NPLY2519, and DPF2534) are referred to as M1, M2, M3, and M4, respectively. Numbering is relative to the full-length 315-HARE protein (13, 15). V5 and His6 epitopes are at the COOH termini of all 190-HARE mutants. B, scheme illustrates the various motif deletion or site-specific 190-HARE CD mutants used. Constructs and stably transfected Flp-In-293 cell lines were prepared as described under “Experimental Procedures.” TM, transmembrane.
Endocytic receptors contain either single or multiple targeting sequences that direct the receptor-ligand complexes into a coated pit pathway (5, 16, 17). There may be multiple coated pit pathways for internalizing and then creating different microenvironments for different receptor-ligand complexes so they are routed or assemble signaling complexes in different ways appropriate for the particular receptor (6, 18, 19). In contrast to single motif receptors, the HARE CD contains five putative endocytic motifs (Fig. 1 and Table 2): YSYF2483, YFRI2485, FQHF2495, NPLY2519, and DPF2534. The first three (YSYF, YFRI, and FQHF) are ΦXXB motifs; where Φ is either Tyr or Phe, X can be any aa, and B is a hydrophobic aa with a bulky side chain. The first two sequences are overlapping, located at the junction between the membrane and CD, and for practical reasons we consider them to be a single motif, YSYFRI2485. The functions of ΦXXB, NPXY, and DP(F/P) motifs in receptor internalization, trafficking, and delivery of ligand to lysosomes have been studied extensively in different receptors (Table 2). Many receptors contain NPXY motifs that facilitate rapid internalization of receptor-ligand complexes from the cell surface.
TABLE 2.
Potential endocytic signals in HARE The candidate endocytic signals (M1–M4) in the HARE CD, their recognition partners, and potential functions are summarized. The first motif contains two overlapping YXXΦ motifs as indicated by the sequences that are underlined or in boldface font.
| HARE sequence | Targeting motif | Proposed recognition protein | Function | Refs. |
|---|---|---|---|---|
| YSYFRI2485 | YXXΦ (ΦXXB) | μ subunits of AP complex | Internalization, basolateral, or lysosomal targeting | 43, 44, 59–62 |
| FQHF2495 | ΦXXB | μ subunits of AP complex | Internalization, basolateral, or lysosomal targeting | 43, 44, 59–62 |
| NPLY2519 | NPXY | μ2 subunit of AP-2, Dab2 Tyr(P) binding domain, clathrin | Internalization, intracellular, routing/signaling | 42, 47, 49, 50 |
| DPF2534 | DPF/DPW | Ear domain of the α subunit of AP-2 complex | Internalization process | 34, 35, 37 |
We recently found that HARE is a signaling receptor able to sense external HA and initiate intracellular activation of the mitogen-activated protein kinase ERK (64). Thus, we are interested in how HARE is targeted for internalization and intracellular routing. In this study, our goal was to determine which of the four candidate targeting motifs in the CD of HARE are important or critical for efficient endocytosis of HA by the coated pit pathway. The results indicate that three motifs are important, and thus provide substantial redundancy, for coated pit targeting and internalization.
EXPERIMENTAL PROCEDURES
Materials and Buffers—Pfu Ultra DNA polymerase for site-directed mutagenesis was from Stratagene (La Jolla, CA), and super-competent DH10B-derived Escherichia coli, TOP10 E. coli, restriction enzymes, cell culture reagents, and hygromycin B were from Invitrogen. Plasmid DNA was purified using Eppendorf Miniprep (Westbury, NY) or Maxiprep plasmid isolation kits (Promega, Madison, WI). Protease inhibitor mixture (catalogue P8340) and alkaline phosphatase conjugated to anti-goat or anti-mouse polyclonal Abs were from Sigma. Anti-V5 Ab was from Bethyl Labs (Montgomery, TX). Horseradish peroxidase conjugated to anti-goat or anti-mouse polyclonal Abs were from Santa Cruz Biotechnology (Santa Cruz, CA). SDS, Tris, acrylamide, bis-acrylamide, TEMED, ammonium persulfate, high range protein molecular weight markers, p-nitro blue tetrazolium, and 5-bromo-4-chloro-3-indolyl phosphate p-toluidine were from Bio-Rad. Solutions for dye binding protein assays (20) and reagents for enhanced chemiluminescence were from Pierce. Autoradiography film was from Molecular Technologies (St. Louis, MO). Na 125I was obtained from GE Healthcare, and 125I-HA was prepared as described previously (21, 22). Nitrocellulose and polyvinylidene difluoride membrane were from Millipore (Billerica, MA). Unless specified otherwise, other chemicals and analytical reagent of the highest purity available were from Sigma or Fisher. Growth Medium is DMEM containing 8% (v/v) fetal bovine serum and 100 μg/ml hygromycin B. TBS contains 20 mm Tris-HCl, pH 7.0, and 150 mm NaCl. TBST is TBS with 0.1% (v/v) Tween 20. TBST/BSA is TBST with 1.0% (w/v) BSA. PBS contains 137 mm NaCl 8 mm Na2HPO4 1.5 mm KH2PO4, 2.7 mm KCl, pH 7.2. HBSS contains 5 mm KCl, 0.4 mm KH2PO4, 0.8 mm MgSO4, 137 mm NaCl, 0.3 mm Na2HPO4, 5.5 mm glucose, 1.26 mm CaCl2, 0.5 mm MgCl2, and 28 μm phenol red; at the time of use, 3.5 g/100 ml of NaHCO3 was added, and the pH was adjusted to 7.2 with HCl. Endocytosis Medium contains DMEM supplemented with 0.05% (w/v) BSA. Lysis Buffer is PBS with 0.5% (v/v) Nonidet P-40, 200 μm PMSF, and 1 μg/ml of protease mixture inhibitor (contains 4-(2-aminoethyl) benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin).
PCR-assisted Site-directed Mutagenesis—Purified plasmid DNA (pSecTag/FRT/V5 His-TOPO; Invitrogen) containing 190-HARE cDNA (4248 bp) ligated into the BamHI site was used as a template to generate the different cDNA mutants (8). Primers (Table 1) were designed for deletion or site-directed mutagenesis of coding regions for individual and multiple potential endocytic motifs in WT 190-HARE plasmid DNA. Mutagenesis PCRs were performed for 18 cycles (94 °C for 20 s, 55–65 °C for 20 s, and 72 °C for 9 min). Parental and newly generated plasmid DNAs were precipitated with 0.1 volume 3 m potassium acetate, pH 5.5, and 2 volumes of 100% ethanol. The DNA pellet was dissolved in 17 μl of water and digested with 5 units of DpnI overnight to eliminate parental DNA. After heat inactivation of DpnI, the DNA solution was used to transform TOP10 E. coli using standard manufacturer protocols. Bacterial colonies were picked, and potential 190-HARE mutant cDNAs were verified by PCR using gene-specific forward (5′-GTTCCATCTACGATCGCCACTGGGCCAG-3′) and vector-specific reverse (5′-CGTAGAATCGAGACCGAGGAG-3′) primers. The complete sequences of the promoter and complete coding regions of cDNAs in the final clones were determined to be correct before use in subsequent experiments. All 190-HARE mutants also contain COOH-terminal V5 and His6 epitopes.
TABLE 1.
Primer sequences used to create mutations in the 190-HARE CD Deletion or substitution mutations were made as described under “Experimental Procedures” using the indicated forward (F) and reverse (R) primers. Deletion of multiple motifs was accomplished by using a single, double, or triple motif deletion mutant HARE cDNA and a set of primers to delete codons specifying another motif.
| 190-HARE mutant | Forward and reverse mutagenic primer sequences |
|---|---|
| (ΔYSYFRI) = ΔM1 | F, 5′-GTTGCCTTGGCTGCTAACCGGAGAAC-3′ |
| R, 5′-GATTGTTCTCCGGTTAGCAGCCAAGG-3′ | |
| (ΔFQHF) = ΔM2 | F, 5′-CGGAGAACAATCGGCGAGTCGGAAGAGGAC-3′ |
| R, 5′-GTCCTCTTCCGACTCGCCGATTGTTCTCCG-3′ | |
| (ΔNPLY) = ΔM3 | F, 5′-CAGCAGCCTGAGAATATCTCGGAGAGCACAACCTCAGCTCCC-3′ |
| R, 5′-GGGAGCTGAGGTTGTGCTCTCCGAGATATTCTCAGGCTGCTG-3′ | |
| (ΔDPF) = ΔM4 | F, 5′-CCAGAACCTTCCTACACGGACTCTGAAGAAC-3′ |
| R, 5′-GTTCTTCAGAGTCCGTGTAGGAAGGTTGCTGG-3′ | |
| Y2519A | F, 5′-GAGAATATCTCGAACTTGGCTGAGAGCACAACC-3′ |
| R, 5′-GGTTGTGCTCTCAGCCAAGTTCGAGATATTCTC-3′ |
Selection and Characterization of Stable Transfectants—Flp-In 293 cells (60–70% confluent in a 100-mm plate with 10 ml of fresh DMEM) were transfected with 750 μl of serum-free DMEM containing 9 μg of pOG44 (which encodes the Flp-In recombinase), 1 μg of mutant pSecTag-190-HARE, and 20 μl of Lipofectamine 2000 (Invitrogen). Cells were incubated overnight in Growth Medium without hygromycin B, and then washed and fed with fresh Growth Medium for the first 3–4 days and then periodically when required. After 2 weeks, individual isolated colonies were collected and grown in 24-well plates until confluent. Clones were tested for HARE protein expression by SDS-PAGE, using 5% gels, followed by Western blot analysis (23) with anti-V5 Ab. Each clone was also tested for correct insertion of the mutant pSecTag-190-kDa hHARE cDNA uniquely into the Flp-In recombination site by the Flp-In recombinase encoded by pOG44, as described previously (7, 8). At least 4–6 clones of each mutant were selected and characterized for further experiments.
125I-HA Binding Assays for Cell Surface and Intracellular Receptors—WT 190-HARE, EV, and mutant 190-HARE cell lines were grown to ∼90% confluence in Growth Medium in 12-well tissue culture plates. Cultured cells were washed with 1 ml of sterile PBS and 1 ml of serum-free DMEM and were incubated at 37 °C in 1 ml of serum-free medium for 1 h. After incubation, medium was removed, and the cells were washed with 2 ml of HBSS and incubated for 2 h at 4 °C in Endocytosis Medium with 1.5 μg/ml 125I-HA with or without a 100-fold excess of unlabeled HA to assess nonspecific binding. To determine total HA binding (by cell surface and intracellular receptors), cells were permeabilized with 0.055% (w/v) digitonin (24, 25) and incubated with 125I-HA ± unlabeled HA as above for 2 h at 4 °C. Media were aspirated, and the cells were washed rapidly three times with HBSS (2 ml per wash) to remove unbound 125I-HA. Cells were then lysed in 1 ml of 0.3 n NaOH, and radioactivity was measured using a Packard Cobra II gamma counter, and protein was determined by the method of Bradford (20) using BSA as standard. Data are expressed as fmol/106 cells ± S.E. and normalized for HARE expression relative to WT HARE. Cell number was calculated based on 398 μg of total protein/106 cells (8). Specific binding was calculated as binding without unlabeled HA (total) minus binding in the presence of unlabeled HA (nonspecific). Experiments were performed with each clone in triplicate.
125I-HA Internalization and Degradation Assays—To assess internalization of 125I-HA, WT 190-HARE, EV, and mutant HARE cell lines were grown to ∼90% confluence, washed, and incubated in serum-free medium as above. Cells were washed with 2 ml of HBSS and incubated at 37 °C in Endocytosis Medium containing 1.5 μg/ml 125I-HA with or without a 100-fold excess of unlabeled HA to assess nonspecific binding, for 2 or 4 h, unless noted otherwise. Medium was aspirated, and cells were washed three times with HBSS (2 ml each), solubilized with 1 ml of 0.3 n NaOH, and radioactivity and protein content were determined as above. Specific HA endocytosis or degradation was calculated as the mean value (as fmol/106 cells/HARE expression relative to WT HARE) without unlabeled HA (total) minus the value in the presence of unlabeled HA (non-specific). Each experiment was repeated 2–3 times with multiple clones.
Degradation of internalized 125I-HA was measured using a cetylpyridinium chloride precipitation assay as described previously (26, 27). Cells were lysed in 0.3 n NaOH, and 100-μl samples were mixed at 22 °C with 47 μl of 0.6 n HCl, 28 μl of distilled water, and 125 μl of 2.0 mg/ml HA in 1.5-ml microcentrifuge tubes. Cetylpyridinium chloride (300 μl of 6%; w/v) in distilled water was added, and the tubes were mixed by vortexing, and after 10 min, the samples were centrifuged (22 °C, 5 min, 9000 rpm) in a microcentrifuge, using a swinging bucket rotor. A supernatant sample (300 μl) was taken to determine radioactivity, and the rest was removed by aspiration. The tube tip containing the precipitated pellet was cut off and put in a gamma counter tube, and radioactivity was determined. Degradation values were calculated as the time-dependent increase of nonprecipitable radioactivity and are expressed as the percent (with WT as 100%) of specifically internalized 125I-HA that was degraded.
Western and Ligand Blot Assays—To analyze and normalize for HARE expression in mutant cell lines, cells at ∼90% confluence in 6-well plates were washed twice with cold PBS and solubilized in Lysis Buffer. Lysates were mixed by vortexing, incubated on ice for 2 h, and centrifuged at 11,250 × g for 5 min to remove debris. Samples (25 μg of protein) were separated in 5% gels by nonreducing SDS-PAGE, followed by electro-transfer to nitrocellulose membranes for 1 h, 4 °C at 110 V in 25 mm Tris-HCl, 192 mm glycine, pH 7.4, and 20% (v/v) methanol. For Western blot analysis of WT and mutant 190-HARE protein, membranes were treated with TBST/BSA for 1 h, washed, and incubated with anti-V5Ab for 1 h at 22°C. Blots were washed three times (5 min each) with TBST/BSA, incubated with goat anti-rabbit Ab conjugated to alkaline phosphatase (or horse-radish peroxidase), washed three times with TBST/BSA, and incubated with p-nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate p-toluidine-conjugated Ab for color development using alkaline phosphatase. Reactions were stopped by washing the membrane with distilled water. For horseradish peroxidase detection, membranes were incubated with enhanced chemiluminescence substrate and exposed to autoradiography film. For comparison of 190-HARE protein expression levels by mutant cell lines (with replicates of 4–6 clones per group), HARE band intensities in Western blots (with equal total cell protein loads) were quantified by densitometry using a FluoroChem8000 Imaging System (Alpha Innotech Corp, San Leandro, CA). Band intensities were normalized to parallel wild type 190-HARE samples and expressed as a percent of WT HARE expression.
For ligand blot assays, the post-transfer membranes were incubated with TBST for 2 h or overnight at 4 °C. To reduce nonspecifically bound material, and thus background, solutions of 1.5 μg/ml 125I-HA in 10 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm EDTA, with or without a 100-fold excess of unlabeled HA, were first exposed to unused nitrocellulose at 22 °C for 2 h. The protein-containing membranes were then incubated with the treated 125I-HA solutions for 2 h at 4°C, washed five times (5 min each) with TBST, air-dried at 22 °C for 30 min, and bound 125I-HA was detected by autoradiography using Kodak BioMax MS film exposed for 1–15 days at -80 °C. The same membrane was washed with TBST/BSA and then probed with anti-V5 Ab to assess HARE expression.
Data Analysis—The assessment of statistical significance for differences between sample sets, based on three independent experiments, was by the unpaired Student's t test.
RESULTS
Whereas many membrane receptors have a single CD sequence that mediates efficient uptake into the coated pit pathway, HARE has multiple candidate motifs for this purpose (Fig. 1A and Table 2). Involvement of multiple motifs might indicate that redundancy is critical, perhaps for multiple functions of HARE, or that different HARE-ligand complexes may follow alternative intracellular pathways. To determine whether any of the four putative endocytic motifs in the HARE CD are active for targeting to coated pits, we created a panel of stable Flp-In 293 cell lines expressing 190-HARE mutants with different endocytic motif deletions or substitution mutations (Fig. 1B). Cells expressing mutant 190-HARE protein that were negative for β-galactosidase and sensitive to Zeocin represented the correct insertion of vector into the single recombinase-specific site in Flp-In 293 cells. We examined multiple clones of the following 190-HARE mutant cell lines containing single motif deletions (ΔYSYFRI (ΔM1), ΔFQHF (ΔM2), ΔNPLY (ΔM3) or ΔDPF (ΔM4)); a double motif deletion (ΔM1M2), triple motif deletions (ΔM1M2M4) without and with a Y2519A substitution in M3, and a quadruple motif deletion (ΔM1M2M3M4).
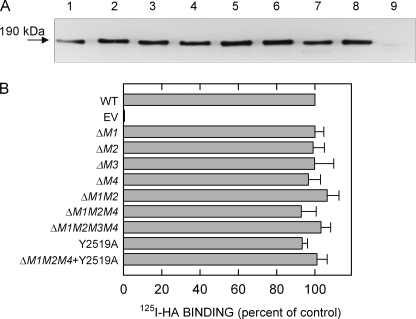
We selected 4–6 individual cell line clones for each mutant and measured HARE expression in replicates of each group of clones by SDS-PAGE, Western analysis, and densitometry using equal amounts of total cell lysate protein as measured by the Bradford assay. Although a single blot is shown (Fig. 2A), the overall results (not shown) demonstrated that HARE expression within each group of mutants was ∼85–100% of the WT 190-HARE protein level. Each HARE CD mutant also contains V5 and His6 epitopes at the COOH terminus. No differences were found in the morphology or cell growth characteristics among the mutant cell lines, compared with 190-HARE cells (data not shown). Ligand blot assays were performed to quantify specific 125I-HA binding to the various HARE mutants after SDS-PAGE and electrotransfer to nitrocellulose (28). Densitometry of autoradiographs followed by Western blot analysis allowed HA binding to be normalized for HARE protein content, relative to WT (Fig. 2B). All of the HARE CD mutants lacking individual or multiple endocytic motifs bound 125I-HA in ligand blot assays essentially at WT levels. The HARE CD mutants also bound comparable amounts of three anti-hHARE mAbs, compared with WT HARE, indicating there were no changes in the folding of the mutant HARE ectodomains (not shown).
FIGURE 2.
Expression and 125I-HA ligand binding of 190-HARE CD mutants. Cells were grown to confluence, washed twice with cold PBS, and centrifuged cell lysates (25 μg of protein) were subjected to SDS-PAGE and electrotransfer. A, HARE protein was detected by Western blot analysis using anti-V5 Ab. The 190-HARE proteins are as follows: lane 1, WT; lane 2, ΔM1; lane 3, ΔM2; lane 4, ΔM3; lane 5, ΔM4; lane 6, ΔM1M2; lane 7, ΔM1M2M4; lane 8, Y2519A; and lane 9, EV. B, ligand blot assays were performed with 125I-HA in multiple experiments using different clones of the indicated mutants, and HARE expression was determined by Western blotting using the same strips after autoradiography. 125I-HA ligand blot signals were normalized to HARE expression using densitometry and to WT (as 100%).
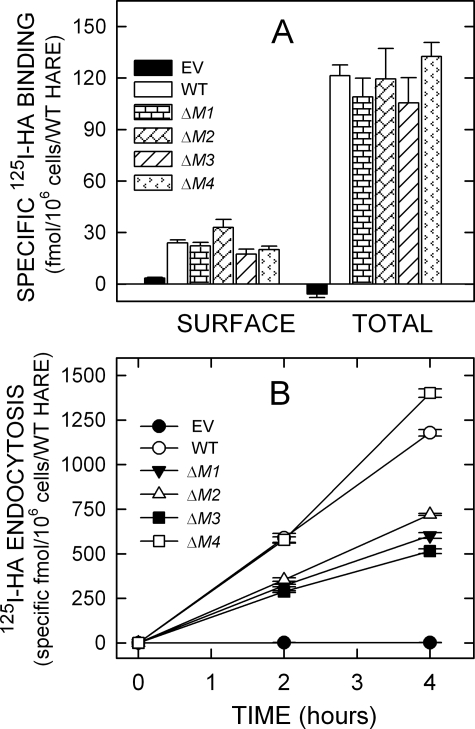
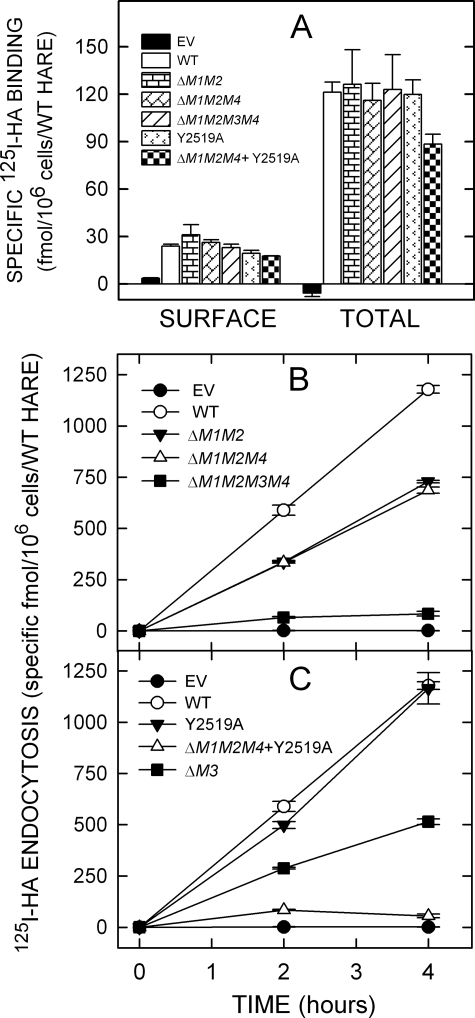
Typically, endocytic recycling receptors constitutively recycle and are found both on the cell surface and in numerous intracellular compartments, representing different stages of the receptor routing and recycling pathway. The majority of such endocytic receptors, including HARE, are intracellular, and this cellular distribution of receptor molecules reflects the kinetic steady-state of the receptor pathway, which is both kinetic and spatial. If mutation of an important motif sequence affected the intracellular routing/recycling kinetics of HARE, then the normal steady-state receptor distribution could also be altered. To test this, cell lines expressing a variety of 190-HARE CD mutants were assessed for cell surface and total (i.e. cell surface and intracellular receptors in digitonin-permeabilized cells) binding of 125I-HA (Fig. 3A and Fig. 4A). All 190-HARE single mutant cells bound 125I-HA at the cell surface and to intracellular sites at WT levels (Fig. 3A). Among the multiple motif deletion mutants, only the ΔM1/M2/M4 + Y2519A HARE mutant displayed significantly less cell surface (p = 0.004) HARE and total (p = 0.02) HARE than WT (Fig. 4A). However, the ΔM1/M2/M4 + Y2519A HARE mutant showed a surface/internal HARE distribution that was similar to WT. Thus, all the 190-HARE CD motif-deletion mutants appear to have normal intracellular receptor trafficking and recycling kinetics.
FIGURE 3.
125I-HA binding and endocytosis by cells expressing single motif 190-HARE CD mutants. Cells expressing EV, WT, or mutant 190-HARE were incubated in serum-free medium and washed, and specific 125I-HA binding at 4 °C or endocytosis at 37 °C was quantified as described under “Experimental Procedures.” A, cell surface and total cellular 125I-HA binding are indicated for cells expressing 190-HARE: WT (white bars), ΔM1 (horizontal rectangles), ΔM2 (diagonal rectangles), ΔM3 (diagonal lines), or ΔM4 (arrowheads) 190-HARE or EV (black). Values from 2 to 3 independent experiments are the means ± S.E. (n = 6–9) specific 125I-HA binding normalized for HARE expression relative to WT 190-HARE. B, specific 125I-HA internalization was determined at 37 °C for 2 and 4 h as described under “Experimental Procedures.” Values from 2 to 3 independent experiments are the mean ± S.E. (n = 6–9) specific 125I-HA endocytosis, normalized for HARE expression relative to WT. The plots show cells expressing WT (○), ΔM1 (▾), ΔM2 (Δ), ΔM3 (▪), or ΔM4 (□) 190-HARE, or EV (•).
FIGURE 4.
125I-HA binding and endocytosis by cells expressing multiple motif HARE CD mutants. Cells expressing EV, WT 190-HARE, or 190-HARE mutants were incubated at 37 °C in serum-free medium for 1 h, washed, incubated, and processed to quantify 125I-HA binding at 4 °C or endocytosis at 37 °C as described in Fig. 3. A, cell surface and total cellular 125I-HA binding are indicated for cells expressing EV (black) or 190-HARE: WT (white bars), ΔM1M2 (horizontal rectangles), ΔM1M2M4 (diagonal rectangles), ΔM1M2M3M4 (diagonal lines), Y2519A (arrowheads), or ΔM1M2M4 + Y2519A (checkerboard). B, endocytosis of 125I-HA by cells expressing 190-HARE: WT (○), ΔM1M2 (▾), ΔM1M2M4 (▵), ΔM1M2M3M4 (▪), or EV (•). C, endocytosis of 125I-HA by cells expressing 190-HARE: WT (○), Y2519A (▾), ΔM1M2M4 + Y2519A (▵), ΔM3 (▪), or EV (•).
To understand the role of putative signal motifs in endocytosis of HA and targeting HARE to coated pits, cells expressing 190-HARE mutants were incubated at 37 °C with 125I-HA with or without excess HA to assess nonspecific uptake (Fig. 3B and Fig. 4B). Cells expressing 190-HARE(ΔM4) did not show any defect in 125I-HA internalization. The specificity of 125I-HA internalization by the WT and all mutant HARE cell lines was >80%. The rates of 125I-HA internalization by HARE ΔM1 and ΔM2 mutant cell lines decreased by ∼49 and ∼39%, respectively (Fig. 3B). Cell lines expressing a double motif (ΔM1M2) or a triple motif (ΔM1M2M4) deletion showed similar endocytic rate decreases of ∼41% (Fig. 4B). The essentially identical rates of HA endocytosis by cells expressing HARE ΔM1M2 or ΔM1M2M4 mutants confirm that the DPF motif does not play an important role in HARE-mediated endocytosis of 125I-HA. Cells expressing HARE lacking the NPLY motif (ΔM3) demonstrated ∼56% decreased endocytosis rates, which indicate that this motif is very important, although not absolutely necessary, for HARE internalization.
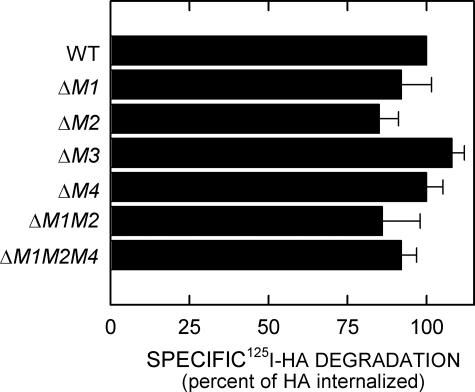
After HA is internalized, it is delivered to lysosomes where it is then degraded by lysosomal hyaluronidase, β-glucuronidase, and β-N-acetylglucosaminidase (10, 26). 125I-HA degradation was quantified by a cetylpyridinium chloride precipitation assay (26). No significant differences in the amounts of 125I-HA degradation were observed for any of the HARE mutants compared with the WT cell lines over 8 h (Fig. 5). The results indicate that deletion of different endocytic motifs in the HARE CD does not affect delivery to and degradation in lysosomes of the 125I-HA internalized by the various mutants.
FIGURE 5.
Degradation of internalized 125I-HA by cells expressing 190-HARE mutants. Confluent cells expressing the indicated WT or HARE mutants were incubated at 37 °C with 1.5 μg/ml 125I-HA with or without 150 μg/ml unlabeled HA for 8 h. Medium samples were taken to assess released 125I-HA degradation products and normalized as described under “Experimental Procedures.” The values plotted are the mean ± S.E. (n = 6) percent degradation of the specifically internalized 125I-HA.
To confirm the redundancy of the multiple motifs in the HARE CD and further assess their role in HARE internalization, we created additional deletion and point mutation mutants of M3. Cells expressing a triple motif deletion HARE CD mutant, Δ(M1M2M4), were compromised in their ability to endocytose HA but still mediated HA uptake at 58% of the WT 190-HARE rate (Fig. 4B). In contrast, a HARE CD mutant with a single Y2519A change in the NPLY signal sequence showed only an ∼10% reduced rate of HA uptake (Fig. 4C); this mutation had little effect compared with deletion of the whole M3 motif. However, when the Y2519A mutation was combined with the triple motif deletion, Δ(M1M2M4), the resulting rate of HARE-mediated 125I-HA endocytosis was reduced by ∼95% (Fig. 4C), to almost the level of background uptake seen in EV cells. The result shows that in the absence of the functional redundancy provided by motifs M1 and M2, Tyr2519 is critical for the function of motif M3. Table 3 summarizes the 125I-HA endocytosis rates for all the 190-HARE mutants used in this study.
TABLE 3.
Rates of 125I-HA endocytosis by 190-HARE CD variants Cells expressing the indicated 190-HARE mutants, WT, or EV were assessed for specific 125I-HA endocytosis and normalized to total protein and then cell number, and HARE expression relative to WT as described under “Experimental Procedures.” Values are the mean ± S.E. (n = 6–9).
| 190-HARE mutant | Specific 125I-HARE | Relative HA uptake |
|---|---|---|
| fmol/106 cells/WT HARE/h | % | |
| WT | 295 ± 5 | 100 |
| EV | 0.5 ± 0.3 | 0.2 |
| ΔM1 | 151 ± 5a,b | 51 |
| ΔM2 | 180 ± 2b | 61 |
| ΔM3 | 129 ± 4a | 44 |
| ΔM4 | 351 ± 6 | 119 |
| ΔM1M2 | 182 ± 2 | 61 |
| ΔM1M2M4 | 172 ± 4 | 58 |
| ΔM1M2M3M4 | 21 ± 3 | 7 |
| Y2519A | 279 ± 19 | 94 |
| ΔM1M2M3 + Y2519A | 14 ± 2 | 5 |
Data indicate that the rates for the ΔM1 and ΔM3 mutants, compared with each other, were significantly different; p < 0.002
Data indicate that the rates for ΔM1 and ΔM2 mutants were significantly different; p < 0.001
DISCUSSION
Receptor-mediated endocytosis via the clathrin-coated pit pathway is a sequential process, involving many cytosolic proteins at different steps, in which plasma membrane receptors are specifically and efficiently internalized into the cell. Based on their interactions with the endocytic cargo (i.e. receptor-ligand complexes), proteins facilitating endocytosis are classified as accessory or adaptor proteins (e.g. AP-2); >30 different adaptor/accessory proteins may be involved in or control endocytic processes (16, 17, 29). Adaptor proteins bind to the endocytic cargo and to coat components, thus linking the two (30). Accessory proteins (e.g. dynamin) are involved in coated vesicle formation without direct interaction with endocytic cargo (31, 32). Although many other proteins are involved in coat-complex formation, clathrin and adaptor proteins, particularly AP-2, are the major proteins in coated vesicles (17, 33). Selective recruitment of cargo proteins into coated pits is mediated by μ2 subunits of AP-2 binding to specific short sequences (3–7 aa) in a receptor CD. These signal sequences are highly, although not exactly, conserved in different organisms, and 2–3 aa of a signal motif are usually crucial for function. Two major classes of endocytic signal motifs are predominantly involved in targeting to the coated pit pathway: those that have Tyr as a critical endocytic aa (e.g. NPXY and YXXΦ) and those with di-Leu (e.g. DEXXX(L/L)I). Accessory proteins implicated in endocytosis process such as Epsin, (34), Eps15 (35, 36), and Dynamin (37) have DPW or DPF signal sequences that bind the α-adaptin appendage domain.
The 72-aa HARE CD contains four potential signal sequence motifs (YSYFRI2485, FQHF2495, NPLY2519, and DPF2534) that could be used to target ligand-HARE complexes to the clathrin-coated pit pathway for rapid endocytosis. As expected, deleting any of these CD motifs had little or no effect on 125I-HA binding by their respective HARE ectodomains or on HARE protein expression levels. All HARE mutants were also recognized by three hHARE-specific mAbs originally raised against rat HARE suggesting that the ecto-domain is not affected by CD mutations (3, 13). However, cells expressing single or multiple deletions of motifs M1, M2, or M3 showed very similar (∼38–52%) decreases in 125I-HA internalization rates. In contrast, deletion of M4 (DPF2534) did not affect 125I-HA endocytosis. Moreover, all of the 190-HARE CD mutants tested showed no defects in the ability to deliver the internalized HA to lysosomes, i.e. deletion of different potential endocytic motifs in HARE did not affect the trafficking pathways that deliver internalized HA from plasma membrane to lysosomes. In all but two of the HARE CD mutants, intracellular receptor routing and recycling appeared to be unaffected. The results do not exclude the possibility that some of the signal sequences are involved in both coated pit targeting and intracellular routing.
Several classes of endocytic motifs present in the CD of membrane proteins have been identified that utilize clathrin-coated pits for endocytosis. Among these signals, Tyr-containing signal motifs such as YXXΦ and NPXY are the best characterized. Cells expressing 190-HARE(ΔM1) showed slower 125I-HA internalization rates (∼49%) compared with WT, indicating that YSYFRI2485, with two overlapping YXXΦ motifs, is important but not essential for endocytosis. Tyr in the CD of endocytic recycling receptors (e.g. cation-independent mannose 6-phosphate, asialoglycoprotein, and transferrin receptors (5, 38)) is essential for binding the μ2 subunit of AP-2 and for accumulation into clathrin-coated pits and rapid internalization (39–41). The dominant signal for β-amyloid precursor protein endocytosis is also a YXXΦ motif, YENP (42). YXXΦ motifs are not limited to only endocytic targeting, they can also help target transmembrane proteins to lysosomes and lysosome-related organelles (43), and sort membrane receptors to the basolateral plasma membrane of polarized epithelial cells (44) or to the trans-Golgi network (5, 30, 45).
Tyr is also critical in NPXY endocytic signal motifs, which are highly conserved from Caenorhabditis elegans and Drosophila melanogaster to mammals, indicating that they participate in evolutionarily conserved sorting mechanisms. The NPXY signal motifs in other type I integral membrane receptors such as the LDL receptor, LDL receptor-related protein, integrin β, and β-amyloid precursor protein mediate rapid endocytosis via coated pits (42, 46–50). Generally, NPXY signals are found in medium length CDs (e.g. ∼40–200 aa), not closer than 10 aa from the membrane domain and not present at the very end of the CD. In hHARE, the NPLY2519 sequence is in the center of the 72-aa-long CD.
This is the first effort to address the functional roles of YXXΦ, DPX, and NPXY motifs in the endocytosis of hHARE-ligand complexes. The hHARE CD actually has three ΦXXB motifs: the overlapping YSYF and YFRI sequences in M1 and the M3 motif FXXF2495. Interestingly, the rat HARE CD has corresponding and nearly identical YSYFRL (M1) and FQRF (M2) sequences but does not contain DP(F/W) or NPXY motifs (13, 14). Our results indicate that although NPLY2519 (M3) is slightly more important for endocytosis by hHARE, the YSYFRI2485 (M1) and FQHF2495 (M2) sequences also contribute substantially to the overall internalization of HA. Thus, no single motif was critical for rapid uptake of HARE, and the three motifs provide, for as yet unknown reasons, unexpected redundancy for internalization. Redundancy may be needed for functions of HARE other than just GAG clearance. One newly discovered function is the recent finding that HARE is a signaling receptor that mediates intracellular activation of ERK1 and -2, in response to the concentration of external HA (64). Perhaps one of the three active endocytic signal sequences is also specifically involved in activation of the ERK signaling pathway.
Some receptors have a single signal motif for internalization. For example, substitution of Tyr807 with Cys or Ala reduces endocytosis by ∼85% in the LDL receptor, whereas substitution with Phe does not abolish rapid internalization. An NPXY motif in the insulin-like growth factor I receptor is the primary motif for internalization and is also involved in the signaling pathway (51). Other receptors possess multiple motifs for internalization, so that mutation or deletion of a single motif often does not completely abolish receptor internalization. For example, two NPXY motifs in integrin β1 are required for recruitment to focal adhesions (52), and the CD of megalin has two NPXY motifs that are both important for coated pit targeting (53). The interleukin-13 receptor α2 contains a diLeu- and a Tyr-based endocytic motif, and substitution or deletion of either motif decreases endocytosis of 125I-interleukin-13 by ∼50%, indicating that both endocytic motifs work together for internalization (54). In some proteins, endocytic motifs are not involved in endocytosis and therefore not signal sequences. The angiotensin II receptor contains four candidate Tyr-based motifs and one diLeu-based motif, but only Tyr319 and LL316 are important for efficient receptor internalization (55). The CD of the receptor for LDL-related protein 1 contains one YXXΦ, two NPXY, and two diLeu motifs. However, only the YXXΦ and one diLeu are dominant endocytic signals for targeting receptor to coated pits (56).
It may not be surprising that multiple endocytic motifs are involved in HARE-mediated internalization of 125I-HA. Although we have characterized HARE most extensively as an HA receptor, it also specifically binds and internalizes many other ligands. In addition to HA and multiple CS types, HARE/Stabilin 2 specifically recognizes advanced glycation end products (e.g. AGE-BSA) and acetylated-LDL (57), which we have confirmed. We also recently discovered that the 190-HARE and 315-HARE proteins are endocytic receptors for heparin (63). Thus, HARE binds and internalizes sulfated (CS, DS, and heparin) and nonsulfated (HA and chondroitin) GAGs and advanced glycation end products. Nonetheless, HARE is specific and does not simply bind any anionic polymer, because heparan sulfate, keratan sulfate, polygalacturonic acid, and nucleic acids are not ligands (2, 7, 8, 58).
In summary, the results show that NPLY2519 is a slightly more active endocytic signal sequence in the CD of 190-HARE than YSYFRI2485 or FQHF2495. In contrast, deletion of the DPF2534 sequence motif did not affect endocytosis. Thus, the maximum rate of HARE endocytosis requires that the three endocytic motifs (YSYFRI, FQHF, and NPLY) work together for efficient coated pit targeting and endocytosis of HARE.
Acknowledgments
We thank Jennifer Washburn for technical assistance and Dr. Svetlana Kyosseva for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM69961 from the NIGMS. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GAG, glycosaminoglycan; aa, amino acids; Ab, antibody (IgG); BSA, bovine serum albumin; CD, cytoplasmic domain; CS, chondroitin sulfate; ECM, extracellular matrix; EV, empty vector; HA, hyaluronic acid, hyaluronate, hyaluronan; HARE, HA receptor for endocytosis; HBSS, Hanks' balanced salts solution; hHARE, human HARE; M1, HARE CD motif 1 (YSYFRI); M2, HARE CD motif 2 (FQHF); M3, HARE CD motif 3 (NPLY); M4, HARE CD motif 4 (DPF); PBS, phosphate-buffered saline; TBS, Tris-buffered saline; 190-HARE, 190-kDa HA receptor for endocytosis; 315-HARE, 315-kDa HA receptor for endocytosis; DMEM, Dulbecco's modified Eagle's medium; TEMED, tetramethylethylenediamine; ERK, extracellular signal-regulated kinase; WT, wild type; LDL, low density lipoprotein.
References
- 1.McDonald, J., and Hascall, V. C. (2002) J. Biol. Chem. 277 4575-4579 [DOI] [PubMed] [Google Scholar]
- 2.McGary, C. T., Raja, R. H., and Weigel, P. H. (1989) Biochem. J. 257 875-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou, B., Weigel, J. A., Fauss, L. A., and Weigel, P. H. (2000) J. Biol. Chem. 275 37733-37741 [DOI] [PubMed] [Google Scholar]
- 4.Smedsrod, B., Malmgren, M., Ericsson, J., and Laurent, T. C. (1988) Cell Tissue Res. 253 39-45 [DOI] [PubMed] [Google Scholar]
- 5.Mellman, I. (1996) Annu. Rev. Cell Dev. Biol. 12 575-625 [DOI] [PubMed] [Google Scholar]
- 6.Weigel, P. H., and Yik, J. H. N. (2002) Biochim. Biophys. Acta 1572 341-363 [DOI] [PubMed] [Google Scholar]
- 7.Harris, E. N., Kyosseva, S. V., Weigel, J. A., and Weigel, P. H. (2007) J. Biol. Chem. 282 2785-2795 [DOI] [PubMed] [Google Scholar]
- 8.Harris, E. N., Weigel, J. A., and Weigel, P. H. (2004) J. Biol. Chem. 279 36201-36209 [DOI] [PubMed] [Google Scholar]
- 9.Tammi, R., Saamanen, A.-M., Maibach, H. I., and Tammi, M. (1991) Investig. Dermatol. 97 126-130 [DOI] [PubMed] [Google Scholar]
- 10.Fraser, J. R. E., Laurent, T. C., and Laurent, U. B. G. (1997) J. Intern. Med. 242 27-33 [DOI] [PubMed] [Google Scholar]
- 11.Fraser, J. R. E., Laurent, T. C., Pertoft, H., and Baxter, E. (1981) Biochem. J. 200 415-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou, B., Oka, J. A., Singh, A., and Weigel, P. H. (1999) J. Biol. Chem. 274 33831-33834 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, B., McGary, C. T., Weigel, J. A., Saxena, A., and Weigel, P. H. (2003) Glycobiology 13 339-349 [DOI] [PubMed] [Google Scholar]
- 14.Zhou, B., Weigel, J. A., Saxena, A., and Weigel, P. H. (2002) Mol. Biol. Cell 13 2853-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Politz, O., Gratchev, A., McCourt, P. A. G., Schledzewski, K., Guillot, P., Johansson, S., Svineng, G., Franke, P., Kannicht, C., Kzhyshkowska, J., Longati, P., Velten, F. W., and Goerdt, S. (2002) Biochem. J. 362 155-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clague, M. J., and Urbe, S. (2001) J. Cell Sci. 114 3075-3081 [DOI] [PubMed] [Google Scholar]
- 17.Sorkin, A. (2004) Curr. Opin. Cell Biol. 16 392-399 [DOI] [PubMed] [Google Scholar]
- 18.Benmerah, A., and Lamaze, C. (2007) Traffic 8 970-982 [DOI] [PubMed] [Google Scholar]
- 19.Mills, I. G. (2007) Semin. Cell Dev. Biol. 18 459-470 [DOI] [PubMed] [Google Scholar]
- 20.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 21.Raja, R. H., LeBoeuf, R. D., Stone, G. W., and Weigel, P. H. (1984) Anal. Biochem. 139 168-177 [DOI] [PubMed] [Google Scholar]
- 22.McGary, C. T., Weigel, J. A., and Weigel, P. H. (2003) Methods Enzymol. 363 354-366 [DOI] [PubMed] [Google Scholar]
- 23.Burnette, W. N. (1981) Anal. Biochem. 112 195-203 [DOI] [PubMed] [Google Scholar]
- 24.Weigel, P. H., Ray, D. A., and Oka, J. A. (1983) Anal. Biochem. 133 437-449 [DOI] [PubMed] [Google Scholar]
- 25.Weigel, P. H., McGary, C. T., and Weigel, J. A. (2003) Methods Enzymol. 363 382-391 [DOI] [PubMed] [Google Scholar]
- 26.McGary, C. T., Yannariello-Brown, J., Kim, D. W., Stinson, T. C., and Weigel, P. H. (1993) Hepatology 18 1465-1476 [PubMed] [Google Scholar]
- 27.Weigel, J. A., and Weigel, P. H. (2003) J. Biol. Chem. 278 42802-42811 [DOI] [PubMed] [Google Scholar]
- 28.Yannariello-Brown, J., Zhou, B., and Weigel, P. H. (1997) Glycobiology 7 15-21 [DOI] [PubMed] [Google Scholar]
- 29.Slepnev, V. I., and De Camilli, P. (2000) Nat. Rev. Neurosci. 1 161-172 [DOI] [PubMed] [Google Scholar]
- 30.Kirchhausen, T., Bonifacino, J. S., and Riezman, H. (1997) Curr. Opin. Cell Biol. 9 488-495 [DOI] [PubMed] [Google Scholar]
- 31.Altschuler, Y., Barbas, S. M., Terlecky, L. J., Tang, K., Hardy, S., Mostov, K. E., and Schmid, S. L. (1998) J. Cell Biol. 143 1871-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Praefcke, G. J., and McMahon, H. T. (2004) Nat. Rev. Mol. Cell Biol. 5 133-147 [DOI] [PubMed] [Google Scholar]
- 33.Brodsky, F. M., Chen, C. Y., Knuehl, C., Towler, M. C., and Wakeham, D. E. (2001) Annu. Rev. Cell Dev. Biol. 17 517-568 [DOI] [PubMed] [Google Scholar]
- 34.Chen, H., Fre, S., Slepnev, V. I., Capua, M. R., Takei, K., Butler, M. H., Di Fiore, P. P., and De Camilli, P. (1998) Nature 394 793-797 [DOI] [PubMed] [Google Scholar]
- 35.Benmerah, A., Begue, B., Dautry-Varsat, A., and Cerf-Bensussan, N. (1996) J. Biol. Chem. 271 12111-12116 [DOI] [PubMed] [Google Scholar]
- 36.Tebar, F., Sorkina, T., Sorkin, A., Ericsson, M., and Kirchhausen, T. (1996) J. Biol. Chem. 271 28727-28730 [DOI] [PubMed] [Google Scholar]
- 37.Wang, L.-H., Südhof, T. C., and Anderson, R. G. W. (1995) J. Biol. Chem. 270 10079-10083 [DOI] [PubMed] [Google Scholar]
- 38.Iacopetta, B. J., Rothenberger, S., and Kuhn, L. C. (1988) Cell 54 485-489 [DOI] [PubMed] [Google Scholar]
- 39.Boll, W., Ohno, H., Zhou, S. Y., Rapoport, I., Cantley, L. C., Bonifacino, J. S., and Kirchhausen, T. (1996) EMBO J. 15 5789-5795 [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno, H., Fournier, M. C., Poy, G., and Bonifacino, J. S. (1996) J. Biol. Chem. 271 29009-29015 [DOI] [PubMed] [Google Scholar]
- 41.Shiratori, T., Miyatake, S., Ohno, H., Nakaseko, C., Isono, K., Bonifacino, J. S., and Saito, T. (1997) Immunity 6 583-589 [DOI] [PubMed] [Google Scholar]
- 42.Perez, R. G., Soriano, S., Hayes, J. D., Ostaszewski, B., Xia, W. M., Selkoe, D. J., Chen, X. H., Stokin, G. B., and Koo, E. H. (1999) J. Biol. Chem. 274 18851-18856 [DOI] [PubMed] [Google Scholar]
- 43.Williams, M. A., and Fukuda, M. (1990) J. Cell Biol. 111 955-966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunziker, W., Harter, C., Matter, K., and Mellman, I. (1991) Cell 66 907-920 [DOI] [PubMed] [Google Scholar]
- 45.Marks, M. S., Woodruff, L., Ohno, H., and Bonifacino, J. S. (1996) J. Cell Biol. 135 341-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filardo, E. J., Brooks, P. C., Deming, S. L., Damsky, C., and Cheresh, D. A. (1995) J. Cell Biol. 130 441-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, W.-J., Goldstein, J. L., and Brown, M. S. (1990) J. Biol. Chem. 265 3116-3123 [PubMed] [Google Scholar]
- 48.Davis, C. G., Lehrman, M. A., Russell, D. W., Anderson, R. G., Brown, M. S., and Goldstein, J. L. (1986) Cell 45 15-24 [DOI] [PubMed] [Google Scholar]
- 49.Yun, M., Keshvara, L., Park, C. G., Zhang, Y. M., Dickerson, J. B., Zheng, J., Rock, C. O., Curran, T., and Park, H. W. (2003) J. Biol. Chem. 278 36572-36581 [DOI] [PubMed] [Google Scholar]
- 50.Keyel, P. A., Mishra, S. K., Roth, R., Heuser, J. E., Watkins, S. C., and Traub, L. M. (2006) Mol. Biol. Cell 17 4300-4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu, D., Knudson, P. E., Zapf, A., Rolband, G. C., and Olefsky, J. M. (1994) Endocrinology 134 744-750 [DOI] [PubMed] [Google Scholar]
- 52.Vignoud, L., Albiges-Rizo, C., Frachet, P., and Block, M. R. (1997) J. Cell Sci. 110 1421-1430 [DOI] [PubMed] [Google Scholar]
- 53.Hjalm, G., Murray, E., Crumley, G., Harazim, W., Lundgren, S., Onyango, I., Ek, B., Larsson, M., Juhlin, C., Hellman, P., Davis, H., Akerstrom, G., Rask, L., and Morse, B. (1996) Eur. J. Biochem. 239 132-137 [DOI] [PubMed] [Google Scholar]
- 54.Kawakami, K., Takeshita, F., and Puri, R. K. (2001) J. Biol. Chem. 276 25114-25120 [DOI] [PubMed] [Google Scholar]
- 55.Thomas, W. G., Baker, K. M., Motel, T. J., and Thekkumkara, T. J. (1995) J. Biol. Chem. 270 22153-22159 [DOI] [PubMed] [Google Scholar]
- 56.Li, Y., Marzolo, M. P., van Kerkhof, P., Strous, G. J., and Bu, G. (2000) J. Biol. Chem. 275 17187-17194 [DOI] [PubMed] [Google Scholar]
- 57.Tamura, Y., Adachi, H., Osuga, J., Ohashi, K., Yahagi, N., Sekiya, M., Okazaki, H., Tomita, S., Iizuka, Y., Shimano, H., Nagai, R., Kimura, S., Tsujimoto, M., and Ishibashi, S. (2003) J. Biol. Chem. 278 12613-12617 [DOI] [PubMed] [Google Scholar]
- 58.Raja, R. H., McGary, C. T., and Weigel, P. H. (1988) J. Biol. Chem. 263 16661-16668 [PubMed] [Google Scholar]
- 59.Rajasekaran, A. K., Humphrey, J. S., Wagner, M., Miesenbock, G., Le Bivic, A., Bonifacino, J. S., and Rodriguez-Boulan, E. (1994) Mol. Biol. Cell 5 1093-1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canfield, W. M., Johnson, K. F., Ye, R. D., Gregory, W., and Kornfeld, S. (1991) J. Biol. Chem. 266 5682-5688 [PubMed] [Google Scholar]
- 61.Jadot, M., Canfield, W. M., Gregory, W., and Kornfeld, S. (1992) J. Biol. Chem. 267 11069-11077 [PubMed] [Google Scholar]
- 62.Blot, V., and McGraw, T. E. (2006) EMBO J. 25 5648-5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris, E. N., Weigel, J. A., and Weigel, P. H. (2008) J. Biol. Chem. 283 17341-17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kyosseva, S. V., Harris, E. N., and Weigel, P. H. (2008) J. Biol. Chem. 283 15047-15055 [DOI] [PMC free article] [PubMed] [Google Scholar]