Abstract
Ubiquitin (Ub) is a sorting signal that targets integral membrane proteins to the interior of the vacuole/lysosome by directing them into lumenal vesicles of multivesicular bodies (MVBs). The Vps27-Hse1 complex, which is homologous to the Hrs-STAM complex in mammalian cells, serves as a Ub-sorting receptor at the surface of early endosomes. We have found that Hse1 interacts with Doa1/Ufd3. Doa1 is known to interact with Cdc48/p97 and Ub and is required for maintaining Ub levels. We find that the Hse1 Src homology 3 domain binds directly to the central PFU domain of Doa1. Mutations in Doa1 that block Hse1 binding but not Ub binding do not alter Ub levels but do result in the missorting of the MVB cargo GFP-Cps1. Loss of Doa1 also causes a synthetic growth defect when combined with loss of Vps27. Unlike the loss of Doa1 alone, the doa1Δ vps27Δ double mutant phenotype is not suppressed by Ub overexpression, demonstrating that the effect is not due to indirect consequence of lowered Ub levels. Loss of Doa1 results in a defect in the accumulation of GFP-Ub within yeast vacuoles, implying that there is a reduction in the flux of ubiquitinated membrane proteins through the MVB pathway. This defect was also reflected by an inability to properly sort Vph1-GFP-Ub, a modified subunit of the multiprotein vacuolar ATPase complex, which carries an inframe fusion of Ub as an MVB sorting signal. These results reveal novel roles for Doa1 in helping to process ubiquitinated membrane proteins for sorting into MVBs.
One of the key sorting steps in sending integral membrane proteins to the lysosome for degradation is their incorporation into vesicles that form from the limiting membrane of early endosomes and bud into the lumen. This budding process results in the formation of a multivesicular body or multivesicular endosome that fuses to lysosomes to deliver its intralumenal vesicles and membrane cargo proteins to the hydrolytic lysosomal lumen (1).
Integral membrane proteins gain access into the MVB sorting pathway by becoming attached to ubiquitin (Ub).3 Ub attachment serves as a sorting signal at the plasma membrane for internalization, at the endosome for incorporation into intralumenal vesicles that comprise MVBs, and at the trans-Golgi network, from where proteins are sorted into vesicles that are delivered to the forming MVB (1, 2). The processing and sorting of Ub cargo at the endosome is in part fulfilled by a set of proteins originally identified in yeast as class E Vps (vacuolar protein sorting) proteins. This set includes 18 proteins whose loss impairs lumenal vesicle formation and protein sorting and results in the accumulation of aberrantly large late endosomal (class E compartment) structures adjacent to the yeast vacuole (lysosome). The majority of the yeast class E proteins have clear functional homologs in mammalian cells and associate in distinct complexes, including ESCRT I, II, III, a Vps4 AAA ATPase complex, and the Vps27-Hse1 (has symptoms of class E) complex, whose mammalian ortholog is the Hrs-STAM complex (1, 2).
Several class E Vps protein components, including Vps27, Vps23 (ESCRT-I), and Vps36 (ESCRT-II), can noncovalently bind Ub via discrete functional domains and are thus believed to have a role in the recognition of Ub cargo. The Vps27-Hse1 complex largely localizes to endosomes via its association with phosphatidylinositiol 3-phosphate, clathrin, and ESCRT-I (3–6). Current models suggest that after Ub cargo is captured by the Vps27-Hse1 complex, it passes through a series of steps that concentrate cargo within areas that will undergo vesicle formation (1, 2). At a late point in the sorting process, Ub is removed from cargo by the deubiquitinating enzyme Ubp4 (ubiquitin peptidase 4)/Doa4 (degradation of MATα2) before incorporation into intralumenal vesicles of the MVB (7). Loss of DOA4 depletes cellular Ub levels, which causes a number of cellular defects, including loss of efficient MVB sorting of a number of integral membrane proteins (8–11). However, despite the wealth of structural data that describe the architecture of these protein complexes and how they interact, little is known about the exact sorting steps they perform or how they are regulated.
The Vps27-Hse1/Hrs-STAM complex has emerged as at least one important focal point for regulation. In mammalian cells, the Hrs-STAM complex interacts with numerous effectors of protein trafficking and is the target of receptor-tyrosine kinases, which can modulate their own down-regulation by tyrosine-phosphorylating Hrs (12–18). This complex also associates with deubiquitinating enzymes and Ub-ligases, and recent studies in yeast suggest that this association serves to regulate the ubiquitination status of membrane protein cargo to either reverse or reinforce cargo trafficking along the MVB pathway (19, 20).
To understand more about how MVB sorting is controlled, we searched for protein machinery that interacts with the Vps27-Hse1 complex and found Ufd3 (ubiquitin fusion degradation 3)/Doa1. Doa1 was first found in genetic screens for mutants defective in degrading model proteasome substrates (21–23). Loss of Doa1 causes many phenotypes in yeast, including sensitivity to DNA-damaging agents, cycloheximide, caffeine, cadmium, canavanine, growth at high temperature, and volatile anesthetics (24–31). Other analysis has shown that loss of Doa1 dramatically decreases Ub levels (21). Many of the phenotypes of doa1Δ mutants can be suppressed by overexpressing Ub, suggesting that many of the noted doa1Δ defects are solely an indirect consequence of lowered Ub levels (21, 26, 28, 31). It is not presently clear how Ub is depleted in the absence of Doa1; however, inhibiting delivery and degradation of ubiquitinated proteins to the proteasome suppresses doa1Δ growth defects, suggesting that more Ub may be degraded by the proteasome in the absence of Doa1 (24, 28, 31).
The N-terminal region of Doa1 contains a seven-bladed WD40 repeat β-propeller, which in general is thought to mediate protein-protein interactions (32). Doa1 also has two other regions dubbed the PFU and PUL domains, based on their shared sequence homology with Doa1 orthologs throughout eukaryota (27, 33). The central PFU domain of Doa1 mediates interaction with Ub, whereas the C-terminal PUL domain mediates interaction with Cdc48 (cell division cycle 48), an AAA ATPase belonging to a family of ATPases associated with various cellular activities (27, 28). Cdc48 (and its mammalian ortholog p97) forms a hexameric ring that acts as a molecular chaperone for a variety of ubiquitinated proteins (34–36). Cdc48 is organized into an N-terminal portion, followed by two AAA ATPase domains. Recent biochemical studies confirm that Cdc48 probably works as a “segregase” that can dissociate aggregates of ubiquitinated proteins, which would help convey them to the proteasome for degradation (37). Cdc48/p97 associates with a variety of “adaptor” proteins that help program it for various functions. Adaptor proteins, such as the UBX family of proteins, bind to the N-terminal portion of Cdc48 (36), whereas Doa1, the Ub E3/E4 ligase Ufd2, and peptide:N-glycanase associate with the C terminus (28, 35, 38). Although the role for some of these Cdc48/adpator complexes is known, little is known about the specific functions executed by a Doa1-Cdc48 complex.
Our analysis indicates that Doa1 plays a role in sorting Ub cargo into MVBs. Part of this activity is directed specifically by association with the Vps27-Hse1 complex, and this association is required for the efficient sorting of particular MVB substrates. However, Doa1 also appears to serve a more general role to help capture and process Ub cargo for concentration at endosomal subdomains and eventual incorporation into MVB lumenal vesicles.
EXPERIMENTAL PROCEDURES
Materials, Yeast Strains, and Plasmids—Synthetic dextrose (SD) medium was made using yeast nitrogen base containing ammonia and 2% glucose. Yeast nitrogen base was purchased from RPI Research Products International Corp. Amino acid supplements were purchased from Bio 101, Inc. (La Jolla, CA). Glutathione-agarose beads were purchased from GE Healthcare. Zymolyase 100T was purchased from Seikagaku Corp. (East Falmouth, MA). Protease inhibitor mixture (Complete™) was purchased from Roche Applied Science. Endocytic tracer dye FM4-64 was purchased from Molecular Probes, Inc. (Eugene, OR). The pCR2.1, pYES2.1, and pET151 TOPO cloning kits were purchased from Invitrogen. Anti-Ub monoclonal antibody P4D1 was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal anti-CPY 10A5-B5 was a kind gift from Tom Stevens (University of Oregon). Anti-HA antibody was purchased from Covance Research Products, Inc. (Berkeley, CA); anti-V5 was from Invitrogen; anti-GFP antibody was from Clontech; and horseradish peroxidase-linked secondary antibodies were from Amersham Biosciences.
Saccharomyces cerevisiae strains used in this study are listed in Table 1. The parental strain was BY4742 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). Gene disruptions were performed by replacing the entire open reading frame with the indicated selectable marker. For disruptions using the Kanr marker, the disruption cassette was amplified from the genomic DNA of the relevant strain from the yeast gene deletion project (39). For disruptions using the URA3 or HIS3 marker, a disruption cassette was made by amplifying the URA3 or His5 gene from Kluyveromyces lactis using loxP-flanked cassettes (40) with oligonucleotides containing 50 bp of flanking DNA homologous to the insertion site.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| SF838-9D | MATα leu2-3,112 ura3-52 his4-519 ade6 pep4-3 | Ref. 69 |
| BY4742 | MATα his3 leu2 lys2 ura3 | Ref. 70 |
| PLY2498 | MATα hse1Δ::Kanr his3, leu2, lys2, ura3 | Yeast Gene Deletion Project |
| PLY3705 | MATα otu1Δ::Kanr his3, leu2, lys2, ura3 | Yeast Gene Deletion Project |
| PLY3706 | MATα ufd2Δ::Kanr his3, leu2, lys2, ura3 | Yeast Gene Deletion Project |
| PLY3704 | MATα ubp6Δ::Kanr his3, leu2, lys2, ura3 | Yeast Gene Deletion Project |
| PLY2490 | MATα vps23Δ::Kanr his3, leu2, lys2, ura3 | Yeast Gene Deletion Project |
| PLY3709 | MATα doa1Δ::HIS3 his3, leu2, lys2, ura3 | This study |
| PLY3462 | MATα doa1Δ::Kanr his3, leu2, lys2, ura3 | This study |
| PLY3694 | MATα doa1Δ::Kanr vps27Δ::LEU2 leu2 lys2, ura3 | This study |
| PLY2498 | MATα hse1Δ::URA3 his3, leu2, lys2, ura3 | Ref. 20 |
| PLY3175 | MATα vps27Δ::Kanr leu2 lys2, ura3 | Ref. 20 |
| PLY3556 | MATα doa1Δ::HIS3 vps27Δ::URA3 leu2 lys2, ura3 | This study |
| PLY3700 | MATα vps27Δ::LEU2 ubp6Δ::Kanr his3, leu2, lys2, ura3 | This study |
| PLY3699 | MATα vps27Δ::LEU2 otu1Δ::Kanr his3, leu2, lys2, ura3 | This study |
| PLY3702 | MATα vps27Δ::LEU2 ufd2Δ::Kanr his3, leu2, lys2, ura3 | This study |
| PLY3707 | MATα doa1Δ::HIS3 ufd2Δ::Kanr his3, leu2, lys2, ura3 | This study |
| PLY3711 | MATα vps27Δ::LEU2 doa1Δ::HIS3 ufd2Δ::Kanr his3, leu2, lys2, ura3 | This study |
The Doa1-V5 expression plasmids used for SH3 binding experiments were made by PCR and subcloning into the pYES2.1-TOPO vector. This vector inserts a V5 and His6 epitope tag on the C-terminal end of the open reading frame and drives expression via the GAL1 promoter. To make the doa1Δ1Hse1 open reading frame, an overlapping PCR strategy was used to replace residues 434–436 and 438–440 with alanines. The doa1Δ2Hse1 open reading frame was made similarly but deleting residues 434–443. The Doa1-ΔPP open reading frame was made by replacing the P399PLKLP motif with A399ALKLA. For bacterial expression of Doa1 residues 1–433 and 1–445, Doa1 fragments were expressed from pYES2.1 expression plasmids in BL21(DE3) bacteria (Invitrogen) due to an upstream T7 promoter capable of driving T7 RNA polymerase-dependent transcription of downstream open reading frames. For expression of isolated PFU domains, PCR fragments encoding the region from residue 301 (which defines the end of the N-terminal β-propeller region) to residue 465 were amplified from wild type DOA1 and the doa1Δ1Hse1 and doa1Δ2Hse1 alleles and expressed from pET151 (Invitrogen), which imparted an N-terminal V5 epitope tag (Invitrogen). For functional studies in yeast, wild type V5 epitope-tagged DOA1 as well as V5 epitope-tagged doa1Δ1Hse1 and doa1Δ2Hse1 open reading frames were cloned into low copy centromere-based plasmids (pRS316). The open reading frames were flanked on the 5′-end by 402 bp of the endogenous DOA1 5′-untranslated region and on the 3′-end with 846 bp of the PHO8 3′-untranslated region. The 2μ UBI4 gene was recovered in this study in a search for multicopy suppressors of doa1Δ mutants using a high copy plasmid library housed in YEp351 and containing genomic DNA isolated from the SEY6210 parental strain. The Ste3-mCherry expression plasmid was made by replacing GFP from p865 encoding Ste3-GFP (41) with mCherry (42). Switching nutritional markers between plasmids was done by using yeast recombination by co-transforming the targeted plasmid with PCR fragments encoding the desired nutritional marker gene, which contained 40 bp of sequence complementary to the insertion site. Table 2 lists the source of other plasmids used in this study.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference/Source |
|---|---|---|
| pGO45 | pRS426 carrying GFP-CPS1 | Ref. 71 |
| pPL1867 | LEU2 conversion of pGO45 | This study |
| pPL3267 | HA-Ub-GFP-CPS1 in pRS316 | Ref. 20 |
| pPL3453 | HA-Ub-GFP-CPS1 in pRS315 | This study |
| pPL1124 | pRS316 containing dominant negative VPS4 | Ref. 20 |
| pGEX-6P-1 | GST expression vector | GenBank™ accession number U78872 |
| pPL2710 | GST-Hse1-SH3 | Ref. 20 |
| pPL2831 | GST-Pex13-SH3 | Ref. 20 |
| pPL3164 | GST-Hse1-SH3*. GST-Hse1-SH3 plasmid containing in which the codons encoding W254A and W255A mutations. | Ref. 20 |
| pPL1978 | GST-Vps27 C terminus | Ref. 3 |
| pPL833 JLU34 | STE3-GFP | Ref. 41 |
| pPL3607 | STE3-mCherry. Derived from pPL833. | This study |
| pGFP-Ub | GFP-Ub expressed from the PRC1 promoter in pRS426. | D. Katzmann Mayo Clinic (Rochester, MN) |
| pPL3601 | LEU2 marker convert of pGFP-Ub. | This study |
| pPL3155 | YEp351 carrying UBI4. | This study |
| pPL3327 | DOA1-GFP downstream of PRC1 promoter and upstream of the PHO8 3′-UT in pRS316. | This study |
| pPL2794 | GAL1 driving full-length DOA1 tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3270 | GAL1 driving truncated DOA1 (residues 1–300) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3273 | GAL1 driving truncated DOA1 (residues 1–470) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3359 | GAL1 driving truncated DOA1 (residues 1–414) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3363 | GAL1 driving truncated DOA1 (residues 1–433) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3341 | GAL1 driving truncated DOA1 (residues 1–445) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3323 | GAL1 driving a fragment of DOA1 (residues 301–470) tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1. | This study |
| pPL3300 | GAL1 driving DOA1 tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1 Contains mutations p399PLKP → A399ALKA. | This study |
| pPL3367 | GAL1 driving DOA1 tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1 Contains mutations F434ILKNTN → A434AAKAAA to make doa1Δ1Hse1. | This study |
| pPL3371 | GAL1 driving DOA1 tagged C-terminally with V5 and His6 tag in 2μ plasmid pYES2.1 Contains deletion of residues 433–445 to make doa1Δ2Hse1. | This study |
| pPL2967 | Expression plasmid for PFU domain from wild type DOA1. Residues 301–466 with N-terminal V5 epitope tag in pET151. | This study |
| pPL3604 | Expression plasmid for PFU domain from Doa1Δ1Hse1. Fragment derived from pPL3367 (F434ILKNTN → A434AAKAAA) residues 301–466 with N-terminal V5 epitope tag in pET151. | This study |
| pPL3603 | Expression plasmid for PFU domain from Doa1Δ2Hse1. PFU domain from pPL3371 (deletion of residues 433–445) with N-terminal V5 epitope tag in pET151. | This study |
| pPL3498 | DOA1-V5 expressed from DOA1 promoter in pRS316. | This study |
| pPL3499 | Doa1Δ1Hse1 (F434ILKNTN → A434AAKAAA) expressed from DOA1 promoter in pRS316. | This study |
| pPL3501 | Doa1Δ2Hse1 (deletion of residues 433–445) expressed from DOA1 promoter in pRS316. | This study |
| pUG72 | Plasmid containing gene disruption cassettes with URA3 (K. lactis) as selection marker. | Ref. 40 |
| pUG27 | Plasmid containing gene disruption cassettes with his5+ (S. pombe) as selection marker. | Ref. 40 |
| pPL2161 | HA-tagged HSE1 in pRS315 | Ref. 3 |
| pPL1124 | VPS4 dominant negative in pRS426 | Ref. 20 |
| pGEX-6P-1 | GST expression vector | GenBank™ accession number U78872 |
| pPL1556 | VPH1-GFP-Ub in pRS316 | Ref. 41 |
| pYES2.1 | High copy (2μ) URA3-containing GAL1 expression plasmid. | Invitrogen |
| pET151 | T7 promoter-based bacterial expression vector. | Invitrogen |
Glutathione-Agarose Affinity Chromatography—GST-fusion proteins were isolated from bacteria using glutathione-Sepharose beads as previously described (43). For binding studies, 250 μg of each isolated GST fusion protein was bound to 50 μl of glutathione-Sepharose in phosphate-buffered saline by rotation for 30 min at 25 °C. Bound GST or GST fusion proteins were then pelleted and washed three times with phosphate-buffered saline. Cell lysate was added to each protein-bead complex and incubated for 2 h at 4 °C. Unbound proteins were removed from beads using four washes of IC buffer (100 mm KAc, 50 mm KCl, 200 mm sorbitol, 20 mm PIPES, pH 6.8), and the bound bead fraction was analyzed by SDS-PAGE and immunoblotting. Yeast lysates were prepared from spheroplasts as previously described (3). Proteins from bacterial lysates for binding studies were prepared as previously described (3).
Fluorescence Microscopy—Cells containing GFP-expressing plasmids were grown in SD medium to midlog phase. Live cells were viewed using an Olympus BX-60 microscope equipped with fluorescein isothiocyanate filters and Nomarski/differential interference contrast (DIC) optics. Images were captured with a Hammamatsu ORCA CCD camera, as previously described (44). GFP-Cps1 (carboxypeptidase 1) was visualized in cells resuspended in 0.2% NaN3, 100 mm Tris, pH 8.0.
Growth Assays—Yeast were first grown in SD medium overnight and serially (1:5) diluted and plated onto an SD plate containing a full complement of nutrients. Plates were grown for 2–3 days at various temperatures.
Western Blotting—Cellular extracts for immunoblotting were made as described (45). Briefly, cells were pelleted, resuspended in 0.1 m NaOH, and incubated for 5 min. Cells were repelleted and resuspended in 5% SDS, 8 m urea, 10% glycerol, 50 mm Tris, pH 6.8, with or without the addition of 2% β-mercaptoethanol. Immunoblots were visualized with horseradish peroxidase-conjugated secondary antibodies and chemiluminescence reagents (Pierce) either indirectly with exposure to film or by capturing luminescent images with a Fluorochem IS-8000 (Alpha Innotech, San Leandro, CA).
RESULTS
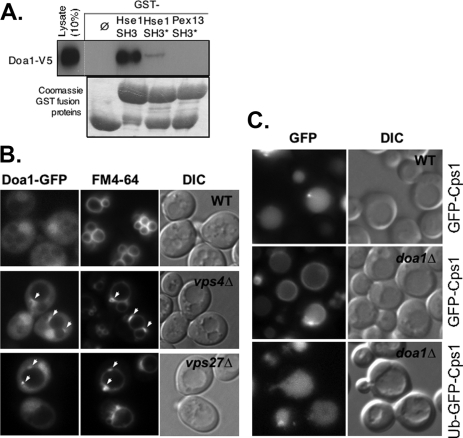
Doa1 Binds Hse1 and Is Required for Sorting MVB Substrates—Previous large scale two-hybrid screens identified Doa1 as potentially interacting with the SH3 domain of Hse1 (46). To confirm this interaction, we produced a C-terminally V5 epitope-tagged Doa1 in yeast and subjected the corresponding lysates to GST pull-down experiments using the SH3 domain of Hse1. Fig. 1A shows that the Doa1 specifically bound the Hse1 SH3 domain. However, Doa1 did not bind the SH3 domain of Pex13. Furthermore, binding of Doa1 was dramatically reduced when a critical tryptophan residue within the predicted hydrophobic binding surface of the Hse1 SH3 domain was altered to alanine.
FIGURE 1.
Association of Doa1 with the MVB sorting machinery. A, lysate from yeast expressing V5 epitope-tagged Doa1 was passed over beads bound with GST only (ø) or GST fused to the SH3 domain of Hse1 (Hse1 SH3), a mutant form of the SH3 domain (Hse1 SH3*), or the SH3 domain from Pex13 (Pex13 SH3). Also shown are the GST fusion proteins used in this analysis. B, Doa1-GFP (expressed from pPL3327) was localized in wild type cells (WT), vps4Δ cells, and vps27Δ cells. Cells were counterlabeled with the endocytic tracer dye FM4-64. C, GFP-Cps1 was correctly localized to the vacuole lumen in wild type cells but not doa1Δ mutant cells, the latter of which showed accumulation of GFP-Cps1 at the limiting membrane of the vacuole. The lower panel shows sorting of Ub-GFP-Cps1 to the vacuole lumen in doa1Δ cells. Also shown are corresponding DIC images.
We next asked whether Doa1 could localize to endosomes. To image Doa1, we fused GFP to its C terminus. The strategy was used previously in a genome-wide effort to C-terminally GFP-tag all yeast open reading frames systematically (47). We first verified that the DOA1-GFP-tagged strain from this collection, in which the only copy of Doa1 is fused to GFP, was able to grow like wild type cells at 37 °C, indicating that the Doa1-GFP was functional (data not shown). We then drove expression of Doa1-GFP from the moderately strong PRC1 promoter, which normally drives the expression of vacuolar carboxypeptidase Y. In wild type cells, Doa1-GFP was localized diffusely to the cytosol, was excluded from the vacuoles, and was slightly concentrated in the nucleus, consistent with previous observations (47). We then localized Doa1-GFP in mutants lacking Vps4, a class E Vps protein whose loss results in the accumulation of aberrantly large endosomes (class E compartments), which can trap a variety of proteins that transiently associate with endosomes (48, 49). Here we saw colocalization of Doa1-GFP within membrane patches adjacent to the vacuole that also accumulated the endocytic tracer dye FM4-64 (Fig. 1B). These data indicated that Doa1-GFP was capable of localizing to endosomes.
We also tested whether loss of DOA1 caused defects in MVB sorting. For this, we used two MVB marker proteins. The first was GFP-tagged Cps1, a vacuolar protease that gains access to the vacuole lumen via the MVB pathway (50). GFP-Cps1 is a Type I membrane protein containing GFP fused to its cytosolic N terminus. Cps1 is ubiquitinated by Rsp5 and Tul1 and is then sorted by the class E Vps machinery into the vacuole lumen (51, 52). The second cargo was Ub-GFP-Cps1, in which GFP-Cps1 is translationally fused to Ub (20). This cargo behaves similarly to GFP-Cps1, except that it does not require Rsp5-dependent ubiquitination to undergo MVB sorting, since it contains its Ub sorting motif constitutively (51). Fig. 1C shows that although wild type cells efficiently sorted GFP-Cps1 into the vacuole lumen, doa1Δ cells did not and accumulated GFP-Cps1 on the limiting membrane of the vacuole. In contrast, Ub-GFP-Cps1, which does not require ubiquitination to undergo Ub-dependent MVB sorting, was sorted correctly to the vacuolar lumen in doa1Δ cells.
These results indicated that Doa1 might indeed provide a function with regard to MVB sorting, presumably by promoting the processing and ubiquitination of cargo proteins like GFP-Cps1. The major caveat to this interpretation is that doa1Δ mutants have lowered levels of Ub, which occurs through an unknown mechanism (21). Lower Ub levels could indirectly affect a number of processes. Therefore, we went on to further characterize the specificity of these effects and examine the functional relevance of the Hse1-Doa1 interaction.
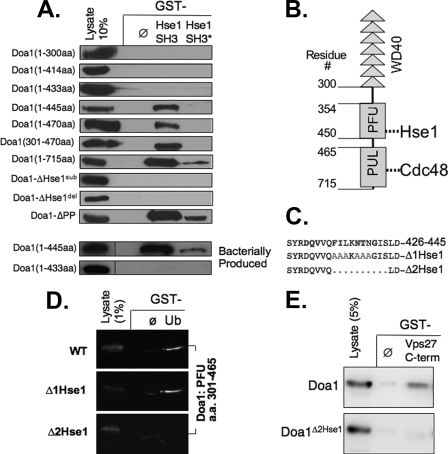
Doa1 Contains a Novel Binding Motif for the Hse1 SH3 Domain—Previous studies indicated that the SH3 domain of Hse1 was the relevant portion of Hse1 that could interact with Doa1 (46). Our previous studies showed that this domain is critical for Hse1 functions but also mediates interactions with other proteins, including Ubp7 and the Rsp5-Ubp2-Rup1-Hua1 complex (20). Thus, phenotypic analysis of Hse1 lacking its SH3 domain would not specifically ablate the Hse1-Doa1 interaction. Therefore, we undertook a series of mapping experiments to find the motif within Doa1 that mediates its interaction with the Hse1 SH3 domain. Fig. 2A shows a set of Doa1 deletion and substitution mutants subjected to GST pull-down experiments using GST alone or GST fused to the SH3 domain of Hse1. As a further control, we also used GST fused to the SH3 domain of Hse1 in which the predicted SH3 ligand interaction surface was mutated (20, 53). We found that full-length Doa1 and Doa1 lacking its C-terminal 270 residues (Doa1 1–445) also specifically bound the Hse1 SH3 domain. Since SH3 domains typically bind to proline-containing motifs, we next mutated the proline residues in the P399P340LKP343 motif, which conforms to a canonical polyproline SH3 ligand. Surprisingly, loss of these prolines had no effect on the interaction of Doa1 with the Hse1 SH3 domain. Further truncation of Doa1 1–433 abolished association with the Hse1 SH3 domain. We then inspected this region (residues 433–445) for residues that were conserved among other yeast species and found the motif F434IXKNTXG441 and either deleted it entirely to make the doa1Δ2Hse1 mutant protein or replaced it with A434AAkAAAG441 to make the doa1Δ1Hse1 mutant. Both of these mutations in the context of full-length Doa1 were no longer able to interact with the Hse1 SH3 domain.
FIGURE 2.
Defining the interaction between Hse1 and Doa1. A, the indicated V5 epitope-tagged fragments of Doa1 were expressed in yeast under the control of the GAL1 promoter, and the corresponding lysates were passed over glutathione-agarose beads bound with GST alone (ø) GST fused to the SH3 domain of Hse1 (Hse1 SH3), or a mutant form of the SH3 domain (Hse1 SH3*). Also shown are full-length Doa1 proteins with the following mutations: Doa1Δ1Hse1 (A434AAKAAA; pPL3367), Doa1Δ2Hse1 (deletion of residues 434–445; pPL3371), and Doa1-ΔPP (A399ALKA; pPL3300). The lower panels show the same analysis of indicated V5 epitope-tagged fragments of Doa1 produced in bacteria. B, schematic of Doa1 showing the N-terminal β-propeller region encoded by seven WD40 repeats, the central PFU domain that contains the SH3-binding motif, and the C-terminal PUL domain that interacts with Cdc48. C, protein sequence of residues 426–445 of Doa1. Conserved residues are shown in boldface type. The alanine substitutions of the Doa1Δ1Hse1 mutant (gray) and the deleted region of the Doa1Δ2Hse1 mutant are shown. D, the V5 epitope-tagged PFU domain derived from wild type (WT) Doa1, the Doa1Δ1Hse1, and the Doa1Δ2Hse1 mutants were produced in bacteria using pPL2967, pPL3604, and pPL3603, respectively. Lysates were passed over beads bound to GST alone (ø) or Ub-GST (Ub). Beads were washed and immunoblotted together with a 1% equivalent of input lysate. Chemiluminescence images were collected directly using a digital camera. E, yeast lysates expressing V5 epitope-tagged wild type Doa1 (pPL2794) or the Doa1Δ2Hse1 mutant (pPL3371) were passed over beads bound with GST alone (ø) or GST fused to the C terminus of Vps27. Samples were immunoblotted with a 5% equivalent of input lysate.
Since our mapping analysis was performed by expressing Doa1 mutants in yeast, we then performed experiments to show that the interaction with the Hse1 SH3 domain was direct and independent of other yeast proteins. We were unable to produce full-length Doa1 in bacteria; however, we were able to produce Doa1 truncation mutants encompassing residues 1–433 and 1–445. Consistent with the above mapping experiments, we found that bacterially produced recombinant Doa1 1–445 was able to interact with the Hse1 SH3 domain, whereas the Doa1 1–433 (lacking the F434IXKNTXG motif) was unable to interact.
The novel SH3 ligand that we found lies within central PFU domain of Doa1. Previous experiments have shown that this domain is required for binding Ub (27, 28). These conclusions were based on deletion analysis showing that both N-terminal and C-terminal fragments of Doa1 could bind to Ub, provided they contain the central PFU domain. However, these studies did not show that the PFU domain itself was sufficient for Ub binding. The PFU domain is not homologous to any other described domain, and the structural basis for how the PFU contributes to Ub binding is unknown. Importantly, Mullally et al. (27) found that when two conserved phenylalanine residues in the PFU domain were altered to alanine, both Ub binding and known Doa1-dependent functions were abolished. These residues were Phe426 and Phe434, the latter of which was altered in both of our Doa1ΔHse1 mutants. Therefore, we wanted to determine if the mutations that abolished SH3 binding had an effect on Ub binding. Fig. 2D shows that bacterially produced V5 epitope-tagged PFU domain alone was able to bind to GST-Ub, demonstrating that this domain is sufficient for Ub binding. Furthermore, the PFU domain from the doa1Δ1Hse1 mutant was able to bind GST-Ub to the same level. In contrast, the PFU domain from the doa1Δ2Hse1 mutant lacking residues 434–443 did not bind GST-Ub. Thus, we have been able to separate the motifs within the PFU Doa1 required for Ub binding and for binding the Hse1 SH3 domain.
To confirm the interaction of Doa1 with Hse1, we exploited our previous observation that previously showed that Hse1 associates with the C-terminal residues (positions 353–622) of Vps27 (3). Therefore, we used GST alone and GST fused to the Vps27 C terminus to see if Doa1 expressed in yeast lysates could indeed associate with a Vps27-Hse1 complex. Fig. 2E shows that although Doa1 could specifically associate with GST-Vps27, the mutant Doa1Δ2Hse1 protein could not.
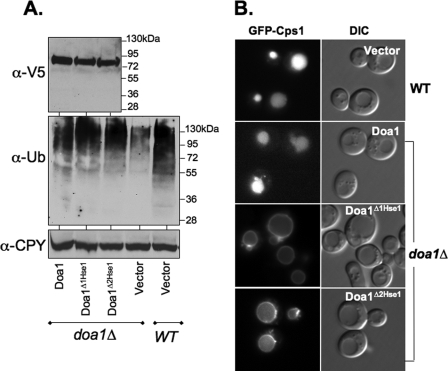
MVB Sorting Defects Caused by Loss of Doa1-Hse1 Association—We next functionally characterized the Doa1Δ1Hse1 and Doa1Δ2Hse1 mutants. Wild type V5 epitope-tagged DOA1 as well as the doa1Δ2Hse1 and doa1Δ1Hse1 alleles were cloned into low copy centromere-based plasmids under the control of the DOA1 promoter and transformed into doa1Δ mutants. Fig. 3A shows that the expression level of the three proteins was comparable, indicating that the overall integrity of the proteins was intact. We next examined Ub levels within these transformants by immunoblotting cell extracts with anti-Ub antibodies and examining the level of Ub conjugates. Consistent with previous studies, loss of Doa1 significantly reduced the level of Ub (21, 27, 31). Using our particular lysis protocol, we found very little unconjugated Ub overall, making the comparison of Ub conjugates more informative for assessing the overall level of Ub. Ub levels were restored with the V5-tagged DOA1 as well as the V5 epitope-tagged doa1Δ1Hse1 and doa1Δ2Hse1 alleles. These data showed that association of Doa1 with the Hse1 SH3 domain was not important for maintaining Ub levels. Furthermore, they suggested that loss of Ub binding by the PFU domain of Doa1 also did not significantly alter cellular Ub levels.
FIGURE 3.
Functional analysis of Doa1 mutants. A, mutant Doa1Δ cells were transformed with low copy plasmids expressing wild type (WT) V5 epitope-tagged Doa1, Doa1Δ1Hse1, Doa1Δ2Hse1 mutants, or vector alone (pPL3498, pPL3499, pPL3501, and pRS316, respectively). Cell lysates were immunoblotted for Doa1 with anti-V5. These cell lysates in addition to lysates from wild type cells were immunoblotted with anti-Ub antibodies to visualize Ub conjugates. Lysates were also immunoblotted for CPY as a loading control. B, localization of GFP-Cps1 in wild type cells and in doa1Δ mutant cells transformed with low copy plasmids expressing V5 epitope-tagged Doa1, Doa1Δ1Hse1, or Doa1Δ2Hse1.
We then examined the effect the doa1ΔHse1 alleles had on sorting GFP-Cps1. Fig. 3B shows that although cells expressing the wild type V5 epitope-tagged Doa1 sorted GFP-Cps1 into the vacuole normally, cells expressing either of the doa1ΔHse1 alleles were defective and accumulated GFP-Cps1 on the limiting membrane of the vacuole. Thus, the persistence of MVB sorting defects in the doa1ΔHse1 cells despite the normal complement of cellular Ub indicated that the Hse1-Doa1 interaction fulfilled a specific function.
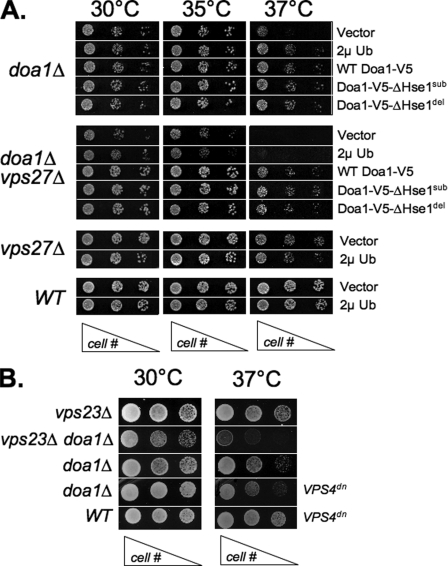
To further confirm that the doa1ΔHse1 alleles could fulfill other Doa1 functions, we tested them for their ability to complement the temperature-sensitive growth defect of doa1Δ mutants. Fig. 4A shows that doa1Δ cells grow poorly at elevated temperature (37 °C), consistent with previous studies. Normal growth was restored by overexpressing Ub using a 2μ multicopy plasmid carrying the UBI4 gene, which encodes a tandem fusion of 5 Ubs. Furthermore, normal growth was also restored by the V5 epitope-tagged wild type DOA1 as well as both doa1ΔHse1 alleles. Thus, these data show that we were able to genetically uncouple the role of Doa1 on Ub levels and generalized stress intolerance from its effect on MVB sorting, which supports the idea that Doa1 may be directly involved in some aspect of MVB sorting.
FIGURE 4.
Complementation of doa1Δ mutants with doa1ΔHse1 alleles. A, the indicated yeast strains were transformed with low copy vector plasmid, low copy plasmids expressing the wild type (WT) and the indicated doa1 alleles, or a high copy plasmid expressing Ub (2μUb; pPL3155). Serial dilutions of cells were plated onto minimal medium plate and grown at the indicated temperature for 2–3 days. The genotype of the yeast is indicated on the left, and plasmids expressing Ub, DOA1 alleles, or vector alone are indicated on the right. B, the synthetic growth defect of the doa1Δ mutation was assessed in combination with vps23Δ mutations or in the presence of a plasmid expressing a dominant-negative VPS4dn allele (pPL1124).
DOA1 Interacts Genetically with VPS27—Although the above experiments indicated that Doa1 performed a specific role by its association with Hse1, we also wondered whether Doa1 might fulfill a more general role in the processing and/or sorting of Ub cargo at the MVB. Indeed, although we observed Doa1-GFP localization to class E compartment endosomes in vps4Δ mutants, we also observed some localization to endosomes in vps27Δ mutants (Fig. 1B). Our previous studies showed that Hse1 associates with class E compartment endosomes by virtue of its association with Vps27 (54). Thus, the fact that Doa1-GFP could still associate with endosomes in the absence of Vps27-Hse1 on endosomes implied that Doa1 can use other means to associate with endosomes and thus perform functions independent of its association with Hse1.
Further genetic analysis revealed that loss of Vps27 in doa1Δ cells caused a severe growth phenotype (Fig. 4A). This was consistent with the idea that Doa1 function helped protect cells from the potentially toxic effects that cells might otherwise endure when MVB formation and degradation of integral membrane proteins in the vacuole is compromised. Importantly, this synthetic effect was not suppressed by overexpressing Ub, suggesting that the synthetic effect reflected a direct contribution of Doa1 function and not an indirect effect of depleted Ub levels compromising the health of vps27Δ mutants. As expected, transforming the doa1Δ vps27Δ double mutant strain with either wild type V5 epitope-tagged DOA1 or the two doa1ΔHse1 alleles restored growth at high temperatures, indicating that even under these more stringent conditions, these mutant alleles largely complemented the functions of DOA1 related to stress tolerance. Furthermore, they demonstrated that the synthetic effect we observe between doa1Δ and vps27Δ reflects a function outside of the interaction between DOA1 and HSE1. We also found that the loss of Vps23, another class E Vps protein that is part of the ESCRT-I complex, as well as expression of dominant negative VPS4 caused a similar synthetic growth defect in combination with loss of Doa1 (Fig. 4B).
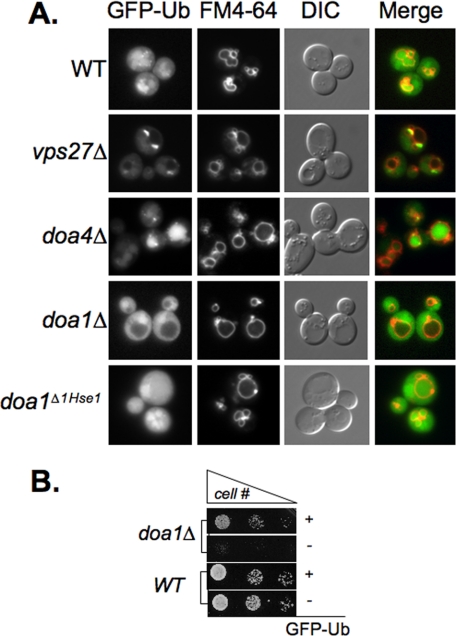
Loss of Doa1 Alters the Flux of Cellular Ubiquitin—The specific synthetic interaction between doa1Δ and class E vps mutants that fail to sort proteins into the MVB prompted us to examine other aspects for how Doa1 could contribute to the process of lysosomal degradation. Our previous experiments focused on a subset of MVB marker proteins, such as GFP-Cps1, which represent an idealized model MVB substrate, which accesses in the MVB pathway as part of its normal mechanism for delivering itself to the vacuole lumen, where it functions (50). Likewise, other MVB marker proteins that are widely used, such as the transporters Gap1, Fur4, and Ste6, as well as the G-protein-coupled receptors Ste3 and Ste2 become rapidly ubiquitinated as part of their natural strategy for down-regulation (55). Moreover, none of these proteins is thought to access the MVB degradative pathway, because they are misfolded or damaged, as would be the case for a proportion of a wide variety of cell surface proteins that might undergo a small but steady rate of vacuolar degradation. Thus, to get a better indication on the flux of Ub cargo in general, we examined the localization of GFP-tagged Ub, which has been used in other systems to monitor the use and distribution of cellular Ub (56, 57). The GFP-Ub construct we used contained GFP fused to the N terminus of wild type Ub replete with its normal C-terminal diglycine tail required for covalent modification of proteins. Production of GFP-Ub was via the moderately strong PRC1 promoter, which was housed on a low copy centomere-based plasmid. Fig. 5B shows that expression of GFP-Ub completely suppressed the temperature-sensitive growth defect of doa1Δ cells, demonstrating that GFP-Ub can functionally substitute for Ub to at least some degree.
FIGURE 5.
Visualization of Ub distribution. A, wild type cells (WT) along with the indicated mutant strains were transformed with low copy plasmid expressing GFP-Ub (pGFP-Ub) and imaged. Cells were co-labeled with the endocytic tracer dye FM4–64. Shown are the green channel (GFP-Ub), red channel (FM4–64), DIC image, and a merge of the fluorescence signals. Also shown are doa1Δ mutant yeast transformed with the doa1Δ1Hse1 allele. B, wild type and doa1Δ mutant cells were transformed with vector only or the GFP-Ub expression plasmid. Serial dilutions of cells were plated and grown at 37 °C for 2 days.
We then examined the distribution of GFP-Ub in wild type and mutant cells. Fig. 5A shows that in wild type cells, GFP-Ub was localized diffusely to the cytosol but could also be found accumulated within the vacuole. These data are consistent with previous studies that showed that although the bulk of Ub is thought to be removed from cargo before its irreversible sorting into the MVB interior, some of it persists and is ultimately delivered to the vacuole interior (7, 8, 54, 58, 59). The significant level of GFP-Ub in the vacuoles of wild type cells implies that the MVB pathway may play an important role in regulating the overall Ub levels within cells. Previous studies on the deubiquitinating enzyme Doa4 have supported a role for this enzyme in the latter stages of sorting Ub cargo into MVB vesicles by removing Ub from MVB cargo (8, 58, 59). Using GFP-Ub, we found direct data supporting this model, since doa4Δ mutants showed a massive accumulation of GFP-Ub within the yeast vacuole with concomitant depletion of Ub in the cytosol. We next examined the distribution of GFP-Ub in vps27Δ cells, which accumulate large endosomes as well as cargo that would otherwise be delivered to the vacuolar lumen. In these mutants we found that the large “class E compartment” endosomes, marked by the endocytic tracer dye FM4-64, also accumulated high levels of GFP-Ub. These data indicated that in vps27Δ mutants, Ub cargo becomes highly concentrated within endosomes and remains ubiquitinated. These results are consistent with similar analysis in mammalian cells, where depletion or disruption of Class E Vps protein function blocks lysosomal degradation of ubiquitinated membrane proteins and leads to the accumulation of ubiquitinated proteins on endosomes (60). Together, these data demonstrated that the GFP-Ub fusion protein reflected the expected distribution of Ub well and supported its use to monitor Ub distribution. A major difference was found when we examined the distribution of GFP-Ub in doa1Δ cells. Here we found that GFP-Ub was excluded from the vacuole, indicating that the bulk of Ub cargo that would otherwise be delivered to the vacuole was blocked by the absence of Doa1. Importantly, these experiments were done where GFP-Ub was exogenously expressed under conditions that suppress the temperature-sensitive growth defect of doa1Δ cells and thus probably do not reflect the nonspecific effect of Ub depletion. The effect of doa1Δ on Ub was not, however, observed in cells expressing the doa1Δ1Hse1 protein, which is unable to associate with Hse1. This is consistent with the idea that the global effects of Doa1 with regard to the overall distribution and flux of GFP-Ub into the vacuole reflect a function independent of its association with Hse1.
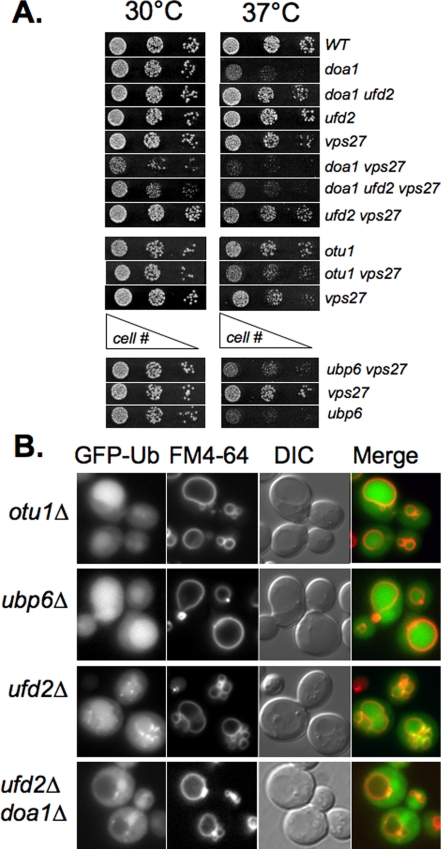
Effects of Doa1 Are Independent of Ufd2 and Otu1—Although Doa1 has been implicated in Ub-dependent processes, specific functions for Doa1 have been difficult to determine. Recent studies have implicated various functions for Doa1 genetically with Cdc48 and its associated co-factors Otu1 and Ufd2 (28). For instance, overexpression of Otu1 suppresses the growth defects of doa1Δ mutants, as does deletion of Ufd2. Otu1 is a Ub-specific cysteine protease, which is a member of the ovarian tumor family of deubiquitinating enzymes (61). Although the precise mechanism for how its overexpression suppresses doa1Δ growth defects are unknown, the most plausible explanation is that it can complex with Cdc48 to deubiquitinate substrates targeted to the proteasome, and increased levels of Otu1 would program more Cdc48 complexes for this purpose and decrease the flux of ubiquitinated substrates to the proteasome. Ufd2 is a “E4” Ub ligase that preferentially extends polyubiquitin chains (62). Overexpression of Ufd2 enhances proteasomal degradation, supporting the idea that loss of Doa1 leads to more Cdc48 complexes programmed with Ufd2, which would increase flux through the proteasome, whereas loss of Ufd2 would decrease flux through the proteasome (28). Thus, the suppression observed by Otu1 overexpression or loss of Ufd2 can be explained by a general inhibition of proteasomal degradation. Under some circumstances, particularly in ubp6 mutants, proteasomes can rapidly degrade much of the cellular Ub, and inhibiting flux to the proteasome in doa1Δ mutants would probably restore Ub levels and viability at high temperature (63). Consistent with this idea are the observations that mutations that compromise proteasome catalytic activity or Cdc48 activity also suppress the growth defect of doa1Δ mutants (24, 27).
To determine whether these alternate interactions of Doa1 affected the phenotypes we observed, we analyzed the effect of deleting UFD2 and OTU1 (ovarian tumor family 1). Fig. 6A confirmed that deletion of UFD2 suppresses the temperature-sensitive phenotype of doa1Δ mutants. However, ufd2Δ mutations were unable to suppress the growth defect of doa1Δ vps27Δ double mutants. These data are consistent with the idea that the synthetic effect of doa1Δ and vps27Δ goes beyond simply the effect of lower Ub levels, since altering Ub levels directly (by overexpressing UBI4) or indirectly (by inhibiting degradation of Ufd2-dependent substrates) does not suppress the growth defect. Similarly, deletion of OTU1 did not have a synthetic effect on vps27Δ mutants. We also examined deletion of UBP6, which encodes a proteasome-associated deubiquitinase whose absence accelerates the degradation of Ub by the proteasome (63). Despite the adverse effects on Ub pools that deletion of UBP6 causes, we found no synthetic effect when ubp6Δ was combined with a vps27Δ mutation. Fig. 6B shows that the distribution of GFP-Ub remained relatively unperturbed in ubp6Δ, otu1Δ, and ufd2Δ mutants. All of these cells showed both cytosolic and intravacuolar GFP-Ub, in contrast to doa1Δ cells, which excluded GFP-Ub from the vacuole. The exclusion of GFP-Ub from the vacuole of doa1Δ was also seen in doa1Δ ufd2Δ cells, showing that UFD2 loss did not suppress this defect. Interestingly, ufd2Δ mutants occasionally showed clumps of ubiquitinated proteins, which may represent aggregates of ubiquitinated cytosolic and/or membrane proteins. However, we did not pursue this observation further.
FIGURE 6.
Specificity of doa1Δ defects. A, cells with the indicated mutations were serially diluted and plated onto minimal media and grown at the indicated temperature. B, the indicated mutants were transformed with the low copy GFP-Ub-expressing plasmid. Transformants were co-labeled with the endocytic tracer dye FM4–64 and imaged. WT, wild type.
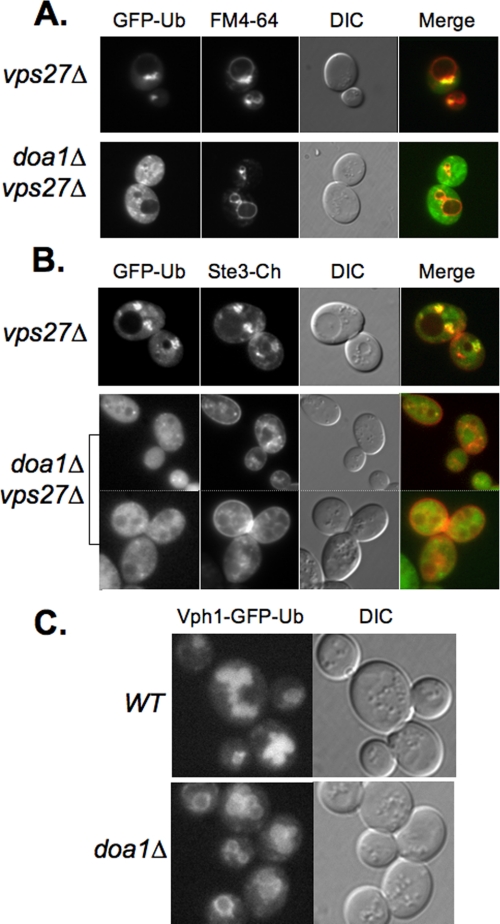
Doa1 Helps Ubiquitinated Cargo Proteins Concentrate on Endosomes—We next investigated how loss of Doa1 was toxic to vps27Δ mutants. Loss of Vps27, like the loss of some other class E Vps proteins, blocks delivery of ubiquitinated membrane proteins to the vacuole interior and blocks the formation of the endosomal intralumenal vesicles themselves. Despite this profound block in degrading membrane proteins, vps27Δ cells are remarkably healthy, and their viability does not significantly suffer from the accumulation of damaged proteins that would otherwise be degraded in the vacuole. Fig. 5A showed that vps27Δ mutants accumulate ubiquitinated proteins in large endosomal class E compartments, which previous studies have found also concentrate proteins destined for vacuolar degradation. Fig. 7 confirmed this by showing that the same compartments that accumulate GFP-Ub in vps27Δ mutants also accumulate Ste3 tagged with the Cherry red fluorescent protein. In stark contrast, we found that loss of DOA1 dramatically blocked the ability of ubiquitinated proteins to be concentrated in an enlarged endosomal compartment. Instead, only small puncta accumulated GFP-Ub, which showed some but not extensive co-distribution with endocytic markers. Likewise, Ste3-Cherry did not accumulate within large perivacuolar structures and was instead found at the cell surface as well as numerous small puncta, which probably corresponded to endosomes. Collectively, these data indicate that in the absence of Vps27 function, where ubiquitinated proteins are blocked from delivery to internal lumenal vesicles, they are still collected and consolidated on endosomal structures. Furthermore, they indicate a specific role for Doa1 in fostering this accumulation and imply that loss of such consolidation may be toxic.
FIGURE 7.
Mislocalization of MVB cargo upon loss of Doa1. A, localization of GFP-Ub in vps27Δ cells and vps27Δ doa1Δ double mutant cells. Cells were counterlabeled with FM4–64. Shown also are the merged fluorescence images and DIC image. B, localization of GFP-Ub (pPL3601) in vps27Δ cells and vps27Δ doa1Δ double mutant cells. Cells were also expressing mCherry-tagged Ste3 (pPL3607). Shown also are the merged fluorescence images and DIC image. C, localization of Vph1-GFP-Ub (pPL1556) in wild type (WT) and doa1Δ mutant cells.
Certain Ubiquitinated Membrane Proteins Are Defective for MVB Sorting in Doa1 Mutants—If Doa1 were somehow required for processing ubiquitinated membrane proteins so that they were capable of moving into intralumenal vesicles, one might expect that specific types of proteins might be affected. These proteins might include damaged or denatured proteins prone to aggregation or bulky multisubunit complexes that need disassembly before sorting. We also searched for particular proteins that could also show such a dependence on Doa1. For this, we examined a GFP-tagged form of Vph1, which is a 100-kDa integral membrane protein of the larger vacuolar ATPase H+ pump complex. Vph1 associates with other membrane proteins that comprise a V0 sector of the pump, which in turn associates with a large soluble multisubunit V1 sector (64). Although GFP-tagged Vph1 localizes exclusively to the limiting membrane of the yeast vacuole, our previous studies have shown that translational fusion of Ub to the C terminus converts this protein into an MVB cargo, which localizes to the vacuolar lumen in wild type cells (41). Thus, this protein gains access to the MVB pathway by virtue of its in-frame C-terminal Ub and is not dependent on cellular ubiquitination for its sorting. Fig. 7C shows that unlike wild type cells, doa1Δ cells were highly defective in properly sorting Vph1-GFP-Ub to the vacuole interior and instead accumulated on the limiting membrane in doa1Δ cells. Upon examining 50 cells, we found that only 4% of wild type cells showed detectable rim fluorescence on the surface of the vacuole. In contrast, 94% of the doa1Δ cells showed rim fluorescence. These data indicate that although Vph1-GFP-Ub carries its own Ub sorting signal, it is inefficiently recognized or processed in doa1Δ mutants so that it fails to be delivered to the vacuole lumen.
DISCUSSION
Doa1 is a Ub-binding Cdc48 adaptor that has been implicated in a number of biological responses to a variety of cellular stresses. Many phenotypes have been found for doa1Δ mutants, but most of these phenotypes probably stem from an indirect effect of lowered Ub levels in the cell (21). The defects we observe with regard to proper MVB sorting of ubiquitinated cargo are independent of lower Ub levels and indicate a more direct role for Doa1 in controlling some aspect(s) of the MVB sorting process. As such, we envisage two roles for Doa1: one that is served by physical connection with the Hse1-Vps27 Ub-sorting receptor and another more general role independent of this association that helps to process Ub cargo so that it can be concentrated on endosomes and efficiently delivered to the ESCRT sorting machinery for incorporation into lumenal vesicles of the forming MVB.
An Hse1-dependent role for Doa1 is mediated by its direct association with the SH3 domain of Hse1. The SH3 ligand we discovered in Doa1 did not conform to a canonical proline-rich motif (65) but instead a novel motif that contained bulky hydrophobic residues that probably mediate interaction with the SH3 domain surface. This region is conserved in not only other yeast orthologs of Doa1 but also the mammalian Doa1 ortholog, PLAP. Previously, we have shown that the Hse1 SH3 domain also interacts with Ubp7, a deubiquitination enzyme, and Hua1, a component of complex containing the Ub ligase Rsp5 (20). These studies indicated that both deubiquitination and Ub ligation activities associate with Hse1 help determine the sorting efficiency of cargo proteins, such as GFP-Cps1, which requires ubiquitination to access the MVB sorting pathway. Furthermore, when association of Hse1 to Rsp5 is severed, MVB sorting of GFP-Cps1 is defective, whereas sorting is restored when Ub is fused in frame to the N terminus of GFP-Cps1. Likewise, we find that mutants of Doa1 lacking their ability to bind Hse1 have a similar defect in sorting GFP-Cps1 but no defect in sorting the Ub-GFP-Cps1 reporter protein. Thus, the function of the Doa1-Hse1 interaction is consistent with a role for Doa1 in helping potentiate the ubiquitination status of GFP-Cps1 rather than affecting the general ability of the Ub-sorting ESCRT apparatus to deliver cargo to the endosomal intralumenal vesicles. These data further show that since binding of Doa1, Hua1, and Ubp7 to the Hse1 SH3 domain is sensitive to the same mutations in the SH3 ligand binding site, all of these activities may compete at some level, implying that there may be a mechanism that regulates access to the Hse1-Vps27 complex by these factors.
Previous studies indicated that the central PFU domain of Doa1 is required for efficient binding to Ub. Rumpf and Jentsch (28) showed that strong Ub binding was contained within residues 1–494 of Doa1, whereas Mullally et al. (27) showed Ub binding with the N-terminal 450 residues as well as a C-terminal fragment encompassing residues 354–715. Combining these overlapping fragments suggests that the central PFU domain defined by residues 354–450 is required for Ub binding. In particular, Mullally et al. (27) altered Phe426 and Phe434 and showed loss of Ub binding by Doa1. We show here that a bacterially expressed PFU domain is indeed sufficient for Ub binding. Since Ub binding is a common feature among many Cdc48-associated proteins, it would be predicted to be a key functional feature of Doa1. However, our data are unclear as to what function Ub binding by the PFU domain itself confers. The SH3 ligand we identified also lies within the PFU domain and contains the one of the residues, Phe434, that Mullally et al. (27) identified as important for Ub binding. We find that although the isolated PFU domain of the Doa1Δ1Hse1 mutant retained its ability to bind Ub, the PFU domain from the Doa1Δ2Hse1 mutant did not. However, both Doa1 mutants were able to complement the growth defect of doa1Δ mutants, in contrast to the F426A/F434A mutant previously analyzed. We do not, however, conclude that Ub binding by Doa1 is dispensable for its function, since we have found other regions within Doa1 that are also sufficient for Ub binding.4 Thus, as Mullally et al. (27) cautioned in their study, the F426A/F434A mutation used may have abrogated other features of the protein.
A broader function of Doa1 at the MVB, outside of its association with Hse1, was first indicated by our observation that doa1Δ cells show a severe synthetic growth defect when combined with loss of VPS27. We also found that the general flux of Ub into the vacuole was defective in doa1Δ cells as was the MVB sorting Vph1-GFP-Ub, a component of the multisubunit V-ATPase complex, which we modified by translationally fusing Ub onto the C terminus. Importantly, all of these defects were largely independent of the nonspecific effect of lowered Ub levels that result from Doa1 loss. For instance, the synthetic growth defect of doa1Δ vps27Δ mutants was not suppressed by Ub overexpression or loss of Ufd2, which competes with Doa1 for Cdc48 binding and may accelerate proteasomal degradation and Ub loss (28). Likewise, the block in GFP-Ub accumulation in the vacuole was performed under conditions where the GFP-Ub suppresses the Ub-dependent temperature-sensitive growth defect of doa1Δ mutants.
One model we favor is that Doa1 is somehow required to help process a range of Ub cargo so that it can be efficiently recognized by the downstream ESCRT machinery and packaged into intralumenal vesicles. This processing would involve cooperation with the Cdc48 ATPase, which could help tease apart ubiquitinated membrane proteins as they are delivered to the endosome. Such activity would not be required for some proteins, such as Ub-GFP-Cps1 or Ste3-GFP, which are reporters based on proteins that access the MVB pathway as part of their natural biological program for down-regulation (55). Instead, we speculate that Doa1-Cdc48 would be required by larger protein complexes as well as damaged or aggregated proteins that are rendered nonfunctional or potentially toxic. Such processing would help make such cargo more accessible to the sorting machinery and help cargo concentrate on endosomal subdomains that mediate formation of lumenal vesicles. A further observation consistent with this model is that Doa1 was required for Ub cargo to accumulate in large endosomal structures in vps27Δ mutants. The doa1Δ vps27Δ mutants showed a dramatic dispersal of Ub cargo throughout the cell. Indeed, the inability to consolidate Ub cargo on endosomal subdomains may explain the severe growth defect we observe in doa1Δ vps27Δ mutants. This effect would be analogous to the toxic effects of aggresomes and other protein aggregates observed in mammalian cells, which accumulate if proteasomal degradation is compromised. Normally, these proteins are localized to the microtubule organizing center by the action of Cdc48 and HDAC6 (66, 67). HDAC6 also binds ubiquitinated proteins and mediates their transport via microtubule motors to the cell center, where they are concentrated (68). Blocking the ability of HDAC6 to consolidate these damaged proteins increases their toxicity perhaps by allowing their aberrant functions to compromise a host of other cellular functions.
Acknowledgments
We thank Scott Moye-Rowley for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM58202 (to R. C. P.). This work was also supported by American Heart Association Predoctoral Fellowship Grant 0515640Z (to J. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Ub, ubiquitin; MVB, multivesicular body; GFP, green fluorescent protein; DIC, differential interference contrast; SH3, Src homology 3; SD, synthetic dextrose; AAA, ATPases associated with various cellular activities; PIPES, 1,4-piperazinediethanesulfonic acid; GST, glutathione S-transferase.
N. Pashkova, S. Winistorfer, L. Gakhar, J. Ren, S. Ramaswamy, and R. Piper, manuscript in preparation.
References
- 1.Piper, R. C., and Luzio, J. P. (2007) Curr. Opin. Cell Biol. 19 459-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piper, R. C., and Katzmann, D. J. (2007) Annu. Rev. Cell Dev. Biol. 23 519-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilodeau, P. S., Winistorfer, S. C., Kearney, W. R., Robertson, A. D., and Piper, R. C. (2003) J. Cell Biol. 163 237-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd, C. G., and Emr, S. D. (1998) Mol. Cell 2 157-162 [DOI] [PubMed] [Google Scholar]
- 5.Katzmann, D. J., Stefan, C. J., Babst, M., and Emr, S. D. (2003) J. Cell Biol. 162 413-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piper, R. C., Cooper, A. A., Yang, H., and Stevens, T. H. (1995) J. Cell Biol. 131 603-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupre, S., and Haguenauer-Tsapis, R. (2001) Mol. Cell. Biol. 21 4482-4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amerik, A. Y., Nowak, J., Swaminathan, S., and Hochstrasser, M. (2000) Mol. Biol. Cell 11 3365-3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochstrasser, M., Johnson, P. R., Arendt, C. S., Amerik, A., Swaminathan, S., Swanson, R., Li, S. J., Laney, J., Pals-Rylaarsdam, R., Nowak, J., and Connerly, P. L. (1999) Phil. Trans. R. Soc. Lond. 354 1513-1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springael, J. Y., Galan, J. M., Haguenauer-Tsapis, R., and Andre, B. (1999) J. Cell Sci. 112 1375-1383 [DOI] [PubMed] [Google Scholar]
- 11.Losko, S., Kopp, F., Kranz, A., and Kolling, R. (2001) Mol. Biol. Cell 12 1047-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, J., Wittekind, S. G., and Barr, M. M. (2007) Mol. Biol. Cell 18 3277-3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullough, J., Row, P. E., Lorenzo, O., Doherty, M., Beynon, R., Clague, M. J., and Urbe, S. (2006) Curr. Biol. 16 160-165 [DOI] [PubMed] [Google Scholar]
- 14.Raiborg, C., Malerod, L., Pedersen, N. M., and Stenmark, H. (2008) Exp. Cell Res. 314 801-813 [DOI] [PubMed] [Google Scholar]
- 15.Rayala, S. K., Hollander, P., Balasenthil, S., Molli, P. R., Bean, A. J., Vadlamudi, R. K., Wang, R. A., and Kumar, R. (2006) J. Biol. Chem. 281 4395-4403 [DOI] [PubMed] [Google Scholar]
- 16.Roxrud, I., Raiborg, C., Pedersen, N. M., Stang, E., and Stenmark, H. (2008) J. Cell Biol. 180 1205-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern, K. A., Visser Smit, G. D., Place, T. L., Winistorfer, S., Piper, R. C., and Lill, N. L. (2007) Mol. Cell. Biol. 27 888-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, Q., Sun, W., Kujala, P., Lotfi, Y., Vida, T. A., and Bean, A. J. (2005) Mol. Biol. Cell 16 2470-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clague, M. J., and Urbe, S. (2006) Trends Cell Biol. 16 551-559 [DOI] [PubMed] [Google Scholar]
- 20.Ren, J., Kee, Y., Huibregtse, J. M., and Piper, R. C. (2007) Mol. Biol. Cell 18 324-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghislain, M., Dohmen, R. J., Levy, F., and Varshavsky, A. (1996) EMBO J. 15 4884-4899 [PMC free article] [PubMed] [Google Scholar]
- 22.Hochstrasser, M., and Varshavsky, A. (1990) Cell 61 697-708 [DOI] [PubMed] [Google Scholar]
- 23.Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995) J. Biol. Chem. 270 17442-17456 [DOI] [PubMed] [Google Scholar]
- 24.Brandina, I., Smirnov, A., Kolesnikova, O., Entelis, N., Krasheninnikov, I. A., Martin, R. P., and Tarassov, I. (2007) FEBS Lett. 581 4248-4254 [DOI] [PubMed] [Google Scholar]
- 25.Kunze, D., MacCallum, D., Odds, F. C., and Hube, B. (2007) Microbiology (Read.) 153 1026-1041 [DOI] [PubMed] [Google Scholar]
- 26.Lis, E. T., and Romesberg, F. E. (2006) Mol. Cell. Biol. 26 4122-4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullally, J. E., Chernova, T., and Wilkinson, K. D. (2006) Mol. Cell. Biol. 26 822-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumpf, S., and Jentsch, S. (2006) Mol. Cell 21 261-269 [DOI] [PubMed] [Google Scholar]
- 29.Wilson, T. E. (2002) Genetics 162 677-688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil, R. L., Wolfe, D., Reiner, T., Peterson, C. J., and Riley, J. L. (1996) Mol. Cell. Biol. 16 3446-3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogiso, Y., Sugiura, R., Kamo, T., Yanagiya, S., Lu, Y., Okazaki, K., Shuntoh, H., and Kuno, T. (2004) Mol. Cell. Biol. 24 2324-2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, T. F., Gaitatzes, C., Saxena, K., and Neer, E. J. (1999) Trends Biochem. Sci. 24 181-185 [DOI] [PubMed] [Google Scholar]
- 33.Iyer, L. M., Koonin, E. V., and Aravind, L. (2004) Cell Cycle 3 1440-1450 [DOI] [PubMed] [Google Scholar]
- 34.Halawani, D., and Latterich, M. (2006) Mol. Cell 22 713-717 [DOI] [PubMed] [Google Scholar]
- 35.Jentsch, S., and Rumpf, S. (2007) Trends Biochem. Sci. 32 6-11 [DOI] [PubMed] [Google Scholar]
- 36.Ye, Y. (2006) J. Struct. Biol. 156 29-40 [DOI] [PubMed] [Google Scholar]
- 37.Shcherbik, N., and Haines, D. S. (2007) Mol. Cell 25 385-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, G., Zhou, X., Wang, L., Li, G., Schindelin, H., and Lennarz, W. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8785-8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., Arkin, A. P., Astromoff, A., El-Bakkoury, M., Bangham, R., Benito, R., Brachat, S., Campanaro, S., Curtiss, M., Davis, K., Deutschbauer, A., Entian, K. D., Flaherty, P., Foury, F., Garfinkel, D. J., Gerstein, M., Gotte, D., Guldener, U., Hegemann, J. H., Hempel, S., Herman, Z., Jaramillo, D. F., Kelly, D. E., Kelly, S. L., Kotter, P., LaBonte, D., Lamb, D. C., Lan, N., Liang, H., Liao, H., Liu, L., Luo, C., Lussier, M., Mao, R., Menard, P., Ooi, S. L., Revuelta, J. L., Roberts, C. J., Rose, M., Ross-Macdonald, P., Scherens, B., Schimmack, G., Shafer, B., Shoemaker, D. D., Sookhai-Mahadeo, S., Storms, R. K., Strathern, J. N., Valle, G., Voet, M., Volckaert, G., Wang, C. Y., Ward, T. R., Wilhelmy, J., Winzeler, E. A., Yang, Y., Yen, G., Youngman, E., Yu, K., Bussey, H., Boeke, J. D., Snyder, M., Philippsen, P., Davis, R. W., and Johnston, M. (2002) Nature 418 387-39112140549 [Google Scholar]
- 40.Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D., and Hegemann, J. H. (2002) Nucleic Acids Res. 30 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urbanowski, J. L., and Piper, R. C. (2001) Traffic 2 622-630 [DOI] [PubMed] [Google Scholar]
- 42.Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., and Tsien, R. Y. (2004) Nat. Biotechnol. 22 1567-1572 [DOI] [PubMed] [Google Scholar]
- 43.Smith, D. B., Rubira, M. R., Simpson, R. J., Davern, K. M., Tiu, W. U., Board, P. G., and Mitchell, G. F. (1988) Mol. Biochem. Parasitol. 27 249-256 [DOI] [PubMed] [Google Scholar]
- 44.Urbanowski, J. L., and Piper, R. C. (1999) J. Biol. Chem. 274 38061-38070 [DOI] [PubMed] [Google Scholar]
- 45.Kushnirov, V. V. (2000) Yeast 16 857-860 [DOI] [PubMed] [Google Scholar]
- 46.Tong, A. H., Drees, B., Nardelli, G., Bader, G. D., Brannetti, B., Castagnoli, L., Evangelista, M., Ferracuti, S., Nelson, B., Paoluzi, S., Quondam, M., Zucconi, A., Hogue, C. W., Fields, S., Boone, C., and Cesareni, G. (2002) Science 295 321-324 [DOI] [PubMed] [Google Scholar]
- 47.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425 686-691 [DOI] [PubMed] [Google Scholar]
- 48.Babst, M., Sato, T. K., Banta, L. M., and Emr, S. D. (1997) EMBO J. 16 1820-1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop, N., and Woodman, P. (2000) Mol. Biol. Cell 11 227-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katzmann, D. J., Babst, M., and Emr, S. D. (2001) Cell 106 145-155 [DOI] [PubMed] [Google Scholar]
- 51.Katzmann, D. J., Sarkar, S., Chu, T., Audhya, A., and Emr, S. D. (2004) Mol. Biol. Cell 15 468-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reggiori, F., and Pelham, H. R. (2002) Nat. Cell Biol. 4 117-123 [DOI] [PubMed] [Google Scholar]
- 53.Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer, A. W., and Schreiber, S. L. (1994) Cell 76 933-945 [DOI] [PubMed] [Google Scholar]
- 54.Bilodeau, P. S., Urbanowski, J. L., Winistorfer, S. C., and Piper, R. C. (2002) Nat. Cell Biol. 4 534-539 [DOI] [PubMed] [Google Scholar]
- 55.Hicke, L., and Dunn, R. (2003) Annu. Rev. Cell Dev. Biol. 19 141-172 [DOI] [PubMed] [Google Scholar]
- 56.Dantuma, N. P., Groothuis, T. A., Salomons, F. A., and Neefjes, J. (2006) J. Cell Biol. 173 19-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian, S. B., Ott, D. E., Schubert, U., Bennink, J. R., and Yewdell, J. W. (2002) J. Biol. Chem. 277 38818-38826 [DOI] [PubMed] [Google Scholar]
- 58.Richter, C., West, M., and Odorizzi, G. (2007) EMBO J. 26 2454-2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amerik, A., Sindhi, N., and Hochstrasser, M. (2006) J. Cell Biol. 175 825-835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bishop, N., Horman, A., and Woodman, P. (2002) J. Cell Biol. 157 91-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balakirev, M. Y., Tcherniuk, S. O., Jaquinod, M., and Chroboczek, J. (2003) EMBO Rep. 4 517-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu, D., Li, W., Ye, Y., and Brunger, A. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15599-15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanna, J., Leggett, D. S., and Finley, D. (2003) Mol. Cell. Biol. 23 9251-9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forgac, M. (2007) Nat. Rev. 8 917-929 [DOI] [PubMed] [Google Scholar]
- 65.Macias, M. J., Wiesner, S., and Sudol, M. (2002) FEBS Lett. 513 30-37 [DOI] [PubMed] [Google Scholar]
- 66.Boyault, C., Gilquin, B., Zhang, Y., Rybin, V., Garman, E., Meyer-Klaucke, W., Matthias, P., Muller, C. W., and Khochbin, S. (2006) EMBO J. 25 3357-3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobayashi, T., Manno, A., and Kakizuka, A. (2007) Genes Cells 12 889-901 [DOI] [PubMed] [Google Scholar]
- 68.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A., and Yao, T. P. (2003) Cell 115 727-738 [DOI] [PubMed] [Google Scholar]
- 69.Raymond, C. K., Howald-Stevenson, I., Vater, C. A., and Stevens, T. H. (1992) Mol. Biol. Cell 3 1389-1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115-132 [DOI] [PubMed] [Google Scholar]
- 71.Odorizzi, G., Babst, M., and Emr, S. D. (1998) Cell 95 847-858 [DOI] [PubMed] [Google Scholar]