Abstract
Mammalian α-defensins, expressed primarily in leukocytes and epithelia, play important roles in innate and adaptive immune responses to microbial infection. Six invariant cysteine residues forming three indispensable disulfide bonds and one Gly residue required structurally for an atypical β-bulge are totally conserved in the otherwise diverse sequences of all known mammalian α-defensins. In addition, a pair of oppositely charged residues (Arg/Glu), forming a salt bridge across a protruding loop in the molecule, is highly conserved. To investigate the structural and functional roles of the conserved Arg6–Glu14 salt bridge in human α-defensin 5 (HD5), we chemically prepared HD5 and its precursor proHD5 as well as their corresponding salt bridge-destabilizing analogs E14Q-HD5 and E57Q-proHD5. The Glu-to-Gln mutation, whereas significantly reducing the oxidative folding efficiency of HD5, had no effect on the folding of proHD5. Bovine trypsin productively and correctly processed proHD5 in vitro but spontaneously degraded E57Q-proHD5. Significantly, HD5 was resistant to trypsin treatment, whereas E14Q-HD5 was highly susceptible. Further, degradation of E14Q-HD5 by trypsin was initiated by the cleavage of the Arg13–Gln14 peptide bond in the loop region, a catastrophic proteolytic event resulting directly in quick digestion of the whole defensin molecule. The E14Q mutation did not alter the bactericidal activity of HD5 against Staphylococcus aureus but substantially enhanced the killing of Escherichia coli. By contrast, proHD5 and E57Q-proHD5 were largely inactive against both strains at the concentrations tested. Our results confirm that the primary function of the conserved salt bridge in HD5 is to ensure correct processing of proHD5 and subsequent stabilization of mature α-defensin in vivo.
Defensins are expressed predominantly in leukocytes and epithelial cells as a family of cationic antimicrobial peptides that play important roles in innate and adaptive immune responses to microbial infection (1–5). In humans, defensins are classified into α- and β-families, differing by amino acid composition, cellular origin, and disulfide connectivity. To date, six human α-defensins have been identified, including four primarily from neutrophils, also known as human neutrophil peptides 1–4 or HNP1–42 (6–10) and two mainly from intestinal Paneth cells, also known as human defensins 5–6 or HD5–6 (11, 12). Although more than 30 putative β-defensin genes have been described using genomics tools (13–15), only a handful of β-defensins have been isolated and characterized at the protein level (16).
Human α-defensins are synthesized in vivo as significantly larger precursor molecules consisting of an N-terminal pro domain of 43–49 amino acid residues and a C-terminal Cys-rich defensin domain of 29–33 amino acid residues. The pro domain in proHNPs, important for correct folding, sorting, and trafficking of defensins (17–20), is proteolytically excised by yet-to-be-identified enzyme(s) prior to the storing of mature HNPs in the azurophilic granules of neutrophils. Immediately after phagocytosis, HNPs along with several antimicrobial enzymes including elastase are discharged into the microbe-containing phagocytic vacuole where microbial killing occurs (21). By contrast, stored in the granules of Paneth cells is unprocessed proHD5, which is secreted, both constitutively and inducibly, into the gut crypts of the small intestine where cleavages by trypsin generate mature HD5 (22).
A fully functional HD5 on the crypt and lumenal surfaces of intestinal epithelia is important for the innate defense against bacterial infection and may also provide antimicrobial protection for stem cells in the gut epithelial layer within the crypts (23). Bevins and colleagues (24) demonstrated in a mouse model that Paneth cells-associated expression of HD5 affords remarkable resistance of transgenic mice to oral challenge with virulent Salmonella typhimurium, providing support for a critical in vivo role of epithelial-derived defensins in mammalian host defense. Notably, the metalloproteinase matrilysin is required for the processing of mouse Paneth cell α-defensins (also known as cryptdins) (25). Mice deficient in matrilysin, thus lacking functional mature cryptdins, are highly susceptible to orally administered Salmonella, indicative of the importance of cryptdins in defense against gut-associated pathogens (25, 26).
Sequence analysis of all known mammalian α-defensins indicates that in addition to the six Cys residues forming three structurally indispensable disulfide bridges and Gly17, the salt bridge between Arg5 and Glu13 (HNP1 numbering) is highly conserved. Xie et al. (27) have previously shown that Gly17 adopting unusual dihedral torsion angles is required for the formation of an atypical β-bulge structure conserved in the α-defensin family and that replacement of Gly17 by any other l-amino acids in HNP2 invariably results in defensin misfolding. In a separate study, Wu et al. (28) demonstrated that destabilization of the conserved Arg5–Glu13 salt bridge in HNP1, whereas functionally inconsequential, dramatically accelerates proteolytic degradation of HNP1 by human neutrophil elastase, suggesting the importance of the salt bridge for defensin in vivo stability. Notably, Rosengren et al. (29) recently reported that destabilization of the conserved salt bridge in mouse cryptdin 4 (Crp4) through a series of mutations of Glu15 (Crp4 numbering) has little impact on the bactericidal activity of the defensin, a result in agreement with that reported by Wu et al. (28). However, in contrast to the findings by Wu et al. (28), E15D-Crp4 and other salt bridge-deficient mutants are resistant to cleavage by the procryptdin processing enzyme matrilysin (29). The authors therefore concluded that the conserved salt bridge in Crp4 is not linked to proteolytic stability of the mature defensin (29).
To better understand why the salt bridge is conserved in α-defensins and to account for the reported discrepancy on its role in defensin stability, we expanded our study to include HD5 whose processing enzyme (trypsin) has been previously identified (22). We chemically synthesized HD5 and proHD5 as well as two corresponding salt bridge-destabilizing analogs: E14Q-HD5 and E57Q-proHD5 (Fig. 1). Detailed biochemical, functional, and structural characterizations of these synthetic defensin/pro defensin molecules help to establish that the conserved Arg6–Glu14 (HD5 numbering) salt bridge is critical not only for correct processing of proHD5 by trypsin but also for proteolytic stability of mature HD5 in the presence of its processing enzyme.
FIGURE 1.
The amino acid sequence of proHD5. The mature domain is in bold typeface, and the maturation site is denoted by an arrow. The boxed residues, Arg49 and Glu57 in proHD5 or Arg6 and Glu14 in HD5, form a salt bridge in the structure and are highly conserved in all known mammalian α-defensins.
MATERIALS AND METHODS
Total Chemical Synthesis of HD5, proHD5, E14Q-HD5, and E57Q-proHD5—The amino acid sequences of HD5 and proHD5 are as follows: 1ATCYCRTGRC10; 11ATRESLSGVC20; 21EISGRLYRLC30; 31CR (HD5); and 1ESLQERADEA10; 11TTQKQSGEDN20; 21QDLAISFAGN30; 31GLSALRTSGS40; 41QARATCYCRT50; 51GRCATRESLS60; 61GVCEISGRLY70; 71RLCCR75 (proHD5). Arg43–Ala44 in proHD5 is the known processing site by trypsin both in vivo and in vitro (22). The conserved salt bridge is formed between Arg6 and Glu14 (in bold typeface) in HD5, as shown in the crystal structure of a chemically synthesized HD5 (30) (PDB code: 1zmp), and, presumably, between Arg49 and Glu57 in proHD5. Preparation of correctly folded synthetic HD5 used for this work was essentially as described previously (31). Similar procedures were used to synthesize E14Q-HD5. Native chemical ligation (32, 33) of the following peptide fragments was used to assemble proHD5 and E57Q-proHD5: (1–47)-proHD5-αCOSR (R = CH2CH2CO-Leu-OH), (5–32)-HD5, and E14Q-(5–32)-HD5. Syntheses of E14Q-HD5, (5–32)-HD5, and E14Q-(5–32)-HD5 on Boc-Arg(Tos)-OCH2-PAM resin and of (1–47)-proHD5-αCOSR on Boc-Leu-OCH2-PAM resin were performed using an in-house chemistry tailored from the N,N-diisopropylethylamine in situ neutralization/2-(1H-benzotriazolyl)-1,1,3,3-tetramethyluroniumhexafluorophosphate activation protocol originally developed by Kent and colleagues (34, 35) for Boc solid phase peptide synthesis. After chain assembly, the peptides were deprotected and cleaved by anhydrous hydrogen fluoride in the presence of 5% p-cresol at 0 °C for 1 h followed by precipitation with cold ether. All crude peptides were purified to homogeneity by preparative C18 reversed phase (RP) HPLC on a Waters Delta Prep 600 system, and their molecular masses were ascertained by a Micromass ZQ-4000 single quadrupole electrospray ionization-mass spectrometer (ESI-MS).
Native chemical ligation between (1–47)-proHD5-αCOSR and (5–32)-HD5 or E14Q-(5–32)-HD5 was carried out at a total peptide concentration of 13 mg/ml in 0.25 m phosphate buffer containing 6 m GuHCl and 2% (v/v) thiophenol, pH 7.5. The ligation reaction went to completion overnight at room temperature, yielding a full-length proHD5 or E57Q-proHD5, which was subsequently purified to homogeneity by preparative RP-HPLC. The determined molecular masses of fully reduced HD5, E14Q-HD5, proHD5, and E57Q-proHD5 were in agreement with the expected values calculated on the basis of the average isotopic compositions of the four synthetic molecules. All defensins were quantified by UV absorbance measurements at 280 nm using molar extinction coefficients calculated according to the algorithm published by Pace et al. (36).
Comparative Oxidative Folding of HD5, proHD5, E14Q-HD5, and E57Q-proHD5—Productive oxidative folding of HD5 was previously established (31). For comparison, reduced peptides were first dissolved at 2 mg/ml in 8 m urea containing 12 mm reduced and 1.2 mm oxidized glutathione followed by a 4-fold dilution with 0.25 m sodium bicarbonate. 30-μl aliquots were withdrawn at different time intervals (0, 0.5, 1, 2, 4, and 8 h), to which an equal volume of 10% acetic acid was added to quench the reaction. The resultant solution (40 μl) was injected for HPLC analysis, which was performed at 40 °C on a Waters Symmetry™ C18 column (5 μm, 4.6 × 150 mm) using a linear gradient of 5–65% acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min over 30 min. For data points collected at time 0, a reducing buffer containing 0.25 m phosphate, 6 m GuHCl, and 0.01 mm dithiothreitol, pH 7.4, was used to dissolve peptide followed by acidification and HPLC quantification. The folding yields were calculated based on the ratio of the integrated areas of the two peaks given by folded defensin and fully reduced starting material.
Obtaining Correctly Folded E14Q-HD5 and R6A-HD5 from Pro Defensin—We also synthesized (R43M, E57Q)-proHD5 and (R43M, R49A)-proHD5 using native chemical ligation. The Met residue was introduced to facilitate chemical release of the C-terminal defensin domain after folding. The pro defensin mutant (R43M, E57Q)-proHD5 or (R43M, R49A)-proHD5 was folded in 2 m urea, 3 mm reduced glutathione, and 0.3 mm oxidized glutathione, pH 8.3. Quantitative cleavage of the Met43–Ala44 peptide bond was achieved after an overnight reaction of the folded (R43M, E57Q)-proHD5 or (R43M, R49A)-proHD5 (1 mg/ml) with CNBr (25 mg/ml) in 2.5% trifluoroacetic acid, and the resultant E14Q-HD5 and R6A-HD5 were purified to homogeneity by RP-HPLC.
In Vitro Processing of proHD5 and E57Q-proHD5 by Trypsin—ProHD5 or E57Q-proHD5 at 0.5 mg/ml was incubated at room temperature with 0.01 mg/ml bovine trypsin (Worthington Biochemical Co.) in 50 mm Tris/HCl buffer containing 20 mm CaCl2 and 0.001% Triton X-100, pH 8.3. Aliquots of 100 μl were withdrawn and quenched with 20 μl of 10% acetic acid at different time intervals (0.5, 1, 2, and 4 h), of which 60 μl was injected for RP-HPLC analysis on a Waters XBridge C18 column (3.5 μm, 4.6 × 150 mm) running a linear gradient of 5–45% acetonitrile containing 0.1% trifluoroacetic acid. ESI-MS was used to identify the cleavage products separated by analytical HPLC.
X-ray Crystallographic Studies of Trypsin-processed HD5—To verify the correct fold of the mature HD5 obtained by the processing of proHD5 with trypsin, we subjected it to the detailed crystallographic analysis. The crystallization and x-ray data collection protocols essentially replicated those described by us earlier for an HD5 preparation obtained by the folding of reduced defensin (30). The x-ray data collected for the single crystal of HD5 extended to the resolution of 2.1 Å. The space group (P6522) and unit cell parameters (a = b = 49.52 Å, c = 254.9 Å) were identical or nearly identical to reported previously (30). Using the program Phaser (37), the structure of HD5 could be easily solved with the model of this protein taken from the Protein Data base (Protein Data Bank entry code: 1zmp). A few steps of refinement with the program Refmac 5 (38), were sufficient to prove that the “new” structure of HD5 is virtually identical to previously reported. Values of the crystallographic R-factor and free-R for the final model were 0.217 and 0.258, respectively, and the root-mean-square deviation calculated for all the equivalent Cα-atoms was less than 0.1 Å. The detail characteristics from the data collection and the structure refinement are shown in supplemental Table S1.
Proteolytic Stability of HD5 and E14Q-HD5—HD5 and E14Q-HD5 at 0.5 mg/ml each were incubated at room temperature with 0.0025 mg/ml bovine trypsin in 50 mm Tris/HCl buffer containing 20 mm CaCl2 and 0.001% Triton X-100, pH 8.3. Aliquots of 80 μl were withdrawn at different time intervals (0, 0.5, 1, 2, 4, and 8 h for HD5; 0, 1, 5, 10, 15, 30, and 60 min for E14Q-HD5), acidified by 20 μl of 10% acetic acid, and injected (50 μl) for analysis on a Waters XBridge C18 column (3.5 μm, 4.6 × 150 mm) running a linear gradient of 5–45% acetonitrile containing 0.1% trifluoroacetic acid. The percentage of residual defensin was calculated based on the ratio of integrated peak areas of the intact molecule at a given time versus at time 0. The percentages of the intermediate produced due to the first cleavage at the Arg13–Gln14 bond were calculated based on the ratio of integrated peak areas of cleaved E14Q-HD5 at a given time versus the intact starting material at time 0. Identical experiments were performed using R6A-HD5 and trypsin. In addition, the proteolytic stability of HD5 and E14Q-HD5 was examined under similar conditions with bovine chymotrypsin (Worthington Biochemical Co.), human leukocyte elastase (Elastin Products Co., Inc.), and human matrix metalloproteinase-7 (MMP-7) (Chemicon).
Antibacterial Activity of HD5, proHD5, E14Q-HD5, and E57Q-proHD5—Antimicrobial assays against Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 (Microbiologics) were conducted using a previously detailed 96-well turbidimetric method dubbed virtual colony counting (39). A 2-fold dilution series of defensin, ranging from 25 to 0.098 μm in 10 mm sodium phosphate, pH 7.4, was incubated at 37 °C for 2 h with E. coli or S. aureus (1 × 106 colony-forming units/ml) followed by the addition of twice-concentrated Mueller-Hinton broth (2×) and 12-h kinetic measurements of bacterial growth at 650 nm.
RESULTS
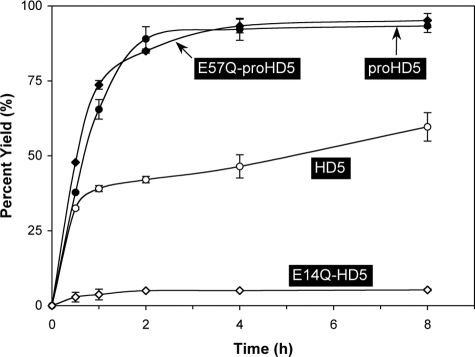
The Arg6–Glu14 Salt Bridge Is Important for In Vitro Oxidative Folding of HD5 but Dispensable for proHD5—We previously developed highly efficient protocols for in vitro oxidative folding of all six human α-defensins and some of their pro forms (20, 28, 31, 40, 41). As was demonstrated with HNP2, destabilization of the conserved salt bridge was detrimental to the folding of reduced HNP2 but had no adverse effect on the folding of reduced proHNP2 (28). To examine the role of the Arg6–Glu14 salt bridge in the folding of HD5 and proHD5, we comparatively monitored time-dependent oxidative folding kinetics of HD5, E14Q-HD5, proHD5, and E57Q-proHD5 using analytical RP-HPLC coupled with ESI-MS, and the results are plotted in Fig. 2. Consistent with our previous finding that the pro peptide catalyzes oxidative folding of α-defensins in vitro (20), proHD5 folded significantly more efficiently than HD5, with nearly 90% of proHD5 folded within 2 h as opposed to ∼40% of HD5 during the same time period. Although the yield of folded proHD5 plateaued at ∼92% after 4 h, the yield of folded HD5 slowly increased over time to ∼60% after 8 h.
FIGURE 2.
Time-dependent folding yields of HD5, proHD5, E14Q-HD5, and E57Q-proHD5. Oxidative folding of each peptide was performed at a concentration of 0.5 mg/ml in 2 m urea, 3 mm reduced and 0.3 mm oxidized glutathione, pH 8.1. Samples were withdrawn at different time intervals for HPLC analysis. Yields, proportional to peak area, were calculated by integration. The data were obtained from two independent experiments, where the error bars represent the average deviation of the measurements.
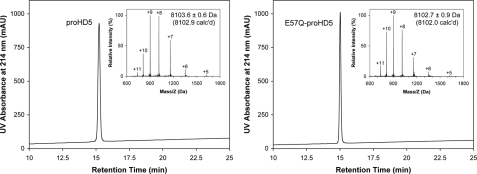
Expectedly, destabilization of the salt bridge dramatically reduced the folding efficiency of HD5, as evidenced by merely a 5% yield of folded E14Q-HD5 during an 8-h time period. However, the folding kinetics of E57Q-proHD5 was nearly identical to that of proHD5, suggesting that the salt bridge-destabilizing mutation (Glu57 → Gln57 in proHD5) had no impact on the folding of the defensin precursor. These results are entirely consistent with our previous findings on HNP2 (28). Shown in Fig. 3 are folded and purified proHD5 and E57Q-proHD5 on analytical RP-HPLC and ESI-MS. Their determined molecular masses of 8103.6 ± 0.6 and 8102.7 ± 0.9 Da are in good agreement with the expected values of 8102.9 and 8102.0 Da, respectively, calculated on the basis of average isotopic compositions of the folded pro defensins.
FIGURE 3.
Folded proHD5 and E57Q-proHD5 analyzed by RP-HPLC and ESI-MS. HPLC analysis was performed at 40 °C on a Waters Symmetry™ 300 C18 column (5 μm, 4.6 × 150 mm) using a linear gradient of 5–65% acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min over 30 min. The measured molecular masses were in agreement with the expected values calculated based on the average isotopic compositions of the folded pro defensins. mAU, milliabsorbance units.
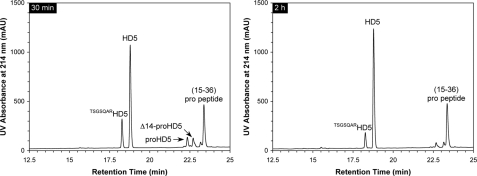
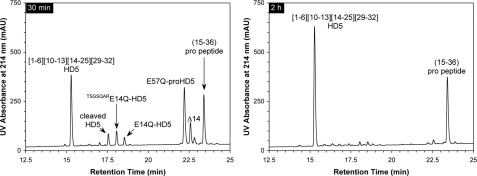
Trypsin Correctly and Efficiently Processes proHD5 in Vitro—Ghosh et al. (22) found that in human Paneth cells, trypsinogen is co-localized with proHD5 in the secretory granules. Upon secretion of proHD5 and trypsinogen, a yet-to-be-identified molecular event activates the enzyme, which in turn removes the pro region of proHD5, yielding predominantly mature HD5 and an N-terminally elongated (by an extra seven amino acid residues) minor processing product, i.e. TSGSQARHD5 (22). To investigate the processing pathway of synthetic proHD5 in vitro, we incubated, at room temperature, 100 μg of proHD5 with 2 μg of bovine pancreatic trypsin in buffer, at pH 8.3, and monitored on HPLC and ESI-MS the cleavage reaction at different time intervals (30 min, 1 h, 2 h, and 4 h). The representative chromatographic traces collected at 30 min and 2 h are shown in Fig. 4. Within 30 min, proHD5 was nearly quantitatively converted to mature HD5 (found 3582.1 Da, expected 3582.2 Da), the minor processing product TSGSQARHD5 (found 4269.9 Da, expected 4269.9 Da), and a major fragment from the pro peptide, 15–36 (found 2263.6 Da, expected 2263.4 Da). Over time, the extra seven amino acid residues at the N terminus of TSGSQARHD5 were slowly excised by trypsin, suggesting that the first and most productive cleavage in proHD5 occurs at the expected maturation site Arg43–Ala44 rather than at the upstream site Arg36–Thr37. Notably, a transient, N-terminally truncated proHD5, Δ14-proHD5 (found 6515.7 Da, expected 6515.3 Da), was also detected during the cleavage reaction. Otherwise, intact HD5 largely remained as the terminally processed product during the entire time period (4 h).
FIGURE 4.
Processing of proHD5 by trypsin at 30 min and 2 h. HPLC analysis was performed at 40 °C on a Waters XBridge C18 column (3.5 μm, 4.6 × 150 mm) using a linear gradient of 5–45% acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min over 30 min. The chromatographic peaks were identified by ESI-MS analyses aided by simulated trypsin digestion. mAU, milliabsorbance units.
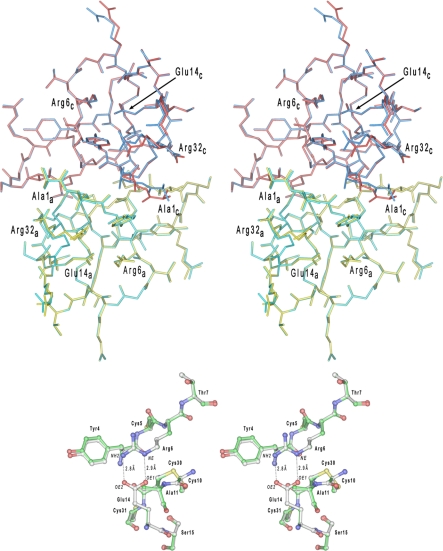
Trypsin-processed HD5 Is Correctly Folded—We previously reported the crystal structure (1zmp) of a synthetic HD5 prepared by stepwise solid phase peptide synthesis (30). To demonstrate that oxidative folding of proHD5 and subsequent cleavage by trypsin yielded correctly folded HD5, we solved the crystal structure of trypsin-processed HD5 at 2.1 Å resolution. Shown in Fig. 5 are dimeric HD5 and its blown-up loop region stabilized by the Arg6–Glu14 salt bridge, where two well defined H-bonds, at an average distance of 2.8 Å, are depicted between Nε and Nη2 of Arg6 and Oε1 and Oε2 of Glu14. The charge-charge interaction, along with a perpendicular disulfide bond (Cys10–Cys30), stabilizes the extruding loop of HD5 encompassing Arg6-His7-Gly8-Arg9-Cys10-Ala11-Thr12-Arg13-Glu14. The overall structure of the trypsin-processed HD5 is virtually identical to the previously determined by us (Protein Data Bank code 1zmp). This result unambiguously shows that the folding and processing procedures utilized here lead to a correctly folded HD5.
FIGURE 5.
Superposition of HD5 dimers in shown in a stereo representation. In the upper panel, a dimer of HD5 colored in yellow and red represents the monomers A and C in the previously determined crystal structure of this defensin and deposited in the Protein Data Bank under the code 1zmp. The second dimer (colored in cyan and blue) is composed of monomers A and C from the current structure. The N- and C-terminal residues, as well as Arg6 and Glu14 (forming the salt bridge), are labeled for both monomers. Both structures are virtually indistinguishable (root mean square deviation for the 64 equivalent Cα-atoms is 0.088 Å) with slight deviations found only for few highly flexible side chains. A region of the Arg6–Glu14 salt bridge is shown in the lower panel for both HD5 structures superimposed. The carbon atoms in the “1zmp” model are painted white, whereas in the current model, they are shown in green.
Trypsin Processes E57Q-proHD5 but Spontaneously Degrades E57Q-proHD5—To investigate the effect of a destabilized salt bridge on the processing of the defensin precursor, we treated E57Q-proHD5 with trypsin the same way as described previously for wild type proHD5, and the chromatographic results obtained after 30 min and 2 h are illustrated in Fig. 6. Trypsin appeared to have correctly processed E57Q-proHD5, as evidenced by brief accumulation in the early stages of several similarly processed products seen in the processing of wild type proHD5, such as Δ14-E57Q-proHD5 (found 6514.3 Da, expected 6514.3 Da), E14Q-HD5 (found 3581.0 Da, expected 3581.2 Da), and TSGSQARE14Q-HD5 (found 4269.2 Da, expected 4268.9 Da). However, E57Q-proHD5 was extremely prone to trypsin digestion. Within 30 min of incubation with trypsin, the majority of E57Q-proHD5 was converted to the terminal degradation product (1–6)(10–13)(14–25)(29–32), a fully cleaved defensin of four peptide fragments interconnected by three native disulfide bridges (Cys1–Cys6, Cys2–Cys4, and Cys3–Cys5)as judged by ESI-MS analyses (found 2889.3 Da, calculated 2889.4 Da). The major degradation product from the pro region, 15–36, in addition to several smaller peptide fragments that eluted early on RP-HPLC (not shown), also resulted. Within 2 h of incubation, E57Q-proHD5 was fully digested by trypsin to (1–6)(10–13)(14–25)(29–32), suggesting that destabilization of the conserved salt bridge is detrimental to proHD5 processing and that the C-terminal defensin domain confers proteolytic susceptibility of E57Q-proHD5.
FIGURE 6.
Processing of E57Q-proHD5 by trypsin at 30 min and 2 h. HPLC analysis was performed at 40 °C on a Waters XBridge C18 column (3.5 μm, 4.6 × 150 mm) using a linear gradient of 5–45% acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min over 30 min. The chromatographic peaks were identified by ESI-MS analyses aided by simulated trypsin digestion. mAU, milliabsorbance units.
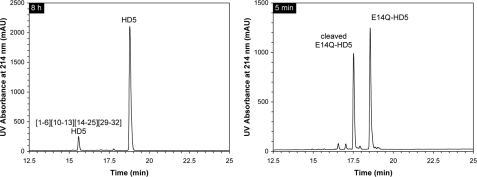
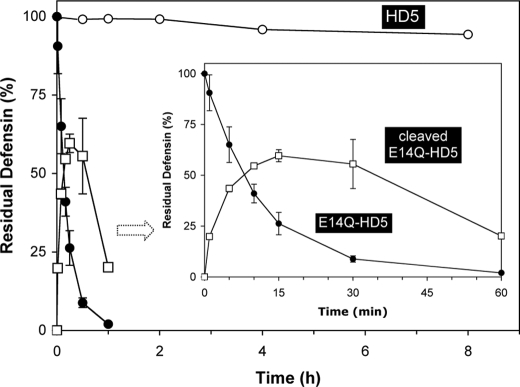
Degradation of E14Q-HD5 by Trypsin Is Initiated by Cleavage of Arg13–Gln14 in the Loop Region—To further examine the proteolytic susceptibility of E14Q-HD5 and to map out the sequence of events in its degradation by trypsin, we measured time-dependent hydrolysis kinetics of E14Q-HD5 on analytical RP-HPLC and ESI-MS, using a defensin-to-enzyme ratio of 200:1 (w/w). Wild type HD5 was used as a control. As shown in Figs. 7 and 8, wild type HD5 was resistant to trypsin treatment, with only 5% degraded to (1ATCYCR6)(10CATR13)(14ESLSGVCEISGR25)(29LCCR32) after 8 h (found 2890.5 Da; calculated 2890.4 Da). In sharp contrast, more than 35% of E14Q-HD5 was hydrolyzed within 5 min, 75% within 15 min, and over 98% within 60 min.
FIGURE 7.
Comparison of proteolytic susceptibility to trypsin between HD5 (8 h) and E14Q-HD5 (5 min). HPLC analysis was performed at 40 °C on a Waters XBridge C18 column (3.5 μm, 4.6 × 150 mm) using a linear gradient of 5–45% acetonitrile containing 0.1% trifluoroacetic acid at a flow rate of 1 ml/min over 30 min. (1–6)(10–13)(14–25)(29–32)-HD5 is the terminal degradation product of HD5, whereas cleaved E14Q-HD5 represents the first intermediate with the internal Arg13–Gln14 peptide bond hydrolyzed by trypsin. mAU, milliabsorbance units.
FIGURE 8.
Hydrolysis kinetics monitored for 8 h for HD5 and 60 min for E14Q-HD5 and cleaved E14Q-HD5. Residual HD5 or E14Q-HD5 was calculated based on the ratio of integrated peak areas of the intact starting material at a given time versus at time 0. The percentage of cleaved E14Q-HD5 relative to the intact molecule was calculated in a similar fashion. The data were obtained from two independent experiments, where the error bars represent the average deviation of the measurements.
A careful examination of the cleavage of E14Q-HD5 by trypsin revealed that the first processing intermediate with only one internal cleavage in the sequence (found 3600.1 Da, expected 3600.2 Da; 18 mass units more than E14Q-HD5) eluted ∼1 min earlier on HPLC than E14Q-HD5 (Fig. 7). The same peak identified by ESI-MS also briefly appeared during the early stages of the processing of E57Q-proHD5 (Fig. 6). As intact E14Q-HD5 disappeared, the percentage of cleaved E14Q-HD5 increased (Fig. 8), reaching ∼60% within 15 min, and then progressively decreased as smaller peptide fragments were generated. Reduction of cleaved E14Q-HD5 by dithiothreitol allowed us to identify two resultant peptide fragments on RP-HPLC whose molecular masses were found to be 1461.8 and 2143.3 Da, which corresponded to (1–13)-HD5 (calculated mass 1461.7 Da) and (14–32)-E14Q-HD5 (calculated mass 2143.5 Da), respectively. This result unequivocally demonstrated that degradation of E14Q-HD5 by trypsin was initiated by cleavage of Arg13–Gln14 in the loop region, which propagated subsequent enzymatic cleavages at different Arg-Xaa sites throughout the molecule.
The salt bridge analog R6A-HD5 was also prone to degradation by trypsin (supplemental Fig. S1). Hydrolysis kinetics analysis indicated that the half-life of R6A-HD5 was ∼30 min, and the enzyme digested more than 80% of the peptide within 4 h. Importantly, the first cleavage, as expected, occurred at the Arg13–Glu14 site in the loop region. As compared with E14Q-HD5, however, R6A-HD5 was degraded less efficiently by trypsin, presumably due to a decreased number of trypsin cleavage sites in the molecule.
E14Q-HD5 Is Correctly Folded—We also attempted to determine the crystal structure of the E14Q-HD5 mutant. Small, irregularly shaped crystals were obtained. However, when subjected to the x-ray experiments, crystals displayed poor diffraction properties, and a very high mosaicity was evident. Consequently, we could not collect the diffraction data necessary for the detailed structural characterization of this mutant. To exclude the possibility that proteolytic susceptibility of E14Q-HD5 was attributable to defensin misfolding, we biochemically determined partial disulfide connectivity in its terminal degradation product, i.e. (1ATCYCR6)(10CATR13)(14QSLSGVCEISGR25)(29LCCR32). The terminally degraded product of E14Q-HD5 was further digested by chymotrypsin at room temperature in 50 mm Tris buffer containing 0.005% Triton X-100 and 20 mm CaCl2, pH 8.3. The reaction was monitored by analytical RP-HPLC and ESI-MS. Chymotrypsin treatment yielded two major fragments on RP-HPLC, which were determined by ESI-MS to be (1ATCY4)(10CATR13)(29LCCR32) (found 1395.3 Da, expected 1395.7 Da) and (5CR6)(17SGVCEI22) (found 881.6 Da, expected 882.0 Da). These results unambiguously demonstrated the presence of one native disulfide bond, Cys5(2)–Cys20(4), and excluded the possibility of disulfide scrambling within the fragment containing, presumably, the remaining two disulfide bridges Cys3(1)–Cys31(6) and Cys10(3)–Cys30(5). Two additional pieces of evidence also supported correct folding of E14Q-HD5. First, the cleavage pattern from complete digestion of E14Q-HD5 by trypsin is entirely consistent with a native disulfide pairing in the mutant defensin. Second, it has been shown that the pro peptide of α-defensins intramolecularly chaperones correct folding of defensin precursors (20). When a defensin molecule contains mutations that incapacitate its ability to fold alone, covalent attachment of its pro peptide invariably rescues misfolding (28). For comparison, we prepared (R43M, E57Q)-proHD5 using native chemical ligation. CNBr cleavage of folded (R43M, E57Q)-proHD5 quantitatively released an E14Q-HD5 that was found identical to the separately folded E14Q-HD5 with respect to their chromatographic, mass spectrometric, and bactericidal properties as well as trypsin susceptibility (data not shown).
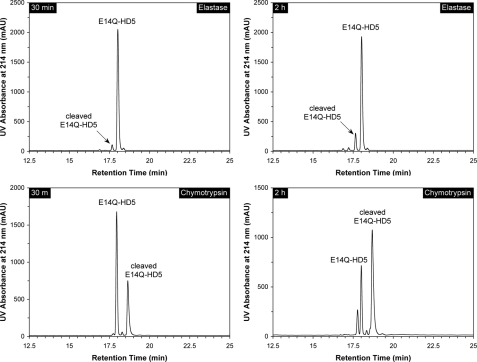
HD5 and E14Q-HD5 versus Other Proteases—Since chymotrypsin is present in the intestinal lumen under physiological conditions and elastase present during acute inflammation, we tested the proteolytic susceptibility of HD5 and E14Q-HD5 against bovine pancreatic chymotrypsin and human neutrophil elastase similarly to trypsin. For the wild type HD5, no detectable cleavage was observed within 4 h of incubation with either enzyme. E14Q-HD5 was relatively stable in the presence of elastase, with only 3% cleaved within 30 min, 13% within 2 h (Fig. 9), and one-third within 8 h. By contrast, E14Q-HD5 was much more susceptible to degradation by chymotrypsin. Within 30 min of incubation, 35% of the peptide was converted to an internally cleaved product (mass found 3600.2 Da), and the percentage increased to 65% within 2 h (Fig. 9). Unlike trypsin, chymotrypsin was inefficient in further processing its first cleavage product of E14Q-HD5. Disulfide reduction coupled with mass mapping unambiguously pinpointed the first chymotrypsin cleavage site to Tyr27–Arg28 (found mass for fragment (1–27) 2955.5 Da, expected mass 2955.4 Da; found mass for fragment (28–32) 649.6 Da, expected mass 649.8 Da), consistent with chymotrypsin specificity. We also examined proteolytic susceptibility of HD5 and E14Q-HD5 to MMP-7 under identical conditions. However, no cleavage was detected on HPLC over a period of 8 h at room temperature.
FIGURE 9.
Degradation of E14Q-HD5 by elastase and chymotrypsin at 30 min and 2 h. Conditions for HPLC analysis were the same as described in the legends for Figs. 4, 6, and 7. For chymotrypsin, cleaved E14Q-HD5 differed from intact E14Q-HD5 by +18 Da in molecular mass, attributable to the cleavage of the Tyr27–Arg28 peptide bond. The minor peak eluted before E14Q-HD5 after 2 h of incubation with the enzyme contained doubly and triply cleaved peptides as indicated by mass spectrometric analysis. mAU, milliabsorbance units.
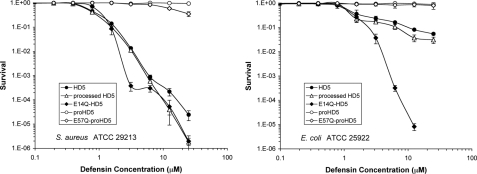
The Functional Importance of the Conserved Salt Bridge in HD5 Appears Strain-specific—We tested the effect of the E14Q mutation in the salt bridge of HD5 on its antimicrobial activity against the Gram-negative bacterium E. coli and the Gram-positive bacterium S. aureus using virtual colony counting as established previously (39). In this assay, bacteria are exposed to a 2-fold serial dilution of peptides for 2 h. Subsequently, the antimicrobial activity of the peptides is neutralized by the addition of Mueller Hinton medium, and optical density is recorded for a period of 12 h as a measure for bacterial recovery. As shown in Fig. 10, HD5 effectively killed both strains of bacteria. Antimicrobial virtual lethal doses (in μm) that kill 50 and 90% of E. coli and S. aureus input viable cells, as inferred using the virtual colony counting procedure, are tabulated in supplemental Table S2. The virtual LD50 and virtual LD90 values of HD5 were comparable with the values reported previously for both strains tested (39). As expected, the antibacterial activity of HD5 derived from proHD5 via trypsin processing was virtually identical to that of a chemically synthesized HD5. Interestingly, although the bactericidal activity of E14Q-HD5 against S. aureus was comparable with that of the wild type peptide, E14Q-HD5 killed E. coli much more effectively at higher concentrations. At the highest peptide concentration tested (25 μm), E. coli did not recover after exposure to the E14Q-HD5 mutant peptide.
FIGURE 10.
Survival curves of E. coli ATCC 25299 (left) and S. aureus ATCC 29213 (right) exposed to HD5-derived peptides as indicated. Strains were exposed to the peptides at concentrations varying 2-fold from 0.097 to 25 μm. Each curve is the mean of three separate experiments, where the error bars represent the standard deviation of the measurements. Points scored as zero survival could not be plotted.
We also tested the effect of the pro domain on the antibacterial activities of these peptides. Only in the case of E57Q-proHD5, we observed modest killing of S. aureus at the two highest concentrations tested. Taken together, these data indicate that replacement of the glutamic acid in the conserved salt bridge of HD5 selectively alters the antibacterial properties of the peptide and that the pro domain acts to neutralize its bactericidal activity. Interestingly, a recombinant proHD5 was reported to be active against S. typhymurium and Listeria monocytogenes (22, 42). The disparity in antibacterial activity between recombinant and synthetic proHD5 may be attributable to the use of different test strains, assay conditions, and concentrations.
DISCUSSION
The Arg6–Glu14 salt bridge found in HD5 is also highly conserved in other mammalian α-defensins except for guinea pig neutrophil α-defensins where the corresponding residues are Thr and Tyr (43). Wu et al. (28) reported that destabilization of the electrostatic interaction between Arg5 and Glu13 in HNPs dramatically accelerated defensin in vitro degradation by elastase, one of several serine proteases co-localized with neutrophil α-defensins in the azurophilic granules, suggesting that the salt bridge in α-defensins is conserved for their in vivo stability. However, Rosengren et al. (29) recently showed that weakening of the corresponding Arg7–Glu15 salt bridge in Crp4 by mutating Glu15 to Asp and several other amino acid residues had no effect on proteolytic susceptibility of the mouse α-defensin to its processing enzyme, matrilysin. It was therefore concluded that the salt bridge in mouse α-defensins is not conserved for their in vivo stability (29).
To help clarify the role of the conserved salt bridge in α-defensin structure and function, we chose human defensin 5 as a model compound because: 1) trypsin is known to be the processing enzyme of proHD5 in vivo and 2) trypsin readily recognizes the Arg-rich loop in HD5 (Arg6-His7-Gly8-Arg9-Cys10-Ala11-Thr12-Arg13-Glu14) stabilized by the Arg6–Glu14 salt bridge. We made the following findings. First, mutation of Glu14 to Gln significantly reduced oxidative folding efficiency of HD5 but had no effect on that of proHD5. Second, bovine trypsin productively and efficiently processed proHD5 in vitro, mimicking the activity of human trypsin in vivo (22), but spontaneously degraded E57Q-proHD5. Third, HD5 was resistant to trypsin treatment, whereas E14Q-HD5 was highly susceptible. Fourth, degradation of E14Q-HD5 by trypsin was initiated by the cleavage of the Arg13–Gln14 peptide bond in the loop region. Finally, the E14Q mutation did not alter the bactericidal activity of HD5 against S. aureus but significantly enhanced the killing of E. coli. By contrast, proHD5 and E57Q-proHD5 were largely inactive against both strains at the concentrations tested. Collectively, these results confirmed that the primary function of the conserved salt bridge in HD5 is to ensure correct processing of proHD5 and to stabilize the defensin in vivo.
How did Rosengren et al. (29) arrive at a conclusion totally different from ours then? The answer appears to lie in the choice of enzyme used in the susceptibility experiments. Although not the processing enzyme of human neutrophil α-defensin precursors, elastase preferably cleaves at multiple sites the extruding loop (Arg5-Ile6-Pro7-Ala8-Cys9-Ile10-Ala11-Gly12-Glu13) stabilized by the Arg5–Glu13 salt bridge in HNPs. By contrast, matrilysin, which prefers small hydrophilic residues at P1 and neutral residues at other subsite positions (44), is incapable of cleaving the heavily cationic loop in mouse cryptdin 4 (Arg7-Lys8-Gly9-His10-Cys11-Lys12-Arg13-Gly14-Glu15) irrespective of the Arg7–Glu15 salt bridge or lack thereof. As was demonstrated by this study using HD5 and HD5 analogs with trypsin, chymotrypsin, elastase, and matrilysin, the outcome is entirely dependent on the choice of enzyme used. Despite its physiological relevance to the processing of pro cryptdins in vivo, matrilysin, lacking the specificity to cleave the loop region, appears in retrospect to be an inappropriate choice of enzyme for the question of why the Arg7–Glu15 salt bridge is conserved in mouse cryptdins. Of note, it was recently brought to our attention that trypsin has been used to test proteolytic susceptibility of Crp4 and a series of its salt bridge destabilizing mutants, and the Arg7–Glu15 salt bridge was indeed found critical for proteolytic stability of the mouse α-defensin, in agreement with our current and earlier reports.3
α-Defensins, due to their small size, lack sufficient hydrophobic packing and are structurally stabilized by the three disulfide bridges Cys1–Cys6, Cys2–Cys4, and Cys3–Cys5 (sequential numbering). The extruding loop connecting the first and second β-strands presents the third Cys residue for disulfide bonding and is stabilized by the conserved Arg6–Glu14 salt bridge perpendicular to Cys3–Cys5 (Fig. 5). Mutation of Glu14 to Gln14 in HD5 dramatically reduced defensin folding efficiency, suggesting the importance of the salt bridge in stabilizing the loop to orient Cys3 for correct disulfide pairings, although loss of such stabilization is well tolerated in the folding of proHD5, where the pro peptide itself can catalyze pro defensin folding through favorable intramolecular interactions (20). It becomes apparent that the β-strand-connecting loop, present in all known mammalian α-defensins, is structurally indispensable for the correct α-defensin fold.
Proteases cleave protein substrates preferably in loops. The reason is 2-fold. First, loops, unlike ordered secondary structures such as α-helices and β-strands, can readily insert into the S1 specificity pocket of the enzyme with minimal steric hindrance. Second, loops are in general flexible, assuming a low free energy barrier to the transition state of the productive enzyme-substrate complex. Thus, conformational rigidity in the loop region of a protein is critical for “slowing down” otherwise efficient proteolytic cleavage (45). Understandably, further hydrolysis of HD5 in the presence of its processing enzyme trypsin was extremely slow due to loop stabilization afforded by the conserved salt bridge. By contrast, destabilization of the salt bridge in HD5 lessened the rigidity of the loop, rendering the loop susceptible to trypsin hydrolysis. The first cleavage by trypsin between Arg13 and Gln14 in the loop of E14Q-HD5 led to global destabilization of the defensin molecule presumably due to the loss of structural support to the Cys3–Cys5 disulfide and to weakened inter-β-strand interactions as well. Consequently, a dramatic acceleration of hydrolysis ensued. Notably, loss of the salt bridge in E14Q-HD5 perpetuated chymotrypsin cleave of Tyr27–Arg28, a site distal to the loop region, further demonstrating a propagated global effect of local structural destabilization on the proteolytic susceptibility of α-defensins.
Mutation of Glu14 to Gln14 in HD5 potentially creates a trypsin cleavage site at the Arg6–Thr7 peptide bond. In the NMR structure of E15D-Crp4, the side chain of Arg7 rotates out and projects into the solution, making itself accessible to the recognition by trypsin (29). Although it is tempting to suggest that a freed Arg6 in E14Q-HD5 directly contributes to defensin degradation, our findings on the initiation of trypsin cleavage indicate otherwise. Trypsin cleavage of E14Q-HD5 did not appear to be a random proteolytic event. Rather, it was specifically linked to loss of the salt bridge, thus destabilization of the protruding loop structure in the molecule. The results on R6A-HD5 clearly supported this conclusion.
Cationic charges functionally contribute to microbial killing by defensins (46–49). Mutation of Glu14 to Gln14 in the salt bridge frees up Arg6, thus increasing the cationicity of HD5. It is therefore reasonable to expect an enhanced bactericidal activity from E14Q-HD5 as compared with wild type HD5. This was clearly the case with E. coli. However, the Glu14-to-Gln14 mutation did not affect the killing of S. aureus. Interestingly, breakage of the salt bridge in HNP2 had little effect on the killing of either E. coli or S. aureus (28). Similar results were also reported for salt bridge-deficient mutants of mouse cryptdin 4 (29), which, as compared with the wild type Crp4, showed little difference in their bactericidal activity against multiple strains. Thus, it appears that the functional ramification of neutralization of the anionic partner in the conserved salt bridge is dependent not only on bacterial strains but also on defensin species. These findings are not entirely surprising in light of the previous reports that bacterial killing by defensins is in general strain- and sequence-specific (39), although the molecular determinants for defensin activity and specificity still remain poorly defined. It is worth pointing out that S. aureus killing by HD5 is structure (disulfide)-dependent, whereas E. coli killing by HD5 is not (50). In contrast, killing of both E. coli and S. aureus by human β-defensin 3 is structure (disulfide)-independent (51–53). Functional disparities seen in the action of a variety of defensins may be a reflection of molecular complexity involving defensin interactions with not just the bacterial membrane but with diverse bacterial cell wall components as well.
Supplementary Material
Acknowledgments
We are grateful to Prof. Andre J. Ouellette for incisive comments and for sharing with us unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grants AI056264 and AI061482 (to W. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental tables and a supplemental figure.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: HNP, human neutrophil peptide; HD5, human α-defensin 5; HPLC, high pressure liquid chromatography; RP-HPLC, reverse phase-HPLC; ESI-MS, electrospray ionization-mass spectrometer; Boc, t-butoxycarbonyl.
A. J. Ouellette, personal communications.
References
- 1.Ganz, T. (2003) Nat. Immunol. 3 710-720 [DOI] [PubMed] [Google Scholar]
- 2.Lehrer, R. I. (2004) Nat. Rev. Microbiol. 2 727-738 [DOI] [PubMed] [Google Scholar]
- 3.Selsted, M. E., and Ouellette, A. J. (2005) Nat. Immunol. 6 551-557 [DOI] [PubMed] [Google Scholar]
- 4.Zasloff, M. (2002) Nature 415 389-395 [DOI] [PubMed] [Google Scholar]
- 5.Yang, D., Biragyn, A., Hoover, D. M., Lubkowski, J., and Oppenheim, J. J. (2004) Annu. Rev. Immunol. 22 181-215 [DOI] [PubMed] [Google Scholar]
- 6.Selsted, M. E., Harwig, S. S., Ganz, T., Schilling, J. W., and Lehrer, R. I. (1985) J. Clin. Investig. 76 1436-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganz, T., Selsted, M. E., Szklarek, D., Harwig, S. S., Daher, K., Bainton, D. F., and Lehrer, R. I. (1985) J. Clin. Investig. 76 1427-1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabay, J. E., Scott, R. W., Campanelli, D., Griffith, J., Wilde, C., Marra, M. N., Seeger, M., and Nathan, C. F. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 5610-5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilde, C. G., Griffith, J. E., Marra, M. N., Snable, J. L., and Scott, R. W. (1989) J. Biol. Chem. 264 11200-11203 [PubMed] [Google Scholar]
- 10.Singh, A., Bateman, A., Zhu, Q. Z., Shimasaki, S., Esch, F., and Solomon, S. (1988) Biochem. Biophys. Res. Commun. 155 524-529 [DOI] [PubMed] [Google Scholar]
- 11.Jones, D. E., and Bevins, C. L. (1992) The J. Biol. Chem. 267 23216-23225 [PubMed] [Google Scholar]
- 12.Jones, D. E., and Bevins, C. L. (1993) FEBS Lett. 315 187-192 [DOI] [PubMed] [Google Scholar]
- 13.Jia, H. P., Schutte, B. C., Schudy, A., Linzmeier, R., Guthmiller, J. M., Johnson, G. K., Tack, B. F., Mitros, J. P., Rosenthal, A., Ganz, T., and McCray, P. B., Jr. (2001) Gene (Amst.) 263 211-218 [DOI] [PubMed] [Google Scholar]
- 14.Schutte, B. C., Mitros, J. P., Bartlett, J. A., Walters, J. D., Jia, H. P., Welsh, M. J., Casavant, T. L., and McCray, P. B., Jr. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2129-2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorin, J. R., and Jackson, I. J. (2007) Science 318 1395. [DOI] [PubMed] [Google Scholar]
- 16.Pazgier, M., Hoover, D. M., Yang, D., Lu, W., and Lubkowski, J. (2006) CMLS Cell Mol. Life Sci. 63 1294-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, L., and Ganz, T. (1995) Blood 85 1095-1103 [PubMed] [Google Scholar]
- 18.Valore, E. V., Martin, E., Harwig, S. S., and Ganz, T. (1996) J. Clin. Investig. 97 1624-1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valore, E. V., and Ganz, T. (1992) Blood 79 1538-1544 [PubMed] [Google Scholar]
- 20.Wu, Z., Li, X., Ericksen, B., de Leeuw, E., Zou, G., Zeng, P., Xie, C., Li, C., Lubkowski, J., Lu, W. Y., and Lu, W. (2007) J. Mol. Biol. 368 537-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal, A. W. (2005) Annu. Rev. Immunol. 23 197-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T., and Bevins, C. L. (2002) Nat. Immunol. 3 583-590 [DOI] [PubMed] [Google Scholar]
- 23.Zasloff, M. (2002) Nat. Immunol. 3 508-510 [DOI] [PubMed] [Google Scholar]
- 24.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y., and Bevins, C. L. (2003) Nature 422 522-526 [DOI] [PubMed] [Google Scholar]
- 25.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M., and Parks, W. C. (1999) Science 286 113-117 [DOI] [PubMed] [Google Scholar]
- 26.Ouellette, A. J. (2006) Curr. Top. Microbiol. Immunol. 306 1-25 [DOI] [PubMed] [Google Scholar]
- 27.Xie, C., Prahl, A., Ericksen, B., Wu, Z., Zeng, P., Li, X., Lu, W. Y., Lubkowski, J., and Lu, W. (2005) J. Biol. Chem. 280 32921-32929 [DOI] [PubMed] [Google Scholar]
- 28.Wu, Z., Li, X., de Leeuw, E., Ericksen, B., and Lu, W. (2005) J. Biol. Chem. 280 43039-43047 [DOI] [PubMed] [Google Scholar]
- 29.Rosengren, K. J., Daly, N. L., Fornander, L. M., Jonsson, L. M., Shirafuji, Y., Qu, X., Vogel, H. J., Ouellette, A. J., and Craik, D. J. (2006) J. Biol. Chem. 281 28068-28078 [DOI] [PubMed] [Google Scholar]
- 30.Szyk, A., Wu, Z., Tucker, K., Yang, D., Lu, W., and Lubkowski, J. (2006) Protein Sci. 15 2749-2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, Z., Ericksen, B., Tucker, K., Lubkowski, J., and Lu, W. (2004) J. Pept. Res. 64 118-125 [DOI] [PubMed] [Google Scholar]
- 32.Dawson, P. E., and Kent, S. B. (2000) Annu. Rev. Biochem. 69 923-960 [DOI] [PubMed] [Google Scholar]
- 33.Dawson, P. E., Muir, T. W., Clark-Lewis, I., and Kent, S. B. (1994) Science 266 776-779 [DOI] [PubMed] [Google Scholar]
- 34.Schnolzer, M., Alewood, P., Jones, A., Alewood, D., and Kent, S. B. (1992) Int. J. Pept. Protein Res. 40 180-193 [DOI] [PubMed] [Google Scholar]
- 35.Kent, S. B. (1988) Annu. Rev. Biochem. 57 957-989 [DOI] [PubMed] [Google Scholar]
- 36.Pace, C. N., Vajdos, F., Fee, L., Grimsley, G., and Gray, T. (1995) Protein Sci. 4 2411-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read, R. J. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 57 1373-1382 [DOI] [PubMed] [Google Scholar]
- 38.Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S., and Dodson, E. J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 247-255 [DOI] [PubMed] [Google Scholar]
- 39.Ericksen, B., Wu, Z., Lu, W., and Lehrer, R. I. (2005) Antimicrob. Agents Chemother. 49 269-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, Z., Powell, R., and Lu, W. (2003) J. Am. Chem. Soc. 125 2402-2403 [DOI] [PubMed] [Google Scholar]
- 41.Wu, Z., Prahl, A., Powell, R., Ericksen, B., Lubkowski, J., and Lu, W. (2003) J. Pept. Res. 62 53-62 [DOI] [PubMed] [Google Scholar]
- 42.Tanabe, H., Ayabe, T., Maemoto, A., Ishikawa, C., Inaba, Y., Sato, R., Moriichi, K., Okamoto, K., Watari, J., Kono, T., Ashida, T., and Kohgo, Y. (2007) Biochem. Biophys. Res. Commun. 358 349-355 [DOI] [PubMed] [Google Scholar]
- 43.Selsted, M. E., and Harwig, S. S. (1987) Infect Immun. 55 2281-2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turk, B. E., Huang, L. L., Piro, E. T., and Cantley, L. C. (2001) Nat. Biotechnol. 19 661-667 [DOI] [PubMed] [Google Scholar]
- 45.Lu, W. Y., Starovasnik, M. A., Dwyer, J. J., Kossiakoff, A. A., Kent, S. B., and Lu, W. (2000) Biochemistry 39 3575-3584 [DOI] [PubMed] [Google Scholar]
- 46.Satchell, D. P., Sheynis, T., Kolusheva, S., Cummings, J., Vanderlick, T. K., Jelinek, R., Selsted, M. E., and Ouellette, A. J. (2003) Peptides 24 1795-1805 [DOI] [PubMed] [Google Scholar]
- 47.Tanabe, H., Qu, X., Weeks, C. S., Cummings, J. E., Kolusheva, S., Walsh, K. B., Jelinek, R., Vanderlick, T. K., Selsted, M. E., and Ouellette, A. J. (2004) J. Biol. Chem. 279 11976-11983 [DOI] [PubMed] [Google Scholar]
- 48.Hancock, R. E. (1997) Lancet 349 418-422 [DOI] [PubMed] [Google Scholar]
- 49.Zou, G., de Leeuw, E., Li, C., Pazgier, M., Zeng, P., Lu, W. Y., Lubkowski, J., and Lu, W. (2007) J. Biol. Chem. 282 19653-19665 [DOI] [PubMed] [Google Scholar]
- 50.de Leeuw, E., Burks, S. R., Li, X., Kao, J. P., and Lu, W. (2007) FEBS Lett. 581 515-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kluver, E., Schulz-Maronde, S., Scheid, S., Meyer, B., Forssmann, W. G., and Adermann, K. (2005) Biochemistry 44 9804-9816 [DOI] [PubMed] [Google Scholar]
- 52.Taylor, K., Clarke, D. J., McCullough, B., Chin, W., Seo, E., Yang, D., Oppenheim, J., Uhrin, D., Govan, J. R., Campopiano, D. J., Macmillan, D., Barran, P. E., and Dorin, J. R. (2008) J. Biol. Chem. 283 6631-6639 [DOI] [PubMed] [Google Scholar]
- 53.Wu, Z., Hoover, D. M., Yang, D., Boulegue, C., Santamaria, F., Oppenheim, J. J., Lubkowski, J., and Lu, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8880-8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.