Abstract
Although the N-methyl-d-aspartate (NMDA) receptor plays a critical role in the central nervous system, many questions remain regarding the relationship between its structure and functional properties. In particular, the involvement of ligand-binding domain closure in determining agonist efficacy, which has been reported in other glutamate receptor subtypes, remains unresolved. To address this question, we designed dual cysteine point mutations spanning the NR1 and NR2 ligand-binding clefts, aiming to stabilize these domains in closed cleft conformations. Two mutants, E522C/I691C in NR1 (EI) and K487C/N687C in NR2 (KN) were found to exhibit significant glycine- and glutamate-independent activation, respectively, and co-expression of the two subunits produced a constitutively active channel. However, both individual mutants could be activated above constitutive levels in a concentration-dependent manner, indicating that cleft closure does not completely prevent agonist association. Interestingly, whereas the NR2 KN disulfide was found to potentiate channel gating and M3 accessibility, NR1 EI exhibited the opposite phenotype, suggesting that the EI disulfide may trap the NR1 ligand-binding domain in a lower efficacy conformation. Furthermore, both mutants affected agonist sensitivity at the opposing subunit, suggesting that closed cleft stabilization may contribute to coupling between the subunits. These results support a correlation between cleft stability and receptor activation, providing compelling evidence for the Venus flytrap mechanism of glutamate receptor domain closure.
Ionotropic glutamate receptors (iGluRs)2 are key mediators of excitatory neurotransmission, contributing to both neuronal development and adult neuroplasticity (1, 2). In particular, the N-methyl-d-aspartate (NMDA) receptor subclass is responsible for initiating activity-dependent changes in synaptic strength, proposed to form the molecular basis of learning and memory (3). Additionally, NMDA receptors have been implicated in the pathogenesis of numerous neurological disorders, including schizophrenia and Alzheimer disease, as well as neurodegeneration following ischemia or brain injury (4–6). Despite their enormous therapeutic potential, clinical use of NMDA receptor antagonists has been limited by significant neurological side effects. However, d-cycloserine (DCS), a well tolerated partial agonist, has shown therapeutic promise as a cognitive enhancer in Alzheimer disease and head trauma recovery (7, 8). Therefore, the mechanism by which glutamate receptors distinguish partial agonists from full agonists is of both scientific and medical interest.
The iGluR family is composed of three subclasses, α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA), kainate, and NMDA receptors, all of which share the same modular domain structure. Two discontinuous segments, S1 and S2, form the ligand binding domain (LBD), whereas the ion channel portion contains three putative transmembrane segments, M1, M3, and M4, and a re-entrant loop, M2. NMDA receptors generally form as tetramers of two NR1 and two NR2 subunits, which bind glycine and glutamate, respectively (9). At present, LBD crystal structures have been obtained for each iGluR subclass, revealing bilobed clamshell-shaped domains arranged as back-to-back dimers (10–12). Within each LBD, domain 1 (D1) is stabilized by cross-subunit interactions, whereas domain 2 (D2) rotates and closes around bound ligand. Extensive studies with the GluR2 AMPA receptor, in complex with a range of full and partial agonists, established a correlation between agonist efficacy and degree of LBD closure, and a similar relationship was observed in kainate receptors (11, 13). These results led to a structural model of receptor activation, in which agonist binding promotes LBD closure by rotating D2 toward D1, separating the linker regions and promoting channel opening. According to this hypothesis, partial agonists produce less domain closure, leading to decreased linker separation and slowing a subunit-specific pregating conformational change (14).
However, crystal structures obtained for the NR1 subunit in complex with full and partial agonists adopt a similar degree of domain closure; thus, it appears that a different structural mechanism is required to account for partial agonist action at the NMDA receptor (15). Structural and molecular dynamics data have implicated the conformation of the hinge region, particularly the second interdomain β strand, in sensing agonist efficacy (15, 16). Increased interpocket motion has also been associated with NR1 site partial agonism, and both glycine and DCS can reportedly move within the pocket without affecting domain closure (16). However, it remains unclear whether this destabilization is transmitted to the ion channel via increased cleft opening, permitting faster agonist dissociation, or a more subtle relay mechanism.
In the NR2 subunit, a recent study has demonstrated that strengthening the D1-D2 interaction can increase open probability, kinetically linking domain closure to channel gating (17). Furthermore, destabilizing the closed cleft state by engineering steric clashes has been shown to reduce agonist efficacy and apparent affinity in NR2, GluR2, and GluR6 (18–20). However, partial agonist crystal structures have not yet been reported for NR2, so it remains to be determined whether the NR2 LBD behaves like NR1 or follows the AMPA/kainate receptor paradigm of partial agonist action.
In the present study, we explored the relationship between cleft closure and activation in the NMDA receptor by stabilizing the NR1 and NR2 subunits in closed-cleft conformations via introduction of cross-cleft disulfide bonds. Both disulfide bonds induced significant constitutive activation, but although the NR2 LBD mutant appeared to be stabilized in a nearly full-agonist conformation, the disulfide-stabilized NR1 LBD displayed characteristics more indicative of a partial agonist. Our results suggest that although cleft closure is an important mediator of agonist efficacy, additional mechanisms are required to achieve full receptor activation, including a potential role for helix F. Additionally, both disulfide mutants increased agonist sensitivity at the opposing subunit, suggesting a possible intersubunit interaction between the NR1 and NR2 binding clefts.
MATERIALS AND METHODS
Molecular Modeling—Residue selection, mutation, and rotamer optimization were performed in the Swiss PDB Viewer (available on the World Wide Web), which was also used to detect hydrogen bonds and measure interatomic distances. The resulting molecular graphics were rendered in POV-Ray version 3.6.
Site-directed Mutagenesis—Mutations in NR1 were generated using the megaprimer PCR method, as described previously (22), and subsequently confirmed by DNA sequencing of the cloned region. cRNA was prepared from linearized plasmids by in vitro transcription (22). Amino acids in both NR1 and NR2 are numbered from the initiator methionine. The NR2 subunit used in all experiments is an NR2A construct with a shortened 5′-untranslated region, previously shown to increase expression (23).
Oocyte Preparation and Injection—Stage V-VI oocytes were surgically removed from Xenopus laevis frogs under tricaine anesthesia, followed by manual defolliculation and treatment with collagenase type I (Invitrogen) (24). All frog work was carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Oocytes were injected with a 1:1 ratio of NR1 and NR2 cRNA (75 nl of a 10–100 ng/μl solution), transferred to SOS buffer, and incubated at 19 °C. SOS buffer consisted of 100 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH-adjusted to 7.6 with NaOH, and supplemented with gentamicin (100 μg/ml).
Two-electrode Voltage Clamp Electrophysiology—Functional expression was assessed 2–6 days after injection using a two-electrode voltage clamp amplifier (OC-725; Warner Instrument, Hamden, CT). The extracellular recording solution consisted of low barium Ringer's solution (Lobar; 100 mm NaCl, 5 mm KCl, 0.5 mm BaCl2, 10 mm HEPES, 10 μm EDTA), pH-adjusted to 7.35 with Tris base and maintained at room temperature. Barium was used as the divalent cation to minimize secondary activation of calcium-activated Cl- currents (25). EDTA was included to chelate trace amounts of the soft metal divalent cations Cd2+ and Zn2+, which have been reported to contaminate buffer solutions and inhibit the NMDA receptor by binding to a high affinity site (26, 27). EDTA also removes a zinc-dependent component of desensitization (28). Maximal current response was elicited by application of 100 μm glycine and 100 μm l-glutamate, and agonists were co-applied unless stated otherwise. Voltage clamp recordings were performed in a perfusion chamber (Warner Instrument Corp., Hamden CT) optimized for laminar flow, and solution changes were accomplished using a gravity-fed, computer-controlled perfusion system, at a flow rate of ∼15 ml/min (24). Oocytes were impaled with low resistance glass microelectrodes (0.5–2.0 megaohms) filled with 3 m KCl and maintained at a holding potential of -60 mV. Data acquisition and voltage control were performed with PClamp hardware and software (Axon Instruments, Burlingame, CA).
Measurement of Contaminating Glycine Concentration—The concentration of glycine in our Lobar solution, both in the presence and absence of glutamate, was evaluated via liquid chromatography-mass spectrometry using an ABI QStar electrospray mass spectrometer. The samples were run in positive mode, using a reverse phase column and 10–95% acetonitrile, 0.1% trifluoroacetic acid gradient. Glycine peaks (m/z = 76) were not observed in the total ion, total wavelength, or extracted ion chromatograms at a detection threshold of 1 pm.
Curve Fitting and Statistics—Concentration-response curves were obtained by fitting data with a modified Hill equation: R/Rmax = Y0 + (1 - Y0)/(1 + (EC50/A)n), where R is the response for the given agonist concentration (A), Rmax is the maximum response, n is the Hill coefficient, EC50 is the concentration midpoint, and Y0 is the y intercept. Parameters were optimized by minimizing the residual sum of squares using the Solver function in Microsoft Excel. Each concentration point represents 4–8 oocytes, and error bars indicate S.E. All other data are presented as mean ± S.E. from 5–12 oocytes and analyzed statistically using one-way analysis of variance. For multiple comparisons, the data were initially subjected to a global analysis of variance incorporating all factors and measurements, and if this test showed a strong interaction between mutant and agonist response (p < 0.001), data were subdivided by agonist for lower order tests. Fischer's protected least significant difference test was then applied to compare the effects of the mutation on each response. Statistical significance is indicated with an asterisk for p < 0.05 (significant) or a double asterisk for p < 0.01 (highly significant). Time courses of methanethiosulfonate (MTSEA) modification and MK-801 block were fitted with first order exponential functions using the Clampfit module in pCLAMP 9.0.
Reagents—All chemicals were purchased from Sigma with the exception of MTSEA (Toronto Research Chemicals), glycine (EMD Biochemicals), and l-glutamate (Invitrogen). DCK stock solutions were initially made in DMSO and diluted to their final concentrations in Lobar. For those experiments, DMSO was added to all recording solutions to maintain a standard concentration.
RESULTS
Previous reports from our laboratory and others have suggested that the M3 domain functions as a transduction element, coupling ligand binding to channel opening, and agonist efficacy has been shown to correlate with M3 accessibility (24). To further explore the activation mechanism of the NMDA receptor, we set out to investigate the role of cleft closure in sensing partial agonist efficacy, specifically focusing on how cleft closure affects the accessibility of M3 and agonist sensitivity of both subunits. Closed cleft stabilization was achieved by engineering disulfide bonds, a technique commonly used to lock proteins in a defined conformation, spanning the NR1 and NR2 ligand-binding clefts.
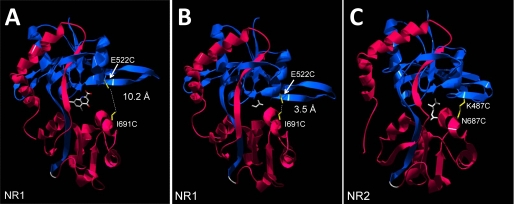
Designing the Disulfide Mutants—Complementary residues in the NR1 subunit were chosen by comparing the glycine- and antagonist-bound S1S2 crystal structures and identifying a cleft-spanning pair that could potentially interact only in the closed cleft conformation. Following cysteine substitution, the S-S distance within the Glu522/Ile691 pair (Fig. 1, A and B) was measured at 10.2 Å in the open cleft state and 3.5 Å after glycine binding, close enough to form a hydrogen bond and potentially a disulfide with a slight increase in cleft closure. Since an open cleft crystal structure for the NR2 S1S2 domain has not yet been published, pairs of NR2 residues were selected from the glutamate-bound structure and compared with full-agonist and antagonist-bound GluR2 crystal structures (29). One set of NR2 residues, K487C and N687C, was predicted to form a disulfide bond in the glutamate-bound NR2 crystal structure (Fig. 1C). The homologous GluR2 substitutions (A452C and S652C) were predicted to form a hydrogen bond in the AMPA-bound structure but moved to 5.66 Å apart in the antagonist-bound structure, suggesting the possibility of a cleft-closing disulfide interaction. The selected residues were then introduced into their respective subunits, both individually and in pairs, and expressed in Xenopus oocytes for functional characterization.
FIGURE 1.
Design of the disulfide mutants. A, structure of the NR1 ligand-binding domain in its open cleft conformation, bound to competitive antagonist DCK. Cysteine substitutions at E522 and I691C, depicted in yellow, are separated by a distance of 10.20 Å. The S1 lobe is shown in blue, S2 in pink, and the GT linker in gray. B, structure of the NR1 ligand-binding domain bound to glycine, representing the closed cleft conformation. Domain closure decreases the distance between E522C and I691C to 3.5 Å. C, the closed cleft conformation of the NR2A ligand-binding domain, shown bound to glutamate. Cysteine substitutions were introduced at positions Lys487 and Asn687, as depicted in yellow, and separated by a distance of 3.3 Å. Measurements were made using Swiss PDB Viewer, based on accession codes 1PBQ, 1PB7, and 2A5S for A–C, respectively.
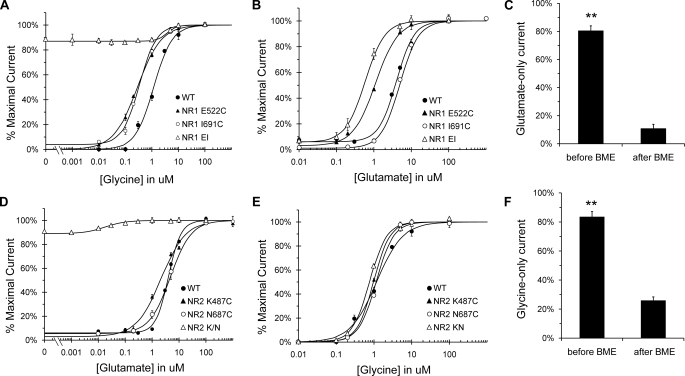
Glycine-independent Activation of the NR1 Subunit—To test for the presence of a cleft-stabilizing interaction, glycine concentration-response curves were performed on the NR1 double mutant and the individual point mutants (Fig. 2A and Table 1). Both E522C and I691C exhibited an increased sensitivity to glycine compared with wild type, as evidenced by the left-shifted curves, but very little glycine-independent current. In contrast, the E522C/I691C double mutant (henceforth referred to as EI) displayed significant glycine-independent current, ∼87% activation in the presence of glutamate alone. A left-shifted concentration-response curve has been previously reported for the I691C mutant, attributed to the stabilizing effects of an interlobe hydrogen bond with Glu522 (30), and replacing Glu522 with the smaller cysteine residue appears to eliminate potential clashes with the cross-cleft isoleucine. However, although each single mutation favorably affects cleft stability, the phenotype of the double mutant is clearly synergistic. Trace amounts of glycine have been reported to contaminate buffer solutions (31, 32), resulting in transient glutamate-only currents, but since NR1 EI also exhibits decreased glycine sensitivity (Fig. 2A), glycine contamination cannot account for the substantial glutamate-only current observed in this mutant. Additionally, our buffer and glutamate-only solutions were assayed via liquid chromatography-mass spectrometry, which determined background glycine levels to be below a detection level of 1 pm.
FIGURE 2.
Agonist-independent activation of NR1 EI and NR2 KN. A, glycine concentration-response curves for NR1 mutants E522C, I691C, and EI in the presence of 100 μm l-glutamate. Glycine sensitivity was increased in all three mutants, but only the EI double mutant exhibited partial activation in the absence of glycine (87%, compared with 4% for E522C and 6% for I691C). B, glutamate concentration-response curves for NR1 E522C, I691C, and EI in the presence of 100 μm glycine. Glutamate sensitivity was increased in both E522C and EI (concentration midpoint = 1.11 and 0.58 μm, respectively, compared with 4.00 μm for wild type) but not I691C. Glutamate-independent current was not significantly different from wild type for any of the mutants. C, BME inhibition of EI glycine-independent response. The ratio of current elicited by 100 μm glutamate to current elicited by full agonist (100 μm glycine plus 100 μm glutamate) was compared before and after a 10-s application of 5% BME. BME treatment was sufficient to inhibit EI glutamate-only current by 70 ± 3%. D, glutamate concentration-response curves for NR2 K487C, N687C, and K487C/N687C (KN) in the presence of 100 μm glycine. K487C displayed a small increase in glutamate sensitivity (midpoint = 2.20 μm), whereas N687C was indistinguishable from wild type (midpoint = 4.00 μm). In contrast, the KN concentration-response curve was significantly left-shifted, exhibiting a concentration midpoint of 0.02 μm. The KN mutant also displayed a significant degree of activation in the absence of glutamate (90%, compared with 5% for either single mutant). E, glycine concentration-response curves for NR2 mutants K487C, N687C, and KN in the presence of 100 μm l-glutamate. K487C and KN displayed slightly increased glycine sensitivity, with the largest increase seen in KN (concentration midpoint = 0.76 μm compared with 1.16 for WT NR2). None of the mutants exhibited any glycine-independent current. F, BME inhibition of KN glutamate-independent response. Same as in C, except the ratio of glycine-only current to full agonist current was measured. BME treatment inhibited NR2 KN glycine-only current by 58 ± 4%.
TABLE 1.
Concentration-response data for NR1 EI and NR2 KN Shown are values obtained from fitting concentration-response curves for glycine, glutamate, DCK, and APV with the Hill equation. Y0 represents the glycine- or glutamate-independent current, nH is the Hill coefficient, and EC50/IC50 is the concentration midpoint in μm. Note that the EI glycine and KN glutamate midpoints are not technically EC50 values, since the receptors are over 50% activated in the absence of agonist.
|
Gly
|
Glu
|
DCK
|
APV
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Y0 | EC50 | nH | Y0 | EC50 | nH | IC50 | nH | IC50 | nH | |
| μm | μm | μm | μm | |||||||
| WT | 0 ± 0% | 1.16 | 1.24 | 7 ± 1% | 4.00 | 1.84 | 41.2 | 1.94 | 18.0 | 1.06 |
| NR1 EI | 87 ± 4% | 3.75 | 2.35 | 7 ± 2% | 0.58 | 1.64 | 250 | 0.38 | 116 | 1.60 |
| NR1 E522C | 4 ± 1% | 0.28 | 0.99 | 2 ± 1% | 1.11 | 1.45 | ||||
| NR1 I691C | 8 ± 1% | 0.34 | 1.35 | 2 ± 1% | 4.64 | 1.94 | ||||
| NR2 KN | 0 ± 0% | 0.76 | 1.90 | 90 ± 1% | 0.02 | 0.90 | 41.5 | 2.07 | 8.81 | 0.54 |
| NR2 K487C | 0 ± 0% | 0.97 | 2.00 | 5 ± 1% | 2.20 | 0.94 | ||||
| NR2 N687C | 0 ± 0% | 1.20 | 2.45 | 5 ± 2% | 4.92 | 1.09 | ||||
To examine whether the altered phenotype of the NR1 LBD had any intersubunit effects, glutamate concentration-response curves were also determined for all three mutants (Fig. 2B and Table 1). Both EI and E522C were significantly left-shifted relative to wild type, suggesting a potential positive coupling between two subunits reported to exhibit negative cooperativity (21). The source of the E522C phenotype is unclear but may be due to the removal of cross-cleft steric clashes observed in silico with the glutamate residue. The presence of a functional disulfide bond was tested initially with 1 and 10 mm dithiothreitol (data not shown), which had little effect on the agonist-independent current. A smaller reducing agent, 2-mercaptoethanol (BME), was successful at reducing the glycine-only current by 70% (Fig. 2C), thus confirming the presence of a cleft-stabilizing disulfide bond. The possibility that some cysteine residues do not form disulfide bonds has been reported for LBD-stabilizing disulfides in the GABA-B receptor (33); however, treatment with 0.1% H2O2, an oxidizing agent, and full agonist test pulses had no significant effect on the glycine-independent response of NR1 EI receptors (data not shown), indicating that unreacted cysteine residues are unlikely to contribute to the phenotype of this mutant.
Glutamate-independent Activation of the NR2 Subunit—The same set of experiments was repeated for the NR2 cysteine mutants, K487C, N687C, and K487C/N687C (KN), to assess the degree of glutamate-independent activation. One of the individual cysteine mutants, K487C, exhibited a small leftward shift of the glutamate concentration-response curve, most likely due to relief of potential clashes observed between lysine and the cross-cleft asparagine (Fig. 2D). The NR2 KN mutant, however, was substantially more sensitive to glutamate, with a concentration midpoint of 21 nm. Furthermore, KN receptors were 90% activated by glycine alone, indicating a significant glutamate-independent response. Glycine concentration-response curves were also performed, in order to gauge the effects of the NR2 phenotype on the NR1 subunit (Fig. 2E). K487C and KN were both slightly left-shifted, with 1.2–1.5-fold decreases in the concentration midpoint, indicating that eliminating the steric clashes between Lys487 and Asn687, and thus facilitating NR2 LBD closure, is the most likely cause of increased glycine sensitivity. Reducing agents were used to probe the existence of a cross-cleft disulfide, which was also resistant to 1 and 10 mm DTT treatment). BME application, however, resulted in a 58% reduction in glycine-only current, strongly suggesting the presence of a cleft-stabilizing disulfide interaction (Fig. 2F). Full agonist test pulses had no effect on the constitutive activation of KN receptors; however, a small degree of potentiation (15%) was observed following H2O2 treatment (data not shown). Thus, we cannot rule out the possibility that unreacted cysteine residues contribute to the phenotype of NR2 KN.
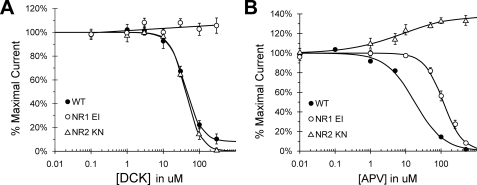
Decreased Sensitivity to Competitive Antagonists—The NR1 EI and NR2 KN mutants were further characterized using concentration-inhibition curves for 5,7-dichlorokynurenic acid (DCK), a competitive glycine-site antagonist, and 2-amino-5-phosphonovalerate (APV), a competitive glutamate site antagonist. NR2 KN displayed similar DCK sensitivity to wild type, whereas NR1 EI was completely immune to inhibition (Fig. 3A). NR2 KN was not only insensitive to APV inhibition; the concentration-response curve revealed a 33% potentiation with the competitive antagonist (Fig. 3B). Both results are consistent with stabilization of the LBDs in a closed cleft state, which could preclude antagonist access to the ligand-binding cleft and/or prevent antagonist-induced cleft opening. Interestingly, NR1 EI also exhibited a 6.5-fold increase in APV IC50, indicating a decreased sensitivity to glutamate site antagonism consistent with the observed increased sensitivity to glutamate.
FIGURE 3.
Decreased antagonist sensitivity in NR1 EI and NR2 KN. A, DCK concentration-inhibition curves for EI and KN, determined in the presence of 100 μm glycine and 100 μm glutamate. The EI double mutant was completely insensitive to DCK inhibition, whereas the KN mutant was indistinguishable from wild type (midpoint = 41.24 μm). B, APV concentration-inhibition curves for EI and KN, also determined in the presence of 100 μm glycine and 100 μm glutamate. The EI curve was right-shifted relative to wild type (concentration midpoint = 115.96 μm compared with 17.96 μm for WT), whereas the KN mutant was completely insensitive to APV inhibition. In contrast, APV treatment activated KN receptors above the response elicited by full agonist, resulting in a 33% potentiation.
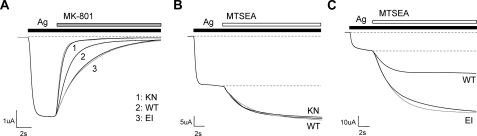
NR1 EI and NR2 KN Have Divergent Effects on Channel Activation—Further experiments were undertaken to determine the effect of our LBD disulfide mutants on the channel gate and the M3 segment, proposed to couple ligand binding to channel gating (24). The rate of inhibition by MK-801, an irreversible NMDA receptor open channel blocker, has been shown to depend on channel open probability (Po) and MK-801 binding rate (34). Given that MK-801 interacts with the channel pore, its binding rate should be unaffected by mutations within the ligand-binding clefts, and we therefore assumed that an altered block rate would reflect a change in Po. Both mutants were treated with 200 nm MK-801 in the presence of saturating glycine and/or glutamate (Fig. 4A); the MK-801 concentration was optimized to yield an inhibition time course of ∼10 s in wild type, facilitating comparison of blocking kinetics. Both disulfide mutants were fully inhibited by 200 nm MK-801 but displayed significant differences in block rate (Table 2). The NR1 EI mutant was found to block ∼2-fold slower than wild type, indicating an unexpected decrease in open probability. In the absence of glycine, the EI block rate decreased even further, but the difference between both agonists and glutamate alone was not statistically significant. Thus, closing the glycine-binding domain via an EI disulfide bond appears to inhibit channel gating, even in the presence of full agonist. In contrast, the MK-801 block rate in the NR2 KN mutant was increased by ∼2-fold relative to wild type, in both the presence and absence of glutamate. These results suggest that the KN disulfide bond has a potentiating effect on channel gating, such that the response elicited from KN with glycine alone has a higher Po than the wild type full agonist response.
FIGURE 4.
Effect of EI and KN mutations on MK-801 block rate and MTSEA potentiation. A, representative whole cell traces illustrating block by 200 nm MK-801, applied in the presence of 20 μm glycine and/or 100 μm glutamate. MK-801 block rates were fitted with first-order exponential functions using the Clampfit module of pCLAMP 9.0. For NR1 EI receptors, the time course of MK-801 inhibition was 2.21-fold slower than WT (with glycine) and 2.82-fold slower (without glycine). NR2 KN receptors, in comparison, were blocked 2.06-fold faster (with glutamate) and 2.05-fold faster (without glutamate). Each mutant trace is numbered, as indicated, and application of agonist (Ag) and MK-801 are depicted by black and gray bars, respectively. The gray traces illustrate block of EI with glutamate only and KN with glycine only. Maximum responses for each trace were normalized to illustrate kinetic differences. B, representative traces depicting MTSEA potentiation of NR2 KN and NR2 WT receptors, co-expressed with the NR1-A7C reporter construct. Currents were elicited with 20 μm glycine and 100 μm glutamate or 20 μm glycine, indicated by the black bar (agonis). Potentiation by 0.5 mm MTSEA (white bar) was not significantly different between WT (1.61-fold) and KN-containing receptors (1.61-fold with glutamate, 1.56-fold without glutamate). KN in the absence of glutamate is represented by a gray trace. In both B and C, maximum current responses were normalized to illustrate differences in degree of potentiation, and dashed gray lines depict normalized response levels in the presence and absence of full agonist. C, MTSEA potentiation of NR1 EI and NR1 WT receptors, co-expressed with the NR2-A7C reporter construct. Currents were elicited with 20 μm glycine and 100 μm glutamate or with 100 μm glutamate (Ag, black bar), and EI in the absence of glycine is represented by a gray trace. The addition of 0.5 mm MTSEA, depicted with a white bar, modified and potentiated EI-containing receptors 4.85-fold (with glycine) and 5.25-fold (without glycine), compared with 2.73-fold for WT.
TABLE 2.
MTSEA modification and MK-801 block rates Shown are the rate and degree of potentation by 0.5 mm MTSEA and inhibition by 200 nm MK-801. MTSEA potentiation values for NR1 EI and NR2 KN indicate modification of the opposing subunit (NR2-A7C and NR1-A7C, respectively). Relative Po values were estimated from the ratio of MTSEA -fold potentiation or MK-801 block, based on a reported peak Po of 0.5 (36).
|
MTSEA
|
MK-801
|
|||||
|---|---|---|---|---|---|---|
| Potentiation | Modified rate | Relative Po | Block | Block rate | Relative Po | |
| -fold | s–1 | % | s–1 | |||
| WT | 96 ± 1 | 0.62 ± 0.05 | 0.50 | |||
| NR2-A7C | 2.73 ± 0.31 | 0.37 ± 0.04 | 0.37 | |||
| NR1 EI | 4.85 ± 0.38 | 0.25 ± 0.03 | 0.21 | 99 ± 1 | 0.28 ± 0.04 | 0.23 |
| NR1 EI –Gly | 5.25 ± 0.40 | 0.24 ± 0.02 | 0.19 | 98 ± 1 | 0.22 ± 0.01 | 0.18 |
| NR1-A7C | 1.61 ± 0.05 | 0.24 ± 0.02 | 0.62 | |||
| NR2 KN | 1.61 ± 0.05 | 0.27 ± 0.03 | 0.62 | 97 ± 1 | 1.28 ± 0.07 | 1.03 |
| NR2 KN –Glu | 1.56 ± 0.07 | 0.19 ± 0.00 | 0.64 | 98 ± 1 | 1.27 ± 0.13 | 1.02 |
This hypothesis was further examined by testing accessibility of the M3 domain in both of our LBD mutants. NR1 EI and NR2 KN were co-expressed with the complementary subunit containing A7C, a well characterized reporter of M3 accessibility (24), and subjected to thiol modification with MTSEA. The NR1 and NR2 reporters are referred to as NR1-A7C and NR2-A7C, respectively. In the absence of the A7C reporter, treatment with MTSEA or 5,5′-dithiobis(2-nitrobenzoic acid), another thiol-modifying reagent, had no effect on the response of either disulfide mutant (data not shown), indicating that the observed MTSEA potentiation results from A7C modification.
NR2-A7C modification proceeded ∼1.5-fold slower when co-expressed with NR1 EI than wild type, indicating that EI causes a decrease in M3 accessibility, and similar results were obtained in the presence and absence of glycine (Fig. 4B). Furthermore, the degree of potentiation observed in the EI mutant was 1.78-fold greater than WT in the presence of glycine and 1.92-fold greater with glutamate alone, although the difference between the full agonist and glutamate-only data was not statistically significant. Previous studies have proposed that thiol-modification of A7C-containing receptors leads to maximum activation, resulting in a Po of ∼1.0 (35). Based on that assumption, the MTSEA modification results agree with our MK-801 data, confirming an inhibition of channel gating in the NR1 EI mutant. Relative Po values were determined for each experiment, calculated as ratios of either the MK-801 block rate, after assigning a wild type Po value of 0.5 (36), or degree of MTSEA potentiation. Although these calculations are estimates, both methods yield quantitatively similar values, resulting in a relative Po of 0.2 (Table 2). In contrast, the time course of NR1-A7C modification was essentially identical between NR2 KN and wild type in the presence of glutamate and only slightly slower in its absence (Fig. 4C), suggesting that the KN mutant causes little effect on M3 accessibility. Furthermore, no difference in degree of potentiation was observed between KN and wild type, implying that channel gating is unaffected by the KN disulfide. These results contrast with the MK-801 rate data, which showed a 2-fold increase in KN block rate. However, previous studies have reported that the NR1-A7C reporter is less sensitive to glutamate site efficacy than the NR2-A7C reporter is to glycine-site efficacy (24), indicating that this method may not be sensitive enough to detect the gating changes observed with MK-801.
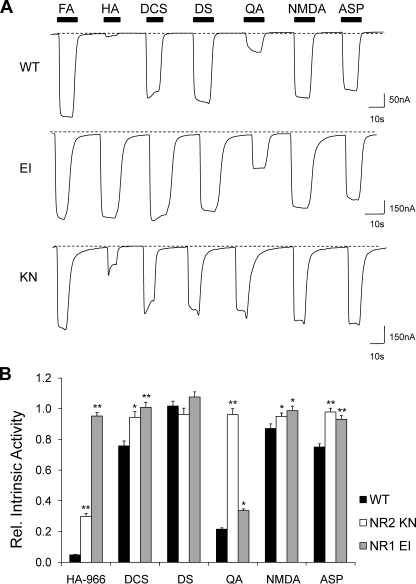
Increased Partial Agonist Efficacy—Since EI and KN exhibit increased and decreased channel gating, respectively, we further characterized the response of our mutants to a selection of NMDA receptor agonists: three glycine site agonists (HA-966, DCS, and d-serine) and three glutamate-site agonists (quinolinic acid, NMDA, and l-aspartate) (Fig. 5A). The EI mutant displayed similar responses to all three glycine site agonists, most notably increasing the intrinsic activity of HA-966 from 0.05 to 0.95 (Fig. 5B). Additionally, smaller increases in efficacy were also observed with all three glutamate-site agonists, but at least one agonist remained a partial agonist (quinolinic acid, α = 0.22 in WT, α = 0.34 in EI). Conversely, the KN mutant was fully activated by all three glutamate-site agonists, a phenotype most evident in the increased intrinsic activity of quinolinic acid from 0.22 to 0.96. KN also displayed increased efficacy in response to both HA-966 and DCS, although both remained partial agonists.
FIGURE 5.
Altered relative efficacy of NR1 and NR2 site partial agonists. A, representative whole cell traces illustrating the response of NR1 EI and NR2 KN to three NR1 agonists (HA-966, DCS, and d-serine) and three NR2 agonists (quinolinic acid, NMDA, and l-aspartate). Agonist concentrations were as follows: 500 μm HA-966 (HA), 1 mm DCS, 100 μm d-serine (DS), 10 mm quinolinic acid (QA), 1 mm NMDA, and 100 μm l-aspartate (ASP). Each agonist was co-applied with a 100 μm concentration of the appropriate co-agonist. Maximal current responses are normalized to facilitate comparison. B, bar graph illustrating the changes in intrinsic activity for each mutant, relative to glutamate. NR1 EI can distinguish partial agonism at the glutamate site but not the glycine site, whereas NR2 displays the opposite phenotype. Increased responses were seen with all partial agonists at both mutants. Statistical significance was determined for each mutant relative to wild type. * and **, p < 0.05 and p < 0.01, respectively. Normalized current responses for NR1 EI were as follows, expressed as mean ± S.E.: HA-966 = 0.95 ± 0.02 (n = 6), DCS = 1.01 ± 0.03 (n = 6), d-serine = 1.08 ± 0.03 (n = 6), quinolinic acid = 0.34 ± 0.01 (n = 6), NMDA = 0.99 ± 0.03 (n = 6), l-aspartate = 0.93 ± 0.02 (n = 6). Normalized current responses for NR2 KN were as follows: HA-966 = 0.30 ± 0.02 (n = 15), DCS = 0.94 ± 0.04 (n = 13), d-serine = 0.96 ± 0.04 (n = 12), quinolinic acid = 0.96 ± 0.04 (n = 12), NMDA = 0.95 ± 0.02 (n = 8), l-aspartate = 0.98 ± 0.02 (n = 7).
To address the issue of whether these agonists bind to the mutant receptors, the agonist-induced responses were compared statistically with constitutive levels (Fig. 2, A and D). For the EI mutant, the responses to d-serine and DCS differed significantly from the glutamate-only responses (p < 0.01), and for the KN mutant, QA and l-aspartate elicited significantly larger responses than glycine alone (p < 0.05). However, the possibility remains that HA-966 and NMDA do not bind to NR1 EI and NR2 KN, respectively, since these responses were not significantly different from constitutive levels.
Taken together, these results indicate that both mutants are capable of sensing partial agonism in the opposing LBD but respond to same-site partial agonists as full agonists. Since intrinsic activity is a relative measurement, dependent upon the “full agonist” used for comparison, the values cannot be directly compared between mutants, since the mutations may have affected glycine or glutamate efficacy (37). The EI results, combined with the observed decrease in channel gating, suggest that the NR1 disulfide may be trapped in a suboptimal or lower efficacy conformation relative to wild type. In contrast, the NR2 KN disulfide appears to be stabilized in a higher efficacy or “full agonist” conformation. Consistent with earlier results, the partial agonist data also indicate increased agonist sensitivity at the opposing subunit in both disulfide mutants.
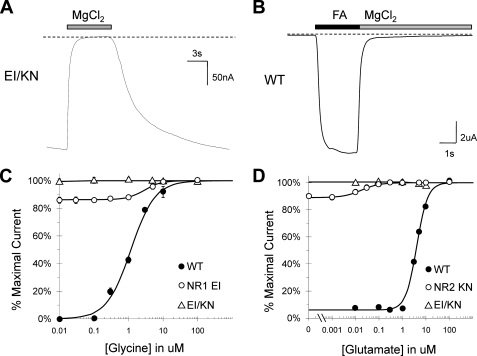
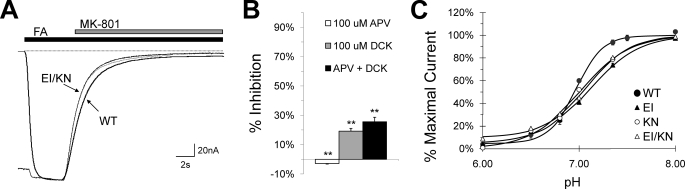
Co-expression of NR1 EI and NR2 KN Results in Constitutive Activation—To study the effect of stabilizing all four LBDs in their closed cleft conformations, the NR1 EI and NR2 KN mutants were co-expressed and tested for constitutive activation. The resulting EI/KN receptor was found to be activated by depolarization (-60 mV) in the absence of both glycine and glutamate (Fig. 6A). EI/KN retained sensitivity to Mg2+ inhibition, which fully blocked agonist-induced wild type currents, and thus 10 mm MgCl2 was used to provide a base-line current level in subsequent experiments. Concentration-response curves were obtained for both glycine and glutamate, revealing very little glycine dependence (y intercept = 99%) and no visible glutamate dependence (Fig. 6, B and C). To further examine channel gating, MK-801 block experiments were performed in both the presence and absence of agonists (Fig. 7A). In contrast to the concentration-response results, the observed MK-801 block rate for EI/KN was comparable with the average of the individual subunit block rates and only slightly faster than wild type (0.71 s-1 compared with 0.62 s-1 for WT). Additionally, the EI/KN block rates with no agonist and full agonist were indistinguishable. As expected from the NR2 KN results, the co-expressed mutant was insensitive to APV inhibition; however, ∼20% inhibition was observed in the presence of 100 m μm DCK, suggesting that DCK can still access the cleft in the absence of competing glycine (Fig. 7B). Both sets of results were significantly different from wild type in the presence of full agonist (Fig. 3).
FIGURE 6.
Co-expression of NR1 EI and NR2 KN yields a constitutively active channel. A, representative whole-cell trace illustrating the response of EI/KN to Mg2+ block, showing activation of EI/KN by depolarization alone (-60 mV) in the absence of glycine or glutamate. Treatment with 10 mm MgCl2, depicted with a gray bar, inhibits the constitutive response. B, whole cell trace showing WT receptors activated by 100 μm glycine and 100 μm glutamate (black bar) and subsequently blocked with 10 mm MgCl2 (gray bar), indicating that Mg2+ completely blocks WT receptors back to base-line levels. Therefore, Mg2+ block was used at the end of each experiment in C and D to provide a base-line current level. C, concentration-response curve for glycine in the presence of 100 μm glutamate, showing very little glycine-dependent activation. A linear fit to the data points yielded a Y0 of 0.99. D, concentration-response curve for glutamate in the presence of 100 μm glycine, showing no visible glutamate-dependent activation. The Y0 value obtained from a linear fit was 1.00.
FIGURE 7.
Inhibition of EI/KN receptors. A, representative trace illustrating block by 200 nm MK-801, performed as in Fig. 4A. The average rate of block (in s-1) for EI/KN receptors was 0.71 ± 0.05 in the presence of 20 μm glycine and 100 μm glutamate and 0.70 ± 0.06 in the absence of agonist, compared with 0.62 ± 0.05 for WT (see Table 2). Thus, these receptors are blocked only slightly faster than WT and very close to the average of the individual EI and KN mutants. B, bar graph showing inhibition by APV and DCK in the absence of agonist. 100 μm APV alone does not block, whereas 100 μm DCK reduces current by 19% ± 2%. Co-application of 100 μm APV and 100 μm DCK increases the degree of block to 26 ± 3%. The inhibition percentage differs significantly from 0 in all three cases, based on 99% confidence intervals (**). 10 mm Mg2+ block was used at the end of each experiment in B and C to provide a base-line current level. C, proton concentration-inhibition curves for EI/KN and both individual disulfide mutants. The concentration midpoint is relatively unaffected (7.09 for EI/KN, 7.11 for EI, and 7.01 for KN, compared with 6.97 for WT), but all three mutations exhibit a decreased Hill slope. Fitted nH values were as follows: WT = 3.15, EI/KN = 1.95, EI = 1.69, KN = 1.68.
Finally, we evaluated the sensitivity of the co-expressed mutant and both mutant subunits to proton inhibition (Fig. 7C). Although the structural determinants of pH sensitivity have not yet been definitively localized in the NMDA receptor, studies have implicated the TM-LBD linker regions of both NR1 and NR2 (38), raising the question of whether closed cleft stabilization of the LBDs would affect sensitivity to proton block. Although both mutants displayed a decreased Hill coefficient, the concentration midpoints were found to be relatively unaffected in all three mutant combinations, consistent with previous findings that inhibition proceeds identically in both the presence and absence of agonist (39). Thus, the proton sensor appears to function independently of agonist binding and cleft closure at both subunits.
DISCUSSION
Previous studies have reported profound effects of LBD destabilization on apparent affinity and agonist efficacy in iGluRs, and recent work in the NR2 subunit showed that the strength of a D1-D2 interaction correlates with gating efficiency (17, 18, 20). In the present study, we engineered novel cleft-spanning disulfide bonds to stabilize the closed cleft conformations of the NR1 and NR2 LBDs, leading to several notable observations regarding the role of LBD stabilization in the activation pathway. Cleft closure appears to be an important mediator of efficacy in both NR1 and NR2, stabilizing the mutant LBDs in partial and full agonist conformations, respectively. Both disulfide-stabilized subunits can be activated above constitutive levels, suggesting that cleft stabilization does not necessarily prevent association or dissociation of agonist. Finally, both disulfide mutants affected agonist sensitivity at the opposing subunit, suggesting a potential intersubunit interaction.
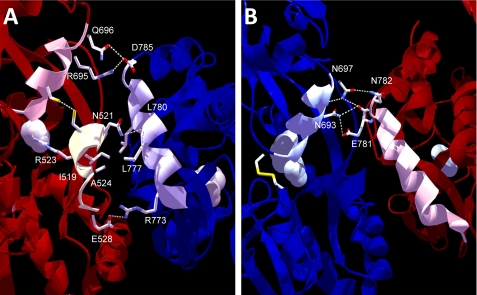
Intersubunit Coupling and the LBD Dimer Interface—Altered cross-subunit phenotypes were observed in the NR1 EI concentration-response curves, the partial agonist results, and the co-expressed EI/KN receptors, which exhibited higher constitutive activity than would be expected based on the individual subunits. Since glycine and glutamate-binding have been reported to exhibit negative cooperativity, the source of the observed phenotype was of particular interest (21); thus, we examined our disulfide mutations in the context of the NR1-NR2A LBD crystal structure (10). As depicted in Fig. 8A, the EI cross-cleft interaction connects helix D (Glu522) and helix F (Ile691) of the NR1 LBD, both of which are predicted to interact with helix J of the NR2 LBD. Several hydrogen bonds and potential hydrophobic interactions on the helix D side would be expected to stabilize the dimer interface, and two additional interactions are observed on the helix F side of the cleft. Helix F comprises part of D2, shown to be the more mobile of the two ligand-binding lobes and proposed to physically couple LBD closure to the linker regions (10, 40). Based on this structural model, NR1 cleft closure could permit increased interaction between NR1 helix D and NR2 helix J, stabilizing the interface and slowing deactivation. Furthermore, previous studies have suggested that cyclothiazide-induced stabilization of the homomeric GluR2 interface also stabilizes cleft closure, consistent with this hypothesis (20).
FIGURE 8.
Predicted interface interactions in the NR1 EI and NR2 KN heterodimer. A, interactions between the EI disulfide region and the NR2 subunit, based on the agonist-bound crystal structure of the NR1-NR2A heterodimer (accession number 2A5T). The NR1 and NR2 subunits are shown in dark red and dark blue, respectively, with NR1 helix F and two C-terminal residues highlighted in pink, helix D and the beginning of sheet 9 in yellow, and NR2 helix J in blue. Glycine and glutamate are shown as gray space-filling molecules. The EI disulfide spans the cleft between helix F and helix D of NR1, both of which are predicted to interact with helix J of the NR2 subunit. Residues shown at the interface are predicted to participate in hydrophobic interactions or hydrogen bonding (dashed lines) with the opposing subunit. Arg523, which is located immediately adjacent to E522C, forms an essential hydrogen bond with the α-carboxyl group of glycine. B, interactions between the KN disulfide regions and the NR1 subunit. The NR2 helix F is highlighted in light blue, and NR1 helix J is highlighted in pink. The KN disulfide spans the cleft between helix F and sheet 6, but only helix F appears to participate in intersubunit interactions. Residues predicted to interact with the NR1 subunit are shown at the interface between NR2 helix F and the C-terminal region of NR1 helix J. Notably fewer interactions are seen than in the EI disulfide dimer.
In contrast, the ligand-binding cleft of the NR2 KN mutant appears to only interact with NR1 via one side of its disulfide bond (Fig. 8B), providing a structural basis for the observation that NR2 KN does not affect glycine sensitivity as strongly as NR1 EI alters glutamate sensitivity. It remains to be determined if this phenomenon is broadly applicable to NMDA receptors, since these mutants are likely to contain structural perturbations not found in wild type. However, our results are consistent with previous reports suggesting that NR1 is more sensitive to changes at the NR2 LBD than vice versa (24). Although speculative, these structural observations implicate the heteromeric interface in coupling agonist binding at opposing LBDs, a hypothesis supported by recent data identifying intersubunit interactions as the source of differences in glycine potency between NR2 subtypes (41).
The previously reported negative cooperativity has been described as a reduction in agonist affinity upon co-agonist binding, mediated by residues Glu427 in NR2A and Lys544 in NR1 (10, 21, 42). However, examination of the crystal structure revealed that these residues are located on the opposite side of the LBD relative to each disulfide linker, and neither residue appears to form or be located near any intersubunit contacts. Thus, these residues may be involved in intrasubunit or interdimer interactions but are unlikely to account for the phenotype observed in our mutants. One possibility is that the NR1 and NR2 LBDs exhibit both positive and negative cooperativity, mediated by separate structural elements. Interestingly, positive cooperativity has also been observed in the C744A/C798A NR1 mutant, which removes an endogenous disulfide linkage and is proposed to facilitate domain closure by increasing helix K flexibility (43).
Cleft Closure and Partial Agonism in the NMDA Receptor—The lack of correlation between agonist efficacy and degree of cleft closure in the NR1 subunit, contrary to results obtained with GluR2 and GluR6, suggests that the NMDA receptor has a distinct structural mechanism for sensing agonist efficacy. One possibility is that stabilization of the closed cleft state, not simply rotation of D2 relative to D1, is the chief determinant of efficacy. This hypothesis finds support in many molecular dynamics studies, which report higher relative domain mobility, decreased hinge stability, and increased agonist flexibility with partial agonists (14, 16). Numerous structure/function studies have documented the effects of destabilizing cross-cleft interactions in NR2, GluR2, and GluR6, which include increased deactivation rates as well as lower apparent affinity and efficacy (18–20). Increasing the strength of D1-D2 interactions or eliminating agonist-cleft clashes has also been reported to increase apparent affinity, slow deactivation, and increase partial agonist efficacy (17, 44). The experiments shown here provide further support for the role of D1-D2 stabilization in NR2 and extend these results to the NR1 subunit. In fact, the NR2 K487C-N687C disulfide bond we describe is very close to the Lys484-Asn687 endogenous interaction reported by Maier et al. (17), indicating that Asn687 is capable of numerous cross-cleft interactions and suggesting that perhaps the exact positioning of these interactions may be less important than the presence of a stabilizing interaction.
Interestingly, the crystal structure data accompanying several cleft stabilization studies do not always follow the LBD closure-agonist efficacy correlation; for example, the GluR2 L650T mutation increases efficacy and degree of domain closure in complex with kainate, but the AMPA-bound structure includes both a partially closed and a fully closed conformation (44). Additionally, introduction of a cleft-stabilizing GluR6 D1D2 interaction into GluR2 dramatically slowed deactivation and increased glutamate sensitivity but had no effect on cleft closure, although GluR6 is almost 6° more closed than GluR2 in the full agonist state (18). Molecular dynamics simulations have also observed ligand movement within the NR1 binding pocket without altering the degree of domain closure (16). It has been suggested that both incomplete and unstable cleft closure have similar effects on single channel gating (20), raising the possibility that the degree of cleft closure may simply be a manifestation of closed cleft stability. Indeed, the discrepancies observed in several GluR2 crystal structure studies raise the possibility that this principle applies to AMPA receptors as well, suggesting that the mechanism of partial agonism may be conserved across glutamate receptor subtypes.
Although we cannot make assumptions about the degree of cleft closure in our LBD mutants, and their disulfide-stabilized structures may not mimic wild type conformational states, the constraint imposed by cross-cleft tethers should prevent the domain reopening observed in GluR2 L650T and facilitate formation of endogenous cross-cleft interactions. Thus, we can conclude that LBD stabilization is an important determinant of agonist efficacy in both NR1 and NR2 and that agonist-mediated stabilizing interactions appear to be necessary for full receptor activation, as demonstrated by the ability of both mutants to be activated above constitutive levels.
Agonist Dissociation from NMDA Receptors—It has been proposed that agonist dissociation from iGluRs only occurs from the open cleft state (20). Support for this observation can be found in muscle nicotinic receptors, which exhibit a 2,500-fold decrease in acetylcholine dissociation rate upon channel opening (45). However, both mutants described here exhibit concentration-dependent activation above their agonist-independent responses, implying that agonist can access and further stabilize the disulfide-bonded LBDs. Disulfide formation may stabilize the cleft in a fully or partially closed conformation; our data do not distinguish between these possibilities. However, assuming that the disulfide bond prevents full LBD reopening, these results suggest that D1 and/or D2 are sufficiently flexible to permit ligand association and dissociation without reverting to a fully open conformation. The possibility of unreacted cysteine residues cannot be discounted and may contribute to the phenotype of these receptors; however, the insensitivity of both mutants to competitive antagonism suggests that unlinked LBDs do not comprise a significant portion of the receptor population. Interestingly, NMR molecular relaxation studies in GluR2 suggested that movement of helix F could potentially permit agonist dissociation without cleft reopening, and this helix has also been implicated in coupling agonist efficacy to receptor activation in NR2A (14, 46). Half of each disulfide mutant pair resides within helix F (NR1 I691C and NR2 N687C), so a prominent role for this region in sensing efficacy and allowing agonist dissociation would be consistent with our experimental results.
Given the wide range of physiological and pathological roles attributed to NMDA receptors, understanding the mechanism of partial agonism holds tremendous potential for structure-based drug design. In addition to promising clinical results with DCS, the recent report of subtype-specific NMDA receptor partial agonists opens the possibility of individually targeting NR2 subtypes, which exhibit both regional and developmental variations in expression (47). The structure/function results reported here and elsewhere emphasize that cleft stability and intersubunit interactions are key structural determinants of agonist efficacy and therefore should be considered in the design of glutamate receptor-specific therapeutic agents.
Acknowledgments
We thank Dr. Timothy Haystead for assistance with the liquid chromatography-mass spectrometry experiments and Dr. John York and Dr. Richard Whorton for helpful suggestions on experimental design.
This work was supported, in whole or in part, by National Institutes of Health Grants F31 NS053030-01 (to M. L. B.) and R01-MH61506 (to A. M. J. V. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: iGluR, ionotropic glutamate receptor; DCK, 5,7-dichlorokynurenic acid; NMDA, N-methyl-d-aspartate; DCS, d-cycloserine; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid; LBD, ligand binding domain; D1 and D2, domain 1 and 2, respectively; MTSEA, methanethiosulfonate; BME, 2-mercaptoethanol; APV, 2-amino-5-phosphonovalerate; EI, E522C/I691C; KN, K487C/N687C; WT, wild type.
References
- 1.Bashir, Z. I., Alford, S., Davies, S. N., Randall, A. D., and Collingridge, G. L. (1991) Nature 349 156-158 [DOI] [PubMed] [Google Scholar]
- 2.Sheng, M., Cummings, J., Roldan, L. A., Jan, Y. N., and Jan, L. Y. (1994) Nature 368 144-147 [DOI] [PubMed] [Google Scholar]
- 3.Kauer, J. A., Malenka, R. C., and Nicoll, R. A. (1988) Nature 334 250-252 [DOI] [PubMed] [Google Scholar]
- 4.Emamian, E. S., Karayiorgou, M., and Gogos, J. A. (2004) J. Neurosci. 24 1561-1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hynd, M. R., Scott, H. L., and Dodd, P. R. (2004) Neurochem. Int. 45 583-595 [DOI] [PubMed] [Google Scholar]
- 6.Mody, I., and MacDonald, J. F. (1995) Trends Pharmacol. Sci. 16 356-359 [DOI] [PubMed] [Google Scholar]
- 7.Yaka, R., Biegon, A., Grigoriadis, N., Simeonidou, C., Grigoriadis, S., Alexandrovich, A. G., Matzner, H., Schumann, J., Trembovler, V., Tsenter, J., and Shohami, E. (2007) FASEB J. 21 2033-2041 [DOI] [PubMed] [Google Scholar]
- 8.Tsai, G. E., Falk, W. E., Gunther, J., and Coyle, J. T. (1999) Am. J. Psychiatry 156 467-469 [DOI] [PubMed] [Google Scholar]
- 9.Schorge, S., and Colquhoun, D. (2003) J. Neurosci. 23 1151-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa, H., Singh, S. K., Mancusso, R., and Gouaux, E. (2005) Nature 438 185-192 [DOI] [PubMed] [Google Scholar]
- 11.Mayer, M. L. (2005) Neuron 45 539-552 [DOI] [PubMed] [Google Scholar]
- 12.Armstrong, N., Sun, Y., Chen, G.-Q., and Gouaux, E. (1998) Nature 395 913-917 [DOI] [PubMed] [Google Scholar]
- 13.Jin, R., Banke, T. G., Mayer, M. L., Traynelis, S. F., and Gouaux, E. (2003) Nat. Neurosci. 6 803-810 [DOI] [PubMed] [Google Scholar]
- 14.Erreger, K., Geballe, M. T., Dravid, S. M., Snyder, J. P., Wyllie, D. J. A., and Traynelis, S. F. (2005) J. Neurosci. 25 7858-7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inanobe, A., Furukawa, H., and Gouaux, E. (2005) Neuron 47 71-84 [DOI] [PubMed] [Google Scholar]
- 16.Kaye, S. L., Sansom, M. S. P., and Biggin, P. C. (2006) J. Biol. Chem. 281 12736-12742 [DOI] [PubMed] [Google Scholar]
- 17.Maier, W., Schemm, R., Grewer, C., and Laube, B. (2007) J. Biol. Chem. 282 1863-1872 [DOI] [PubMed] [Google Scholar]
- 18.Weston, M. C., Gertler, C., Mayer, M. L., and Rosenmund, C. (2006) J. Neurosci. 26 7650-7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, K. B., Clausen, R. P., Bjerrum, E. J., Bechmann, C., Greenwood, J. R., Christensen, C., Kristensen, J. L., Egebjerg, J., and Brauner-Osborne, H. (2005) Mol. Pharmacol. 68 1510-1523 [DOI] [PubMed] [Google Scholar]
- 20.Robert, A., Armstrong, N., Gouaux, J. E., and Howe, J. R. (2005) J. Neurosci. 25 3752-3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regalado, M. P., Villarroel, A., and Lerma, J. (2001) Neuron 32 1085-1096 [DOI] [PubMed] [Google Scholar]
- 22.Wood, M. W., VanDongen, H. M. A., and VanDongen, A. M. J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4882-4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood, M. W., VanDongen, H. M. A., and VanDongen, A. M. J. (1996) J. Biol. Chem. 271 8115-8120 [DOI] [PubMed] [Google Scholar]
- 24.Jones, K. S., VanDongen, H. M. A., and VanDongen, A. M. J. (2002) J. Neurosci. 22 2044-2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard, J. P., and Kelso, S. R. (1990) Neuron 4 53-60 [DOI] [PubMed] [Google Scholar]
- 26.Low, C.-M., Zheng, F., Lyuboslavsky, P., and Traynelis, S. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 11062-11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paoletti, P., Perin-Dureau, F., Fayyazuddin, A., Le Goff, A., Callebaut, I., and Neyton, J. (2000) Neuron 28 911-925 [DOI] [PubMed] [Google Scholar]
- 28.Zheng, F., Erreger, K., Low, C. M., Banke, T., Lee, C. J., Conn, P. J., and Traynelis, S. F. (2001) Nat. Neurosci. 4 894-901 [DOI] [PubMed] [Google Scholar]
- 29.Armstrong, N., and Gouaux, E. (2000) Neuron 28 165-181 [DOI] [PubMed] [Google Scholar]
- 30.Kalbaugh, T. L., VanDongen, H. M. A., and VanDongen, A. M. J. (2004) Mol. Pharmacol. 66 209-219 [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L., Peoples, R. W., Oz, M., Harvey-White, J., Weight, F. F., and Brauneis, U. (1997) J. Neurophysiol. 78 582-590 [DOI] [PubMed] [Google Scholar]
- 32.Lerma, J., Zukin, R. S., and Bennett, M. V. L. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 2354-2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kniazeff, J., Saintot, P.-P., Goudet, C., Liu, J., Charnet, A., Guillon, G., and Pin, J.-P. (2004) J. Neurosci. 24 370-377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenmund, C., Feltz, A., and Westbrook, G. L. (1995) J. Neurosci. 15 2788-2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, P. E., Geballe, M. T., Stansfeld, P. J., Johnston, A. R., Yuan, H., Jacob, A. L., Snyder, J. P., Traynelis, S. F., and Wyllie, D. J. A. (2005) Mol. Pharmacol. 67 1470-1484 [DOI] [PubMed] [Google Scholar]
- 36.Erreger, K., Dravid, S. M., Banke, T. G., Wyllie, D. J. A., and Traynelis, S. F. (2005) J. Physiol. 563 345-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colquhoun, D. (1998) Br. J. Pharmacol. 125 924-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Low, C.-M., Lyuboslavsky, P., French, A., Le, P., Wyatte, K., Thiel, W. H., Marchan, E. M., Igarashi, K., Kashiwagi, K., Gernert, K., Williams, K., Traynelis, S. F., and Zheng, F. (2003) Mol. Pharmacol. 63 1212-1222 [DOI] [PubMed] [Google Scholar]
- 39.Banke, T. G., Dravid, S. M., and Traynelis, S. F. (2005) J. Neurosci. 25 42-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arinaminpathy, Y., Sansom, M. S. P., and Biggin, P. C. (2002) Biophys. J. 82 676-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, P. E., Geballe, M. T., Katz, E., Erreger, K., Livesey, M. R., O'Toole, K. K., Le, P., Lee, C. J., Snyder, J. P., Traynelis, S. F., and Wyllie, D. J. A. (2008) J. Physiol. 586 227-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benveniste, M., Clements, J., Vyklicky, L., Jr., and Mayer, M. L. (1990) J. Physiol. 428 333-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaye, S. L., Sansom, M. S. P., and Biggin, P. C. (2007) Biochemistry 46 2136-2145 [DOI] [PubMed] [Google Scholar]
- 44.Armstrong, N., Mayer, M., and Gouaux, E. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5736-5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosman, C., and Auerbach, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14102-14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFeeters, R. L., and Oswald, R. E. (2002) Biochemistry 41 10472-10481 [DOI] [PubMed] [Google Scholar]
- 47.Erreger, K., Geballe, M. T., Kristensen, A., Chen, P. E., Hansen, K. B., Lee, C. J., Yuan, H., Le, P., Lyuboslavsky, P. N., Micale, N., Jorgensen, L., Clausen, R. P., Wyllie, D. J. A., Snyder, J. P., and Traynelis, S. F. (2007) Mol. Pharmacol. 72 907-920 [DOI] [PubMed] [Google Scholar]