Abstract
Hydrogels have been commonly used as model systems for 3-dimensional (3-D) cell biology, as they have material properties that resemble natural extracellular matrices (ECMs), and their cell-interactive properties can be readily adapted in order to address a particular hypothesis. Natural and synthetic hydrogels have been used to gain fundamental insights into virtually all aspects of cell behavior, including cell adhesion, migration, and differentiated function. However, cell responses to complex 3-D environments are difficult to adequately explore due to the large number of variables that must be controlled simultaneously. Here we describe an adaptable, automated approach for 3-D cell culture within hydrogel arrays. Our initial results demonstrate that the hydrogel network chemistry (both natural and synthetic), cell type, cell density, cell adhesion ligand density, and degradability within each array spot can be systematically varied to screen for environments that promote cell viability in a 3-D context. In a testbed application we then demonstrate that a hydrogel array format can be used to identify environments that promote viability of HL-1 cardiomyocytes, a cell line that has not been cultured previously in 3-D hydrogel matrices. Results demonstrate that the fibronectin-derived cell adhesion ligand RGDSP improves HL-1 viability in a dose-dependent manner, and that the effect of RGDSP is particularly pronounced in degrading hydrogel arrays. Importantly, in the presence of 70µM RGDSP, HL-1 cardiomyocyte viability does not decrease even after 7 days of culture in PEG hydrogels. Taken together, our results indicate that the adaptable, array-based format developed in this study may be useful as an enhanced throughput platform for 3-D culture of a variety of cell types.
Keywords: extracellular matrix, degradation, RGD peptide, tissue engineering, cardiomyocyte
Introduction
A variety of recent studies demonstrate the importance of 3-dimensional (3-D) cell culture formats in fundamental cell biology[1–3]. Hydrogels have been particularly prevalent model systems, as they have material properties that resemble natural extracellular matrices (ECMs), and their cell-interactive properties can be readily adapted to address a particular hypothesis. Therefore, natural and synthetic hydrogels have been used to gain fundamental insights into virtually all aspects of cell behavior, including cell adhesion[4], migration[5], and differentiated function[6, 7]. A broad conclusion of these previous studies is that cell behavior in a 3-D context is strongly dependent on local ECM cues, particularly cell adhesion ligands, soluble or immobilized growth factors, mechanical properties, and degradability. In addition, cellular responses to ECM signals are context-dependent, as signals presented in a 3-D context differ significantly from those presented in traditional 2-D cell culture. For example, Yamada and coworkers have shown that focal adhesion formation during cell-ECM adhesion is mechanistically different in naturally-derived 3-D hydrogels when compared to protein-coated tissue culture polystyrene (TCP) substrates[4]. Other recent studies specifically implicate 3-D ECM signals as key regulators of carcinogenesis[8, 9], hematopoiesis[10], and neurogenesis[11]. Based on these studies and others, there is a clear need for hydrogel matrix formats in which cells can be exposed to controllable 3-D signaling environments.
Hydrogel-based biomaterials have also emerged as an important component of several tissue engineering strategies, which aim to regenerate natural tissue structure and function using combinations of materials, cells, and soluble signals[12, 13]. Recent hydrogel design strategies have focused on exposing cells to specific ECM-derived signals, such as cell adhesion ligands, and characterizing their influence on cell behavior[14, 15]. A subset of these approaches has attempted to achieve control over the cell’s environment by using a “blank slate” base material that can present specific biological moieties while avoiding non-specific interactions with other molecules such as serum proteins. Notable examples of blank slate materials include poly(ethylene glycol) (PEG)[14, 16, 17], agarose[18], and alginate[12] hydrogels. PEG chains derivatized on each end with vinyl-containing functional groups have been a particularly prevalent base polymer, as they can be readily processed into a cell-laden hydrogel network via photo-crosslinking[7, 19] and can be derivatized via simple chemistries to generate networks that contain ECM-derived signals [20, 21]. These properties have led to widespread use of PEG hydrogels as matrices to support tissue formation by various cell types, including osteoblasts[22], chondrocytes[23, 24], neural cells[6], and multiple stem cell types[7, 17, 25, 26]. For example, in a series of studies Anseth and coworkers have used PEG-based hydrogels to demonstrate that 3-D cartilage formation is influenced by ECM chemistry[27], mechanics[23], and degradation[28, 29]. Based on these studies it is clear that ECM signals strongly influence tissue formation in 3-D matrices, and that control over the identity and quantity of ECM signals is vital in tissue engineering approaches.
Although investigators have gained valuable insights from cell culture and tissue engineering studies in 3-D hydrogels, previous approaches have typically been capable of exploring the effects of a limited range of ECM signals. It is impractical to characterize the effects of widely varying quantities of a particular ECM signal or to determine the effects of several simultaneous signals due to the extensive analysis time and material costs required. This is a significant limitation, because broad ranges of ECM signal quantities and complex signal combinations are routinely presented to cells in vivo[14] and are likely to be important in both 3-D cell culture and functional tissue engineering applications in vitro. Investigators have recently begun to address this limitation by creating micro-scale hydrogels containing viable cells[30]. Photolithographic methods have been used to generate spatially patterned hydrogel structures, in which distinct regions contain specific cell types[31], cell adhesion ligands[32] or ECM chemistries[33, 34]. For example, Pishko and coworkers have used photopolymerization within channels[35] or spots[32] to generate PEG microstructures, and demonstrated that multiple mammalian cell types remain viable in spatially patterned hydrogels for up to 7 days. Other recent studies have used microfluidic systems to generate cell-laden peptide-based hydrogels[36]. Taken together, these studies have demonstrated that it is possible to generate spatially patterned, cell-laden hydrogel “arrays”, and these array formats provide a promising platform to enhance the throughput of 3-D cell culture studies. However, previous approaches often involved complex or specialized methods, which are not readily adaptable to explore broad ranges of ECM properties. For example, the aforementioned approaches each used photopolymerization and/or laminar flow to generate hydrogel arrays, and the ECM chemistries and size scales that can be explored using these techniques may be limited. An ideal 3-D cell culture system would allow for simple, systematic variation of important cell culture parameters, which include: ECM chemistry, ECM mechanical properties, cell adhesion ligand type and density, cell type and seeding density, soluble signal type and concentration, and ECM degradability.
Here we describe an adaptable, automated approach for 3-D cell culture within hydrogel arrays. To assemble arrays we first generate a hydrogel “background” containing cylindrical spots, which are then filled with combinations of cells, materials, and biological molecules using an automated liquid handling robot. Relative to previous methods, this approach is simple, rapid, and readily adaptable. Results presented here demonstrate that the cell type, cell density, cell adhesion ligand density, ECM chemistry, and ECM degradability in each array spot can each be varied independently. As a test-bed application of this approach, we explored the influence of broad ranges of 3-D ECM signals on viability of HL-1 cardiomyocytes. These cells represent a well-characterized model for cardiomyocyte behavior[37], but they have not been cultured in 3-D hydrogel matrices in previous studies. Therefore, results of these studies provide new insights into the factors that may influence cardiomyocyte viability in 3-D matrices. The hydrogel arrays described herein may be useful as a general, adaptable platform to study the influence of various ECM parameters on cell behavior with enhanced throughput.
Materials and Methods
Preparation of synthetic PEG hydrogel arrays
Poly(ethylene glycol) with molecular weights of 8kDa, and 20kDa were purchased from Sigma-Aldrich. The synthesis of PEG-diacrylate (PEGDA) was performed as described elsewhere[38, 39]. PEGDA hydrogels were prepared by first mixing 10wt% PEGDA and 0.05%w/v photoinitiator Irgacure 2959 (I2959, BASF, Ludwigshafen, Germany) in PBS (Fisher), to create the hydrogel “precursor solution”. This solution was added to a Teflon mold containing 16 cylindrical posts (1 mm diameter, 1.25 mm depth), and crosslinked via exposure to UV radiation (λ=365 nm, intensity=4.5 mW/cm2) for 5 min (Fig. 1) to form the hydrogel “background”. Upon removal from the mold, the resulting PEG hydrogel networks contained an array of 16 spots. These spots were then filled with the aforementioned precursor solution, along with various cell types included at a concentration of 1×106cells/ml unless otherwise stated. In experiments in which cells were included, PBS was replaced by serum free medium during hydrogel preparation and all solutions were passed through a 0.22 µm filter (Fisher) for sterilization.
Figure 1.
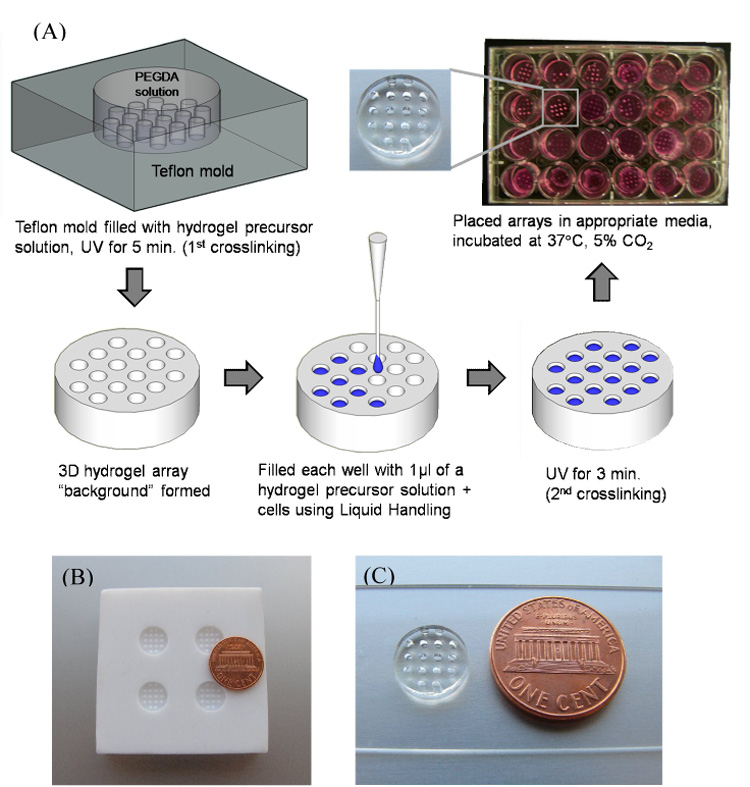
A) Schematic representation of fabrication of 3D hydrogel array. B) Teflon mold used to create hydrogel “background”. C) PEGDA 8K hydrogel array background.
In some experiments PEG hydrogel arrays were designed to degrade hydrolytically over time using an approach described elsewhere[39]. Briefly, 8kDa PEGDA chains were reacted with variable amounts of dithiothreitol (DTT, Fisher Scientific) to form water-soluble, acrylate-terminated (PEG-DTT)n-PEG conjugates. These conjugates were then incorporated into the photocrosslinking procedure described above to create PEG hydrogels with “DTT bridges” included. The ester bonds proximal to thioether linkages in these DTT bridges degrade hydrolytically. Thus, the degradability of (PEG-DTT)n-PEG networks is dictated by the total amount of DTT incorporated, which can be readily varied as demonstrated in a previous study[39]. Hydrogels containing DTT bridges are referred to as “degradable” networks in subsequent sections of this manuscript, while hydrogels without DTT are referred to as “non-degradable”.
In some experiments, acrylate-terminated PEG chains or (PEG-DTT)n-PEG conjugates were reacted with a fibronectin-derived cell adhesion ligand, CGGRGDSP, to generate cell-interactive hydrogel networks. In these experiments a 10X excess of acrylate-terminated PEG chains (Mw=8kDa) or (PEG-DTT)n-PEG conjugates were incubated with a CGGRGDSP peptide at 37°C for 90 minutes in PBS to allow for Michael-type addition of the cysteine sulfhydryl group to the acrylate group, as described previously[39–41]. The resulting solutions contained PEG-diacrylate and acrylate-PEG-CGGRGDSP molecules that were subsequently photo-crosslinked to form cell-interactive hydrogel networks.
Preparation of natural hydrogel arrays
PEG hydrogel arrays containing wells loaded with type I collagen hydrogels were prepared manually in a similar manner to the manually-prepared PEG arrays. Briefly, PEG hydrogel backgrounds prepared with 8kDa PEGDA (10wt%) were pre-swelled in Dulbecco’s Modification of Eagle Medium (DMEM). Cell laden collagen gels were created using an established protocol, altered only to produce a final concentration of collagen of 2mg/ml[42]. Medium was first removed from PEG hydrogel background wells via aspiration, and type I collagen solutions containing 1×106 cells/ml (unless otherwise noted) were then added to array wells and allowed to gel at 37°C for 2 hours. Cell-containing type I collagen solutions were kept on ice prior to formation of array spots to avoid spontaneous gelation.
Physicochemical characterization of hydrogel arrays
(i) Fluorescein-labeled PEGDA interpenetration into the hydrogel array background
In order to characterize the linkage between the array spots and the background hydrogel we first characterized the extent of interpenetration of PEGDA into the array background during formation of an array spot. We first synthesized PEG chains covalently linked to cysteine-fluorescein conjugates. These conjugates were prepared using standard Fmoc solid phase peptide synthesis techniques on a C36s automated peptide synthesizer (CS Bio, Menlo Park, CA) to link 5(6)-carboxyfluorescein to the N-terminus of an Mtt-protected cysteine residue (Novabiochem, San Diego, CA), followed by cysteine deprotection and resin cleavage. The resulting cysteine-fluorescein conjugates were dried under a nitrogen stream, lyophilized, and treated with tris(2-carboxyethyl)phosphine (TCEP) (Pierce, Rockford, IL), 150mM NaCl and 20mM NaPO4 (pH 8.0) at 50°C for 30 minutes to reduce disulfide linkages between cysteine residues. The cysteine-fluorescein conjugates were incubated with 8kDa PEGDA at a 1:10 molar ratio in PBS for 30 minutes to achieve Michael-type addition of sulfhydryl groups on cysteine to terminal acrylate groups on PEGDA chains. The product was then placed in 3500 MWCO SnakeSkin dialysis tubing (Pierce, Rockford, IL) and dialyzed to remove TCEP and unreacted cysteine-fluorescein conjugates. PEGDA-cysteine-fluorescein conjugation was then confirmed via MALDI-TOF Mass Spectrometry (Bruker, Billerica, MA) (see supplementary information, Fig. S1).
We next used acrylate-terminated PEG-cysteine-fluorescein conjugates to measure the extent of interpenetration of PEGDA chains into the hydrogel array background during formation of an individual array spot. Conjugates were suspended in PBS with photoinitiator (0.05%w/v I2959), pipetted manually into a well in a PEG hydrogel array background prepared with 8kDa PEGDA, allowed to interpenetrate for a typical array preparation timeframe (2.5 minutes), and the polymer solution was removed from the well. The background arrays were then exposed to UV light for 5 minutes to cross-link the interpenetrated polymer, placed in PBS, and stored in a cell culture incubator. Fluorescent micrographs were captured 0.5, 1, 2, 4, 8, and 15 days post interpenetration and analyzed using ImageJ software (NIH Freeware, Bethesda, Maryland).
(ii) Swelling ratio and mesh size determination
After polymerization, gels were placed in a 2 ml PBS solution and incubated at 37°C for 24 hr. Gels were weighed (wet weight, ws), then incubated in DI water to remove buffer salts, lyophilized for 48 hrs and weighed again (dry weight, wd). Three replicates of each PEG molecular weight were used. The mass equilibrium swelling ratio (Qm) was calculated according to the equation:
| (1) |
Degradation of degradable PEG hydrogel arrays was characterized by measuring their equilibrium swelling ratio after various incubation times in PBS, as described previously[39].
Biological characterization of hydrogel arrays
(i) Cell culture
hMSCs (Cambrex Bio Science, Walkersville, MD, passage 5–6) were cultured in Mesenchymal Stem Cell Growth Medium (Cambrex). HUVECs (Cambrex, Walkersville, MD, passage 11–12) were cultured in an endothelial cell growth medium (Cambrex). NIH/3T3 mouse fibroblasts (passages 24–26) were cultured in DMEM (Mediatech, VA) supplemented with 5% cosmic calf serum (HyClone) and 3.7 g/l NaHCO3 (ACROS Organics, NJ). HL-1 cells were cultured in Complete Claycomb Medium (SAFC Biosciences, KS). All media were supplemented with 100 units/ml of penicillin/streptomycin. Cell cultures were maintained at 37°C/5% CO2 and media was replaced every 3–4 days.
(ii) Cell-seeding within PEG hydrogel arrays
Cells were photoencapsulated in a 10 wt% polymer solution (final concentration) in serum free media at a seeding density of 1×106 cells/ml, unless otherwise stated. The cell/polymer solution (1µl) was pipetted in the wells of the hydrogel array either manually using a 0.2–10µl pipetteman (Eppendorff, Hamburg, Germany) or automatically using a Gilson automated liquid handler (Model: 223 Sample Changer) and ‘Trilution LH version 1.2’ control software (Gilson, Inc.). Upon ultraviolet light exposure for 3 min, PEG-based hydrogels were cross-linked and cells were physically entrapped within the networks. The arrays were placed in media appropriate for each cell type (described above) and cultured at 37°C and 5% CO2, replacing media every 2–3 days.
(iii) Cell viability within hydrogel arrays
After photoencapsulation, the arrays were removed from culture at various time points (1–14 days) and were stained using the LIVE/DEAD assay (Molecular Probes) according to the manufacturer’s instructions. This assay identifies esterase activity in live cells via green fluorescence emission from calcein AM and nuclear permeability in dead cells via red fluorescence emission from ethidium homodimer-1. Arrays were analyzed using an inverted, compound fluorescence microscope (IX51, Olympus). Cell viability was visualized and the total percentage of viable cells was determined by manual analysis of live and dead cells in photomicrographs for at least 10 images per condition. In some conditions, cell morphology in hydrogel array spots was also characterized qualitatively using the same inverted microscope operating in brightfield mode.
Statistical analysis
The results are expressed as mean ± standard deviations for at least 4 samples per condition. Differences between data sets were assessed by one-way ANOVA analysis. In some cases, Tukey’s two-way analysis was performed using the R software package (Freeware, http://www.r-project.org/). Regardless of the test, a p-value less than 0.05 was considered a significantly significant difference.
Results
Formation of synthetic hydrogel arrays
Hydrogel arrays can be formed via both manual and automated processes, and the physical properties of hydrogels in each spot in the hydrogel array can be varied. To explore the connectivity of synthetic hydrogel array spots to the background hydrogel, we first performed a set of experiments with fluorescently labeled 8kDa PEG-acrylate. This fluorescently tagged molecule was included in a 10% (w/v) solution of PEGDA with photoinitiator, added to well of a hydrogel array, and crosslinked via exposure to UV radiation, as detailed in the Methods section. Results indicate that PEG chains added to the array spot interpenetrate into the array background, indicating that the hydrogel network in the array spot is intertwined with the network in the array background (Fig. 2). This interpenetration was still apparent 15 days after crosslinking, indicating that a stable interpenetrated network exists at the interface between each array spot and the array background. These data support our observation (not shown) that hydrogels formed within array spots do not dissociate from the hydrogel array during vigorous handling.
Figure 2.
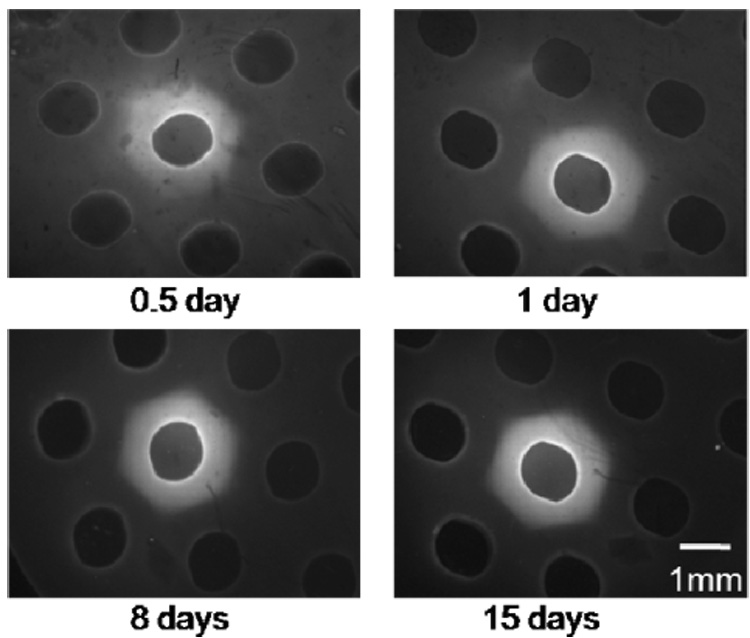
Incorporation of fluorescently-labeled PEG-acrylate molecules into the background hydrogel during formation of an array spot. Fluorescein-cysteine-PEG-acrylate molecules diffuse into the hydrogel background and form a stable, interpenetrated network between the array spot and the hydrogel background.
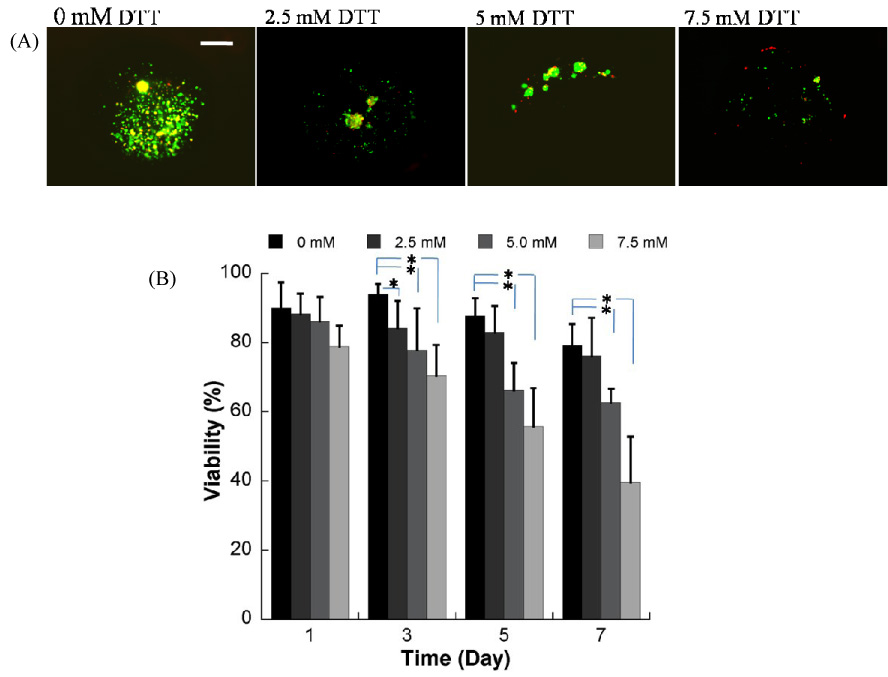
Hydrogel networks containing degradable DTT bridges can also be formed within array wells via photo-crosslinking. The presence of DTT has a significant influence on initial hydrogel swelling and degradation over time (Fig. 3). Hydrogels were prepared with 10% (w/v) PEGDA (Mw=8kDa) and 0mM≤[DTT]≤12.5mM. The equilibrium swelling ratio of hydrogels containing no DTT (22.2±1.3) increases substantially to 25.5±1.1, 29.7±1.0, 32.8±1.9, 37.3±3.5, and 46.8±2.1 with 2.5mM, 5mM, 7.5mM, 10mM and 12.5mM DTT included, respectively (Fig. 3). This trend of increasing equilibrium swelling ratio can be attributed to an increase in the average molecular weight of chains at the outset of photo-crosslinking, which is due to step growth polymerization of PEGDA in the presence of DTT, as described in our previous study[39]. Similarly, degradation of hydrogel arrays proceeds in a spatially homogeneous manner, with array spots degrading in concert with the background hydrogel. All hydrogels that include DTT bridges increased their equilibrium swelling ratio with incubation time in PBS (Fig. 3). In contrast, the Qm of hydrogel arrays without DTT does not significantly increase with time, as expected. The degradation process in these hydrogel arrays is similar to degradable hydrogels formed without array spots described in our previous work[39].
Figure 3.
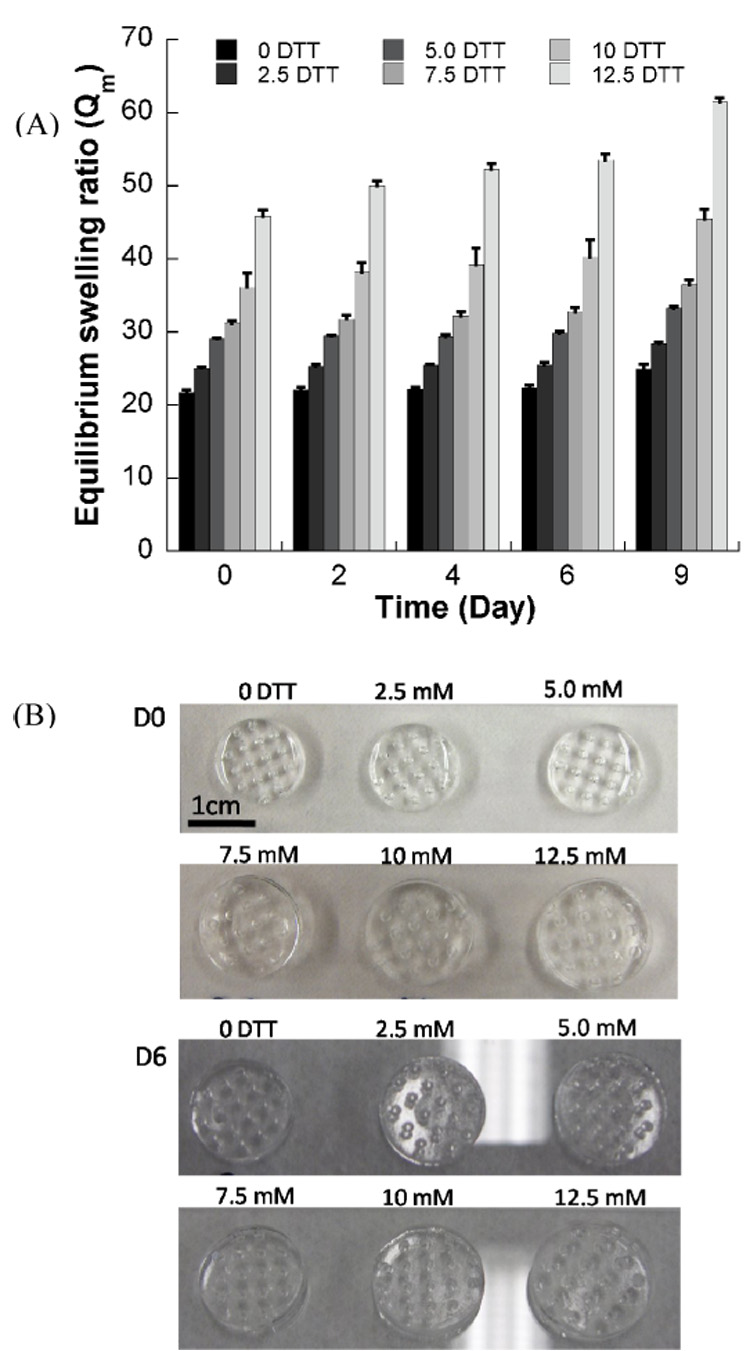
Degradable PEG hydrogel arrays containing DTT have varying mass equilibrium swelling ratios (Qm) over time. A) Changes in Qm of PEG hydrogels with various DTT concentrations during incubation in PBS, pH=7.4, T=37°C. B) Images of hydrogel arrays prepared with different concentration of DTT at day 0 and day 6 after hydrogel preparation.
Cell culture in 3-D PEG hydrogel arrays
Multiple cell types can be cultured within non-modified PEG hydrogel arrays, and cell viability is dependent on cell type and array preparation technique (automated versus manual) (Fig. 4). Viability of primary cell types, HUVECs (17.3±7.6% viability at 7 days) and hMSCs (17±3.5% at 7 days), was significantly lower when compared with the NIH-3T3 fibroblast line (38±5.4% at 7 days) in array spots formed manually (Fig. 4). Similarly, NIH-3T3 cells in arrays formed via an automated process showed higher viability at 7 days (54.7±3.3%) than HUVECs (52.6±2.7%) or hMSCs (29.5±3.4%). In addition, 7 days after array preparation the NIH-3T3 viability was higher in the hydrogel array spots formed via an automated liquid handler when compared with NIH-3T3 cells in array spots formed manually. Viability of HUVECs and hMSCs was significantly enhanced in hydrogel array spots formed via an automated process at 3, 5, and 7 days post-preparation when compared with these same cell types in array spots formed manually. It is noteworthy that the loss of viability in non-modified PEG hydrogels is consistent with previously published studies of cells in PEG hydrogels formed via conventional approaches[7, 17, 32].
Figure 4.
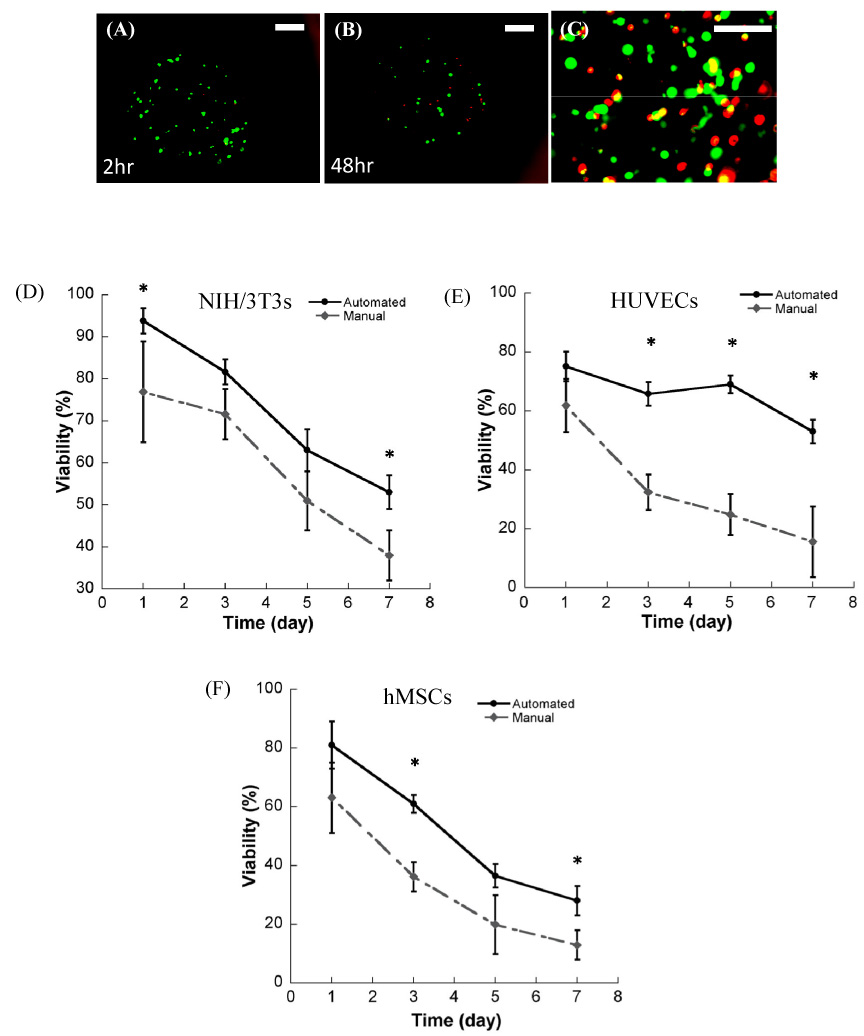
Representative images of NIH/3T3 cells incorporation into PEG hydrogel arrays A) 2 hours after encapsulation, and B) 48 hours after encapsulation. ((A) and (B) were taken at 40x magnification, scale bar = 100 µm). Representative image at higher magnification (100x magnification, scale bar = 50 µm) 2 hours after encapsulation. (D–F) Viability of three cell types in hydrogel arrays filled manually (by hand) and filled automatically (via a liquid handling robot) at 3 min UV exposure time at various time point; D) NIH/3T3 fibroblasts, E) HUVEC, and F) hMSC. (* Significant difference between manual and automated array processing methods within the same time points, p<0.05, t test).
The cell seeding density in hydrogel array spots can be readily controlled and varied over a wide range (Fig. 5). NIH-3T3 cells were incorporated into a PEG hydrogel array at concentrations of 1×105– 2×106 cells/ml, and there was a direct correlation between the number of cells initially incorporated during array preparation, and the number of cells quantitatively measured within array spots at 24 hours after cell seeding, as expected (Fig. 5F, R2=0.99). This control over cell density allowed us to characterize the influence of cell density on the viability of NIH-3T3 fibroblasts. An increase in cell seeding density from 5×105– 2×106 cells/ml resulted in a slight but not significant increase in NIH-3T3 viability 3 and 5 days after initial cell seeding. Seven days after cell seeding this effect was more pronounced, and there was a significant increase in cell viability with increasing NIH-3T3 seeding density (Fig. 5G, ANOVA p<0.05).
Figure 5.
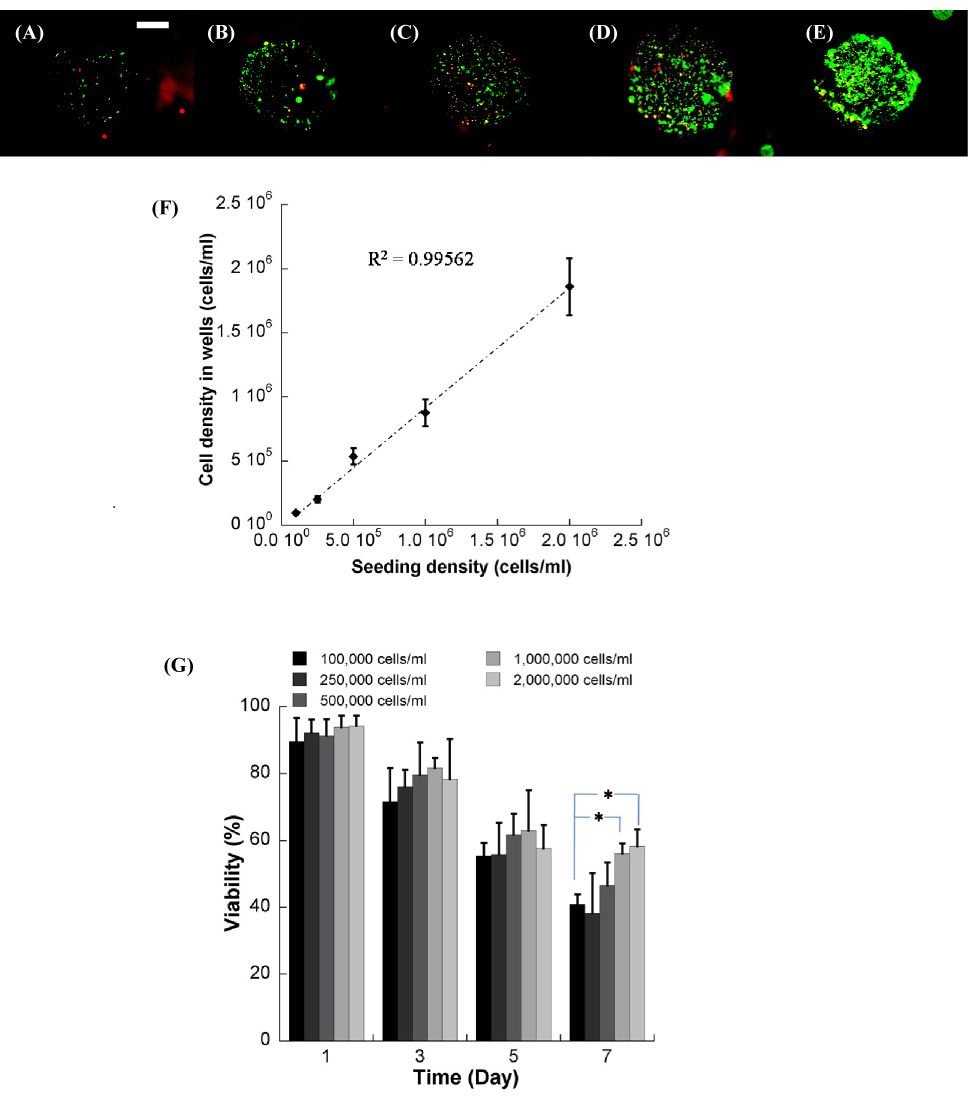
NIH/3T3 cells seeded in the well of the hydrogel array at various cell seeding densities, (A) 1.0×105, (B) 2.5×105,(C) 5.0×105, (D) 1.0×106, and (E) 2.0×106 cells/ml respectively. F) Relationship between cell density in the hydrogel precursor solution and cell density measured in the wells. G) Viability of NIH/3T3 cells seeded at various densities over the range of 7 days. * Significant difference within the same time points, p<0.05. (40x magnification, scale bar = 100 µm).
Cell culture in 3-D natural hydrogel arrays
Viable NIH-3T3 fibroblasts could be incorporated in a controllable manner into type I collagen hydrogels within hydrogel arrays (Fig. 6A). The cell seeding density could be readily varied (Fig. 6B) in a manner similar to the synthetic hydrogel array spots described above (Fig. 5). Virtually all cells observed in these collagen hydrogel spots were viable at all time points and cell densities examined (Fig. 6). Cells demonstrated spreading (Fig. 6C–D) and formation of localized cell aggregates (Fig. 6B) over time in culture. The aggregation of fibroblasts seeded within collagen hydrogels could possibly be caused by contraction of collagen gel in the array spot. However, there is no direct evidence of this contraction, as array spots remain intact for the duration of the experiment.
Figure 6.
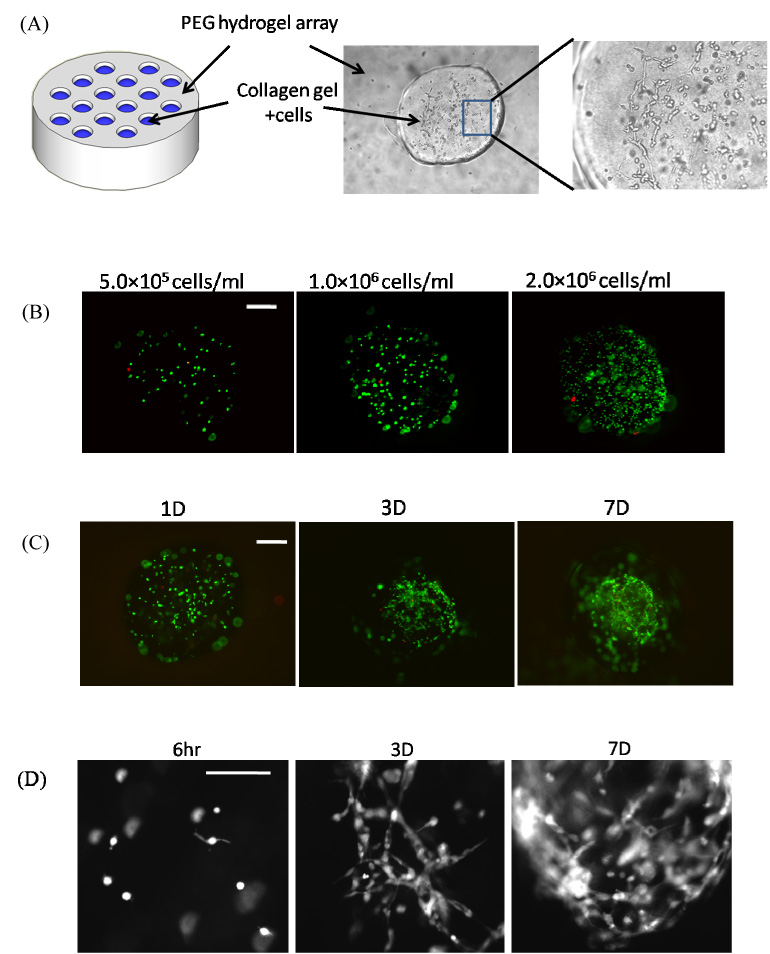
A) Schematic representation of PEG hydrogel array filled with collagen hydrogels in spots. B) Seeding of NIH/3T3 fibroblasts into collagen hydrogel spots with various cell seeding densities. C) Live/dead images of NIH/3T3 fibroblasts cultured in collagen hydrogel spots for various times. D) Higher magnification (100X) images of NIH/3T3 fibroblasts spreading over time in collagen hydrogel array spots.
Screening for the influence of ECM parameters on HL-1 cardiomyocyte viability
Hydrogel arrays were next used as an enhanced throughput culture system to screen for the effects of multiple ECM parameters on the viability of HL-1 cardiomyocytes, a cell type that has not been previously cultured in a 3-D context. In PEG hydrogels prepared without a cell adhesion ligand, ECM degradation led to significant decreases HL-1 viability at 3, 5, and 7 days after initial cell seeding (Fig. 7). In contrast, covalent incorporation of the fibronectin-derived cell adhesion peptide RGDSP into PEG hydrogel networks led to a significant enhancement in viability in both degradable and non-degradable hydrogels (Fig. 8, 9). Fourteen days after initial cell seeding HL-1 viability was 42.7±5.2% in non-degradable array spots without RGDSP, while viability increased to 54.8±3.5%, 66.9±2.5%, and 69.3±3.3% in hydrogel arrays with 7, 35, and 70µM RGDSP, respectively.
Figure 7.
A) Live/Dead images demonstrating the influence of matrix degradation on viability of HL-1 cells encapsulated in PEG hydrogel arrays with varying degradability, 7 days after hydrogel preparation (seeding density=1.0×106 cells/ml) (40x magnification, scale bar = 100 µm). B) Quantitative analysis of HL-1 cardiomyocyte viability in hydrogel arrays with varying degradability. (* Significant difference between conditions with varying DTT content, within the same time points, p<0.05).
Figure 8.
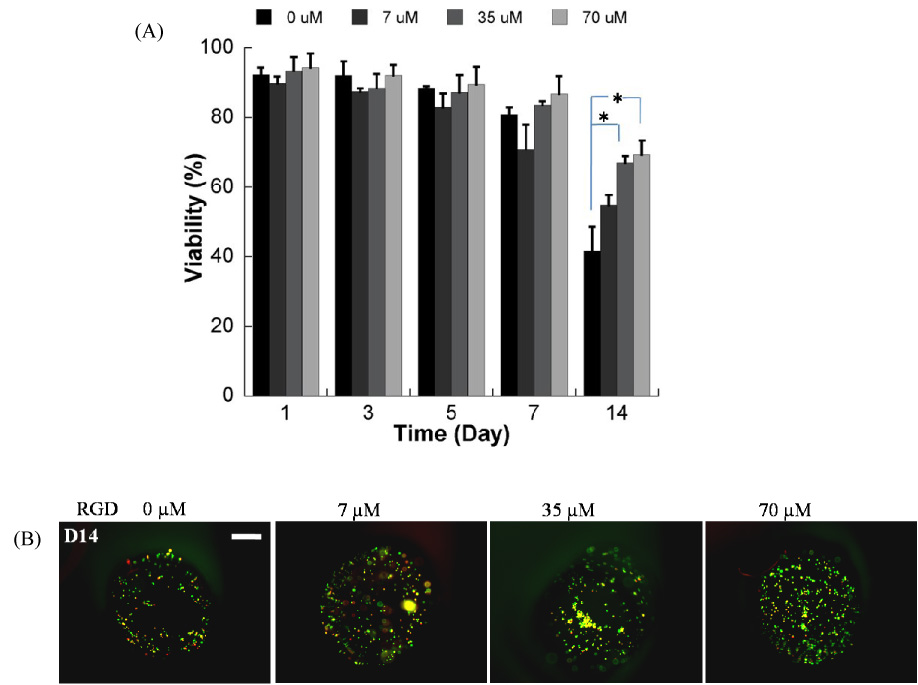
A) Quantitative analysis of HL-1 cardiomyocyte viability within non-degradable PEG hydrogel arrays containing variable RGDSP concentrations (* Significant difference within the same time points, p<0.05); B) Representative images of live/dead staining of cells in array spots 14 days after array preparation demonstrating the influence of RGDSP on HL-1 cardiomyocyte viability. (40x magnification, scale bar = 100 µm).
Figure 9.
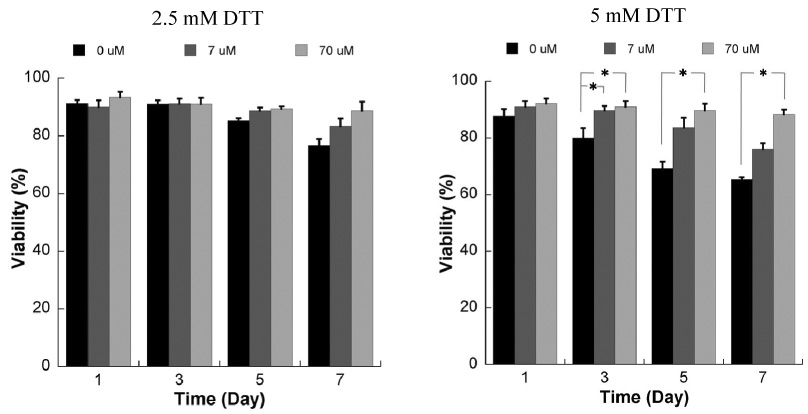
Influence of RGD incorporation on viability of HL-1 cells encapsulated within degradable PEG hydrogels using the live/dead assay. Data are shown for hydrogels prepared with 2.5mM (left) or 5mM DTT (right), and variable concentrations of the RGDSP peptide * Significant difference within the same time points, p<0.05.
Interestingly, the influence of RGDSP on HL-1 was more pronounced and was evident earlier in degradable hydrogel arrays when compared to non-degradable hydrogel arrays (Fig. 9). The presence of RGDSP appears to enhance HL-1 viability 7 days after cell seeding in degradable hydrogels prepared with 2.5mM DTT bridges, but the effect is not statistically significant. In contrast, RGDSP improved HL-1 viability in a dose-dependent manner 5 and 7 days after cell seeding in degradable hydrogels prepared with 5mM DTT bridges. It is noteworthy that in degrading hydrogel arrays that included the maximum amount of RGDSP, there was no significant decrease in HL-1 viability between day 1 and day 7 after cell seeding (Fig. 8, 9), indicating that the presence of RGDSP promotes survival of HL-1 cells during ECM degradation. Taken together, these data validate that hydrogel arrays can be used to explore the impact of multiple quantities of multiple synthetic ECM signals on cell behavior.
Discussion
We have developed an automated method to create adaptable hydrogel arrays for cell culture and tissue engineering. Results indicate that the hydrogel network chemistry (natural and synthetic), cell type, cell density, cell adhesion ligand density, and degradability within each array spot can be systematically varied. These arrays can therefore be used to explore the influence of synthetic and natural ECM parameters on the viability of multiple cell types, including immortalized cell lines (e.g. NIH-3T3 fibroblasts, HL-1 cardiomyocytes) and primary human cells (e.g. HUVECs, hMSCs). A particular value of this approach is the ability to screen for the influence of ECM characteristics on behavior of important cell types that have not been previously cultured in 3-D matrices. We demonstrate this capability herein with HL-1 cardiomyocytes, an immortalized cell line that has been shown to maintain differentiated adult cardiomyocyte phenotype[37]. In principle, this automated approach can be used to explore the influence of a broad range of ECM signals on the behavior of virtually any cell type of interest in a 3-dimensional context.
Our data indicate that specific physical properties of spots in hydrogel arrays can be readily controlled. The cross-linking density and degradability of array spots are dictated by the molecular weight of PEG-diacrylate precursor molecules in PEG networks and the amount of degradable DTT bridges incorporated into (PEG-DTT)n-PEG networks (Fig. 3), respectively. An inherent challenge in previously developed hydrogel arrays has been the propensity of hydrogels to change their physical properties over time[43]. For example, hydrogels swell in aqueous solution and their swollen volume changes during degradation and erosion. When hydrogels are synthesized within the confines of a rigid mold (e.g. PDMS, silicon, polystyrene multi-well plates) they are not able to undergo isotropic swelling to equilibrium due to the surrounding rigid material, and their physical properties during swelling, degradation, and erosion are likely to differ from the properties of a free-standing hydrogel or a hydrogel placed into an in vivo location. The approach described herein creates a background material and wells that degrades in a controllable manner (Fig. 3). Therefore, the background can be designed to swell and degrade in concert with array spots to explore the influence of degradation on cell behavior (Fig. 7). Therefore, the entire array can change its physical properties (e.g. swelling, mechanics, degradation, erosion) in concert, allowing one to explore ECM materials with various equilibrium swelling ratios and degradability. It is also possible to generate arrays in which the array spots are degradable but the background does not degrade (See supplementary data, Fig. S2), as well as arrays in which the background and spots degrade at variable rates (See supplementary data, Fig. S3). These experimental conditions result in confinement of degrading array spots if they degrade faster than the background hydrogel, or strain on array spots if they degrade slower than the background hydrogel. The ability to tailor the dynamic properties of hydrogel array spots and the background hydrogel independently may ultimately allow for more direct mimicry of in vivo applications, in which hydrogel matrices are injected or implanted into environments that are inherently changing their physical properties over time.
Our data also indicate that specific biological properties of spots in hydrogel arrays can be readily controlled. The cell density in each array spot is simply defined by the cell number added in the hydrogel precursor solution, and results demonstrate that the density can be varied over a wide range (1×105–2×106 cells/ml). This range is arbitrarily chosen and could likely be expanded by increasing or decreasing the cell density in the hydrogel precursor solution. The ability to vary cell density is important in view of previous studies, which have shown that changes in cell density in 3-D PEG hydrogels strongly influence cell survival and differentiated function[44]. Herein we found that NIH-3T3 cell viability is improved after 7 days of culture at high cell seeding densities when compared with culture at lower seeding densities (Fig. 5). When NIH-3T3 fibroblasts, which require an adherent substrate to attach and grow, were encapsulated within non-modified hydrogels, which are resistant to cell adhesion, viable cells tended to exist as cell aggregates (e.g. Fig. 5E). Based on these observations, we speculate that at higher cell seeding density the formation of cell aggregates was more prevalent, leading to greater cell viability. The ECM can also be modified to include peptides, such as the fibronectin-derived cell adhesion ligand (RGDSP), and the peptide concentration can be varied (7µM<[RGDSP]<70µM). Importantly, both synthetic and natural hydrogels can be explored as cell culture matrices in this format, as demonstrated by formation of collagen array spots (Fig. 6). Therefore, in contrast to the previous methods used to generate patterned hydrogel networks, our approach for hydrogel array synthesis does not require light-sensitive precursor materials, which may improve accessibility of this approach by those interested in naturally-derived ECMs.
Our results validate that 3-D hydrogel arrays can be used as platforms for enhanced throughput, 3-D culture of cell types with a long history of 3-D culture, including HUVECs[45], hMSCs[7, 17, 26], and NIH-3T3 cells[46]. Importantly, the array format described here can also be used to screen for the influence of 3-D ECM cues on the behavior of key cell types that do not yet have a history of 3-D culture. This capability is demonstrated here via culture of HL-1 cardiomyocytes, a cell line that has been previously shown to be an appropriate model cell type for cardiomyocyte apoptosis, cell cycle regulation, electrophysiology, oxidative stress, signal transduction, transcriptional regulation, and cell transplantation (reviewed in [37]). Our results indicate that the fibronectin-derived cell adhesion ligand RGDSP has a significant, dose-dependent influence on HL-1 cardiomyocyte viability within 3-D PEG hydrogels. This result is consistent with previous 2-dimensional culture studies of HL-1 cardiomyocytes, in which fibronectin was required for cell growth while maintaining differentiated function[47]. Furthermore, the influence of RGDSP is more pronounced within degradable PEG hydrogels when compared to non-degradable PEG hydrogels, particularly at early time points. HL-1 cell viability decreases with hydrogel degradability in the absence of RGDSP, but RGDSP promotes increased viability in a dose dependent manner in both non-degradable and degradable PEG hydrogels. This dose-dependent influence is more pronounced, and apparent at earlier timepoints in more highly degradable networks (Fig. 9, days 3, 5, and 7) when compared to non-degradable networks (Fig. 8, day 14). Interestingly, we found that HL-1 cell viability was maintained at high levels (>85%, Fig. 9) in degradable hydrogels in which the maximum [RGDSP] was included (70µM). One likely reason for these dependencies is that as the hydrogel degrades, the swollen volume increases, which leads to decreases in the effective RGDSP concentration. Therefore, higher initial RGDSP ligand concentrations are needed to maintain a high ligand presentation to HL-1cells within a degrading network. Taken together, these results suggest that the RGDSP ligand may be particularly important in cardiac tissue engineering applications in which the ECM surrounding cardiomyocytes is often designed to degrade over time.
The mechanism for array synthesis described herein is simple, rapid, and adaptable relative to hydrogel arrays described in previous studies. Previous studies in 2-D and 3-D culture work have used photolithographic techniques[31, 33–35], laminar flow[36, 49–52], and combinations thereof[32, 53, 54], to generate microarrays of photo-reactive polymers. Although these previous studies provide valuable insights into the effects of ECM signals on cell behavior, photolithographic techniques are typically limited to the use of photo-reactive polymers, and laminar flow is dimensionally limited to micro-scale materials. In contrast, the approach described here can be used to explore multiple natural and synthetic base materials without regard to the polymerization mechanism or spatial dimension. The use of an automated liquid handler also enables a large number of separate array spots to be synthesized rapidly. In the current format, sixteen 1µl array spots can be processed in 4 minutes, a production rate of 15 seconds/array spot, which implies that hundreds of 3-D environments can be practically analyzed. In principle, this technique can also be scaled to smaller array spots, as liquid handler systems are routinely able to pipette ≤0.1µl. Although the manual analysis of viable cell number performed in this manuscript is not high-throughput, there are several high-content image analysis techniques and software tools available [17, 48] that could be readily used in conjunction with this array processing approach to enhance analysis throughput.
It is noteworthy that viability of primary cell types (HUVECs and hMSCs) was significantly enhanced in hydrogel arrays produced using an automated liquid handler when compared to cells in arrays produced by manual pipetting (Fig. 4). It is possible that automated array synthesis decreases cell death by decreasing the amount of time cells must remain in suspension prior to photo-crosslinking, as the manual array processing rate (90 seconds/spot) was approximately 6-fold longer than the automated processing rate (15 seconds/spot). Indeed, our results (See supplementary data, Fig. S4) indicate that there is a decrease in viable cell number per well as the amount of time cells are suspended in PEGDA solution prior to photo-crosslinking is increased. Cell viability also decreases further after the cells are subjected to ultraviolet light exposure for 3 min, as expected.
Supplementary Material
Acknowledgements
The authors acknowledge financial support from the National Institutes of Health (R21EB005374 and R21HL084547 to W.L.M), the American Heart Association (0650032Z to G.E.L), the National Library of Medicine (5T15LM007359 to J.W.W), and the National Science Foundation for a graduate fellowship to W.J.K. The authors would like to thank Jim Molenda for collecting data for cells suspended in polymer solution.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edelman DB, Keefer EW. A cultural renaissance: in vitro cell biology embraces three-dimensional context. Exp Neurol. 2005;192(1):1–6. doi: 10.1016/j.expneurol.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14(5):633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 3.Green JA, Yamada KM. Three-dimensional microenvironments modulate fibroblast signaling responses. Adv Drug Deliv Rev. 2007;59(13):1293–1298. doi: 10.1016/j.addr.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 5.Vaalamo M, Mattila L, Johansson N, Kariniemi AL, Karjalainen-Lindsberg ML, Kahari VM, et al. Distinct populations of stromal cells express collagenase-3 (MMP-13) and collagenase-1 (MMP-1) in chronic ulcers but not in normally healing wounds. J Invest Dermatol. 1997;109(1):96–101. doi: 10.1111/1523-1747.ep12276722. [DOI] [PubMed] [Google Scholar]
- 6.Mahoney MJ, Anseth KS. Three-dimensional growth and function of neural tissue in degradable polyethylene glycol hydrogels. Biomaterials. 2006;27(10):2265–2274. doi: 10.1016/j.biomaterials.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Nuttelman CR, Tripodi MC, Anseth KS. In vitro osteogenic differentiation of human mesenchymal stem cells photoencapsulated in PEG hydrogels. J Biomed Mater Res. 2004;68A(4):773–782. doi: 10.1002/jbm.a.20112. [DOI] [PubMed] [Google Scholar]
- 8.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5(8):711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 9.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89(19):9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki T, Takagi M, Soma T, Yoshida T. 3D culture of murine hematopoietic cells with spatial development of stromal cells in nonwoven fabrics. Cytotherapy. 2002;4(3):285–291. doi: 10.1080/146532402320219808. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, Fitzgerald W, Liu QY, O'Shaughnessy TJ, Maric D, Lin HJ, et al. CNS stem and progenitor cell differentiation into functional neuronal circuits in three-dimensional collagen gels. Exp Neurol. 2004;190(2):276–288. doi: 10.1016/j.expneurol.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Drury JL, Mooney DJ. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24(24):4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 13.Tsang VL, Bhatia SN. Three-dimensional tissue fabrication. Adv Drug Deliv Rev. 2004;56(11):1635–1647. doi: 10.1016/j.addr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 15.Saltzman WM, Olbricht WL. Building drug delivery into tissue engineering. Nat Rev Drug Discov. 2002;1(3):177–186. doi: 10.1038/nrd744. [DOI] [PubMed] [Google Scholar]
- 16.Castner DGRBD. Biomedical surface science: Foundations to frontiers. Surface Science. 2002;500:28–60. [Google Scholar]
- 17.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol. 2005;24(3):208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Uludag H, De Vos P, Tresco PA. Technology of mammalian cell encapsulation. Adv Drug Deliv Rev. 2000;42(1–2):29–64. doi: 10.1016/s0169-409x(00)00053-3. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez AL, Gobin AS, West JL, McIntire LV, Smith CW. Integrin interactions with immobilized peptides in polyethylene glycol diacrylate hydrogels. Tissue Eng. 2004;10(11–12):1775–1786. doi: 10.1089/ten.2004.10.1775. [DOI] [PubMed] [Google Scholar]
- 20.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4(3):713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 21.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Systematic modulation of Michael-type reactivity of thiols through the use of charged amino acids. Bioconjug Chem. 2001;12(6):1051–1056. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 22.Benoit DS, Durney AR, Anseth KS. Manipulations in hydrogel degradation behavior enhance osteoblast function and mineralized tissue formation. Tissue Eng. 2006;12(6):1663–1673. doi: 10.1089/ten.2006.12.1663. [DOI] [PubMed] [Google Scholar]
- 23.Bryant SJ, Bender RJ, Durand KL, Anseth KS. Encapsulating chondrocytes in degrading PEG hydrogels with high modulus: engineering gel structural changes to facilitate cartilaginous tissue production. Biotechnol Bioeng. 2004;86(7):747–755. doi: 10.1002/bit.20160. [DOI] [PubMed] [Google Scholar]
- 24.Rice MA, Anseth KS. Encapsulating chondrocytes in copolymer gels: bimodal degradation kinetics influence cell phenotype and extracellular matrix development. J Biomed Mater Res A. 2004;70(4):560–568. doi: 10.1002/jbm.a.30106. [DOI] [PubMed] [Google Scholar]
- 25.Hwang NS, Kim MS, Sampattavanich S, Baek JH, Zhang Z, Elisseeff J. Effects of three-dimensional culture and growth factors on the chondrogenic differentiation of murine embryonic stem cells. Stem Cells. 2006;24(2):284–291. doi: 10.1634/stemcells.2005-0024. [DOI] [PubMed] [Google Scholar]
- 26.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26(30):5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Bryant SJ, Arthur JA, Anseth KS. Incorporation of tissue-specific molecules alters chondrocyte metabolism and gene expression in photocrosslinked hydrogels. Acta Biomater. 2005;1(2):243–252. doi: 10.1016/j.actbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Bryant SJ, Durand KL, Anseth KS. Manipulations in hydrogel chemistry control photoencapsulated chondrocyte behavior and their extracellular matrix production. J Biomed Mater Res A. 2003;67(4):1430–1436. doi: 10.1002/jbm.a.20003. [DOI] [PubMed] [Google Scholar]
- 29.Rice MA, Anseth KS. Controlling cartilaginous matrix evolution in hydrogels with degradation triggered by exogenous addition of an enzyme. Tissue Eng. 2007;13(4):683–691. doi: 10.1089/ten.2006.0142. [DOI] [PubMed] [Google Scholar]
- 30.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103(8):2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Photo- and electropatterning of hydrogel-encapsulated living cell arrays. Lab Chip. 2005;5(1):111–118. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 32.Koh WG, Revzin A, Pishko MV. Poly(ethylene glycol) hydrogel microstructures encapsulating living cells. Langmuir. 2002;18(7):2459–2462. doi: 10.1021/la0115740. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda J, Khademhosseini A, Yeo Y, Yang X, Yeh J, Eng G, et al. Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials. 2006;27(30):5259–5267. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 34.Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, 3rd, Langer R, et al. Micromolding of photocrosslinkable hyaluronic acid for cell encapsulation and entrapment. J Biomed Mater Res A. 2006;79(3):522–532. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 35.Koh WG, Itle LJ, Pishko MV. Molding of hydrogel microstructures to create multiphenotype cell microarrays. Anal Chem. 2003;75(21):5783–5789. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- 36.Kim MS, Yeon JH, Park JK. A microfluidic platform for 3-dimensional cell culture and cell-based assays. Biomed Microdevices. 2007;9(1):25–34. doi: 10.1007/s10544-006-9016-4. [DOI] [PubMed] [Google Scholar]
- 37.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286(3):H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 38.Cruise GM, Scharp DS, Hubbell JA. Characterization of permeability and network structure of interfacially photopolymerized poly(ethylene glycol) diacrylate hydrogels. Biomaterials. 1998;19(14):1287–1294. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 39.Hudalla GA, Eng TS, Murphy WL. An approach to modulate degradation and mesenchymal stem cell behavior in poly(ethylene glycol) networks. Biomacromolecules. 200;9:842–849. doi: 10.1021/bm701179s. [DOI] [PubMed] [Google Scholar]
- 40.Murphy WL, Dillmore WS, Modica J, Mrksich M. Dynamic hydrogels: Translating a protein conformational change into a macroscopic motion. Angewandte Chemie Int Ed. 2007;46:1–5. doi: 10.1002/anie.200604808. [DOI] [PubMed] [Google Scholar]
- 41.Sui Z, King WJ, Murphy WL. Dynamic materials based on a protein conformational change. Advanced Materials. 2007;19(20):3377–3380. [Google Scholar]
- 42.Wozniak M, Keely P. Use of three-dimensional collagen gels to study mechanotransduction in T47D breast epithelial cells. Biol Proced Online. 2005;7(1):144–161. doi: 10.1251/bpo112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flory P. Principles of Polymer Chemistry. Ithaca, NY: Cornell University Press; 1953. [Google Scholar]
- 44.Troken A, Marion N, Hollister S, Mao J. Tissue engineering of the synovial joint: the role of cell density. Proc Inst Mech Eng [H] 2007;221(5):429–440. doi: 10.1243/09544119JEIM288. [DOI] [PubMed] [Google Scholar]
- 45.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, et al. Micro-well arrays for 3D shape control and high resolution analysis of single cells. Lab Chip. 2007;7(8):1074–1077. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
- 46.Yeh J, Ling Y, Karp JM, Gantz J, Chandawarkar A, Eng G, et al. Micromolding of shape-controlled, harvestable cell-laden hydrogels. Biomaterials. 2006;27(31):5391–5398. doi: 10.1016/j.biomaterials.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Claycomb WC, Lanson NA, Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abraham VC, Taylor DL, Haskins JR. High content screening applied to large scale cell biology. Trends Biotechnol. 2004;22:15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Burdick JA, Khademhosseini A, Langer R. Fabrication of gradient hydrogels using a microfluidics/photopolymerization process. Langmuir. 2004;20(13):5153–5156. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 50.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Microfluidic scaffolds for tissue engineering. Nat Mater. 2007;6(11):908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 51.Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, et al. A cell-laden microfluidic hydrogel. Lab Chip. 2007;7(6):756–762. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- 52.Zaari NR P, Kim SK, Engler AJ, Wong JY. Photopolymerization in microfluidic gradient generators: microscale control of substrate compliance to manipulate cell response. Advanced Materials. 2004;16(23):2133–2137. [Google Scholar]
- 53.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nature Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 54.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embronic stem cells. Nature Biotechnology. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


