Abstract
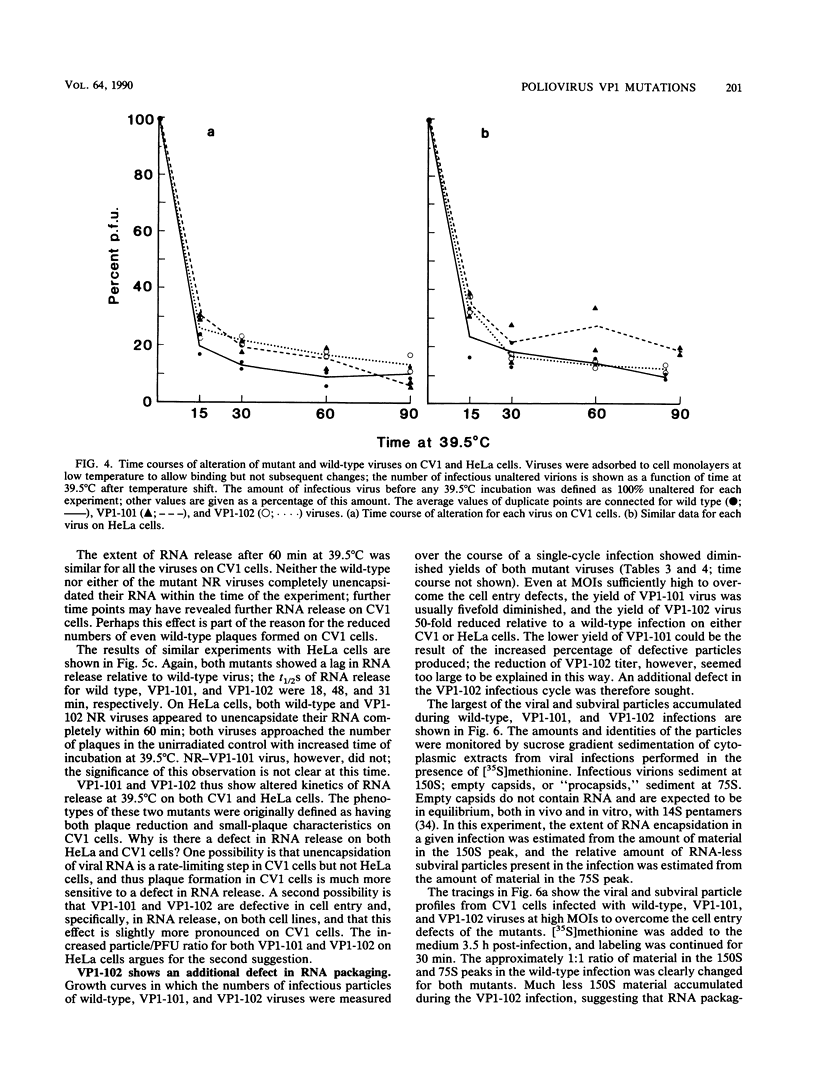
The phenotypic defects of two type 1 Mahoney poliovirus mutants, termed VP1-101 and VP1-102, were caused by two different small deletions in the region of the RNA genome encoding the amino terminus of the capsid protein VP1. This portion of VP1 was unresolved in the three-dimensional structure of the poliovirion, buried within the virion, and likely to interact with the viral RNA. Both VP1-101 and VP1-102 showed a diminished ability to enter CV1 but not HeLa cells; both mutants formed plaques on CV1 and HeLa cells that were smaller than wild type. Neither the rate of binding to cells nor the rate of subsequent receptor-dependent conformational change of the mutant poliovirions was affected. However, both mutants displayed delayed kinetics of RNA release compared with wild-type virus. One of the mutants, VP1-102, also displayed a defect in viral morphogenesis: 75S empty capsids formed normally, but 150S particles that contained RNA accumulated much more slowly. We suggest that the VP1-102 mutation affects RNA encapsidation as well as RNA release, whereas the VP1-101 mutation affects only RNA release. Therefore, RNA packaging and RNA release are genetically linked but can be mutated separately in different VP1 alleles, and both processes involve the amino terminus of VP1.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold E., Luo M., Vriend G., Rossmann M. G., Palmenberg A. C., Parks G. D., Nicklin M. J., Wimmer E. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci U S A. 1987 Jan;84(1):21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J., Minor I., Kremer M. J., Oliveira M. A., Smith T. J., Griffith J. P., Guerin D. M., Krishnaswamy S., Luo M., Rossmann M. G. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc Natl Acad Sci U S A. 1988 May;85(10):3304–3308. doi: 10.1073/pnas.85.10.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D., Girard M., Darnell J. E. Aspects of the synthesis of poliovirus RNA and the formation of virus particles. Virology. 1966 Jun;29(2):179–189. doi: 10.1016/0042-6822(66)90024-9. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Picornaviruses are no longer black boxes. Science. 1985 Sep 27;229(4720):1366–1367. doi: 10.1126/science.2994219. [DOI] [PubMed] [Google Scholar]
- Baron M. H., Baltimore D. Antibodies against the chemically synthesized genome-linked protein of poliovirus react with native virus-specific proteins. Cell. 1982 Feb;28(2):395–404. doi: 10.1016/0092-8674(82)90357-9. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Sonenberg N., Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985 Nov;5(11):2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROWTHER D., MELNICK J. L. The incorporation of neutral red and acridine orange into developing poliovirus particles making them photosensitive. Virology. 1961 May;14:11–21. doi: 10.1016/0042-6822(61)90127-1. [DOI] [PubMed] [Google Scholar]
- Caliguiri L. A., McSharry J. J., Lawrence G. W. Effect of arildone on modifications of poliovirus in vitro. Virology. 1980 Aug;105(1):86–93. doi: 10.1016/0042-6822(80)90158-0. [DOI] [PubMed] [Google Scholar]
- Chow M., Newman J. F., Filman D., Hogle J. M., Rowlands D. J., Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987 Jun 11;327(6122):482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- Colonno R. J., Condra J. H., Mizutani S., Callahan P. L., Davies M. E., Murcko M. A. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5449–5453. doi: 10.1073/pnas.85.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sena J., Mandel B. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology. 1977 May 15;78(2):554–566. doi: 10.1016/0042-6822(77)90130-1. [DOI] [PubMed] [Google Scholar]
- Eggers H. J. Selective inhibiton of uncoating of echovirus 12 by rhodanine. A study on early virus-cell interactions. Virology. 1977 May 1;78(1):241–252. doi: 10.1016/0042-6822(77)90095-2. [DOI] [PubMed] [Google Scholar]
- Filman D. J., Syed R., Chow M., Macadam A. J., Minor P. D., Hogle J. M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989 May;8(5):1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve J. M., Davis G., Meyer A. M., Forte C. P., Yost S. C., Marlor C. W., Kamarck M. E., McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989 Mar 10;56(5):839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- Guttman N., Baltimore D. A plasma membrane component able to bind and alter virions of poliovirus type 1: studies on cell-free alteration using a simplified assay. Virology. 1977 Oct 1;82(1):25–36. doi: 10.1016/0042-6822(77)90029-0. [DOI] [PubMed] [Google Scholar]
- HOLLAND J. J. Irreversible eclipse of poliovirus by HeLa cells. Virology. 1962 Feb;16:163–176. doi: 10.1016/0042-6822(62)90292-1. [DOI] [PubMed] [Google Scholar]
- Hogle J. M., Chow M., Filman D. J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985 Sep 27;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Nelsen B. Conditional poliovirus mutants made by random deletion mutagenesis of infectious cDNA. J Virol. 1990 Jan;64(1):185–194. doi: 10.1128/jvi.64.1.185-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Lonberg-Holm K., Gosser L. B., Shimshick E. J. Interaction of liposomes with subviral particles of poliovirus type 2 and rhinovirus type 2. J Virol. 1976 Aug;19(2):746–749. doi: 10.1128/jvi.19.2.746-749.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANDEL B. THE FATE OF THE INOCULUM IN HELA CELLS INFECTED WITH POLIOVIRUS. Virology. 1965 Jan;25:152–154. doi: 10.1016/0042-6822(65)90264-3. [DOI] [PubMed] [Google Scholar]
- Madshus I. H., Olsnes S., Sandvig K. Mechanism of entry into the cytosol of poliovirus type 1: requirement for low pH. J Cell Biol. 1984 Apr;98(4):1194–1200. doi: 10.1083/jcb.98.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSharry J. J., Caliguiri L. A., Eggers H. J. Inhibition of uncoating of poliovirus by arildone, a new antiviral drug. Virology. 1979 Sep;97(2):307–315. doi: 10.1016/0042-6822(79)90342-8. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C. L., Wimmer E., Racaniello V. R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989 Mar 10;56(5):855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Bradley J., Yang X. F., Wimmer E., Moss E. G., Racaniello V. R. Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science. 1988 Jul 8;241(4862):213–215. doi: 10.1126/science.2838906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C., Frasel L., Kuechler E., Blaas D. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology. 1987 May;158(1):255–258. doi: 10.1016/0042-6822(87)90264-9. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y., Ohsawa C., Aoyama M., Umeda I., Suhara Y., Ishitsuka H. Antivirus agent, Ro 09-0410, binds to rhinovirus specifically and stabilizes the virus conformation. Virology. 1984 Apr 30;134(2):269–276. doi: 10.1016/0042-6822(84)90296-4. [DOI] [PubMed] [Google Scholar]
- Putnak J. R., Phillips B. A. Picornaviral structure and assembly. Microbiol Rev. 1981 Jun;45(2):287–315. doi: 10.1128/mr.45.2.287-315.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981 Nov 20;214(4523):916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Arnold E., Erickson J. W., Frankenberger E. A., Griffith J. P., Hecht H. J., Johnson J. E., Kamer G., Luo M., Mosser A. G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985 Sep 12;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Sarnow P., Bernstein H. D., Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci U S A. 1986 Feb;83(3):571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P. Role of 3'-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989 Jan;63(1):467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. J., Kremer M. J., Luo M., Vriend G., Arnold E., Kamer G., Rossmann M. G., McKinlay M. A., Diana G. D., Otto M. J. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986 Sep 19;233(4770):1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- Staunton D. E., Merluzzi V. J., Rothlein R., Barton R., Marlin S. D., Springer T. A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989 Mar 10;56(5):849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- WILSON J. N., COOPER P. D. ASPECTS OF THE GROWTH OF POLIOVIRUS AS REVEALED BY THE PHOTODYNAMIC EFFECTS OF NEUTRAL RED AND ACRIDINE ORANGE. Virology. 1963 Oct;21:135–145. doi: 10.1016/0042-6822(63)90249-6. [DOI] [PubMed] [Google Scholar]
- Zeichhardt H., Otto M. J., McKinlay M. A., Willingmann P., Habermehl K. O. Inhibition of poliovirus uncoating by disoxaril (WIN 51711). Virology. 1987 Sep;160(1):281–285. doi: 10.1016/0042-6822(87)90075-4. [DOI] [PubMed] [Google Scholar]
- van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]






