Abstract
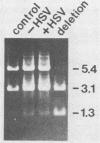
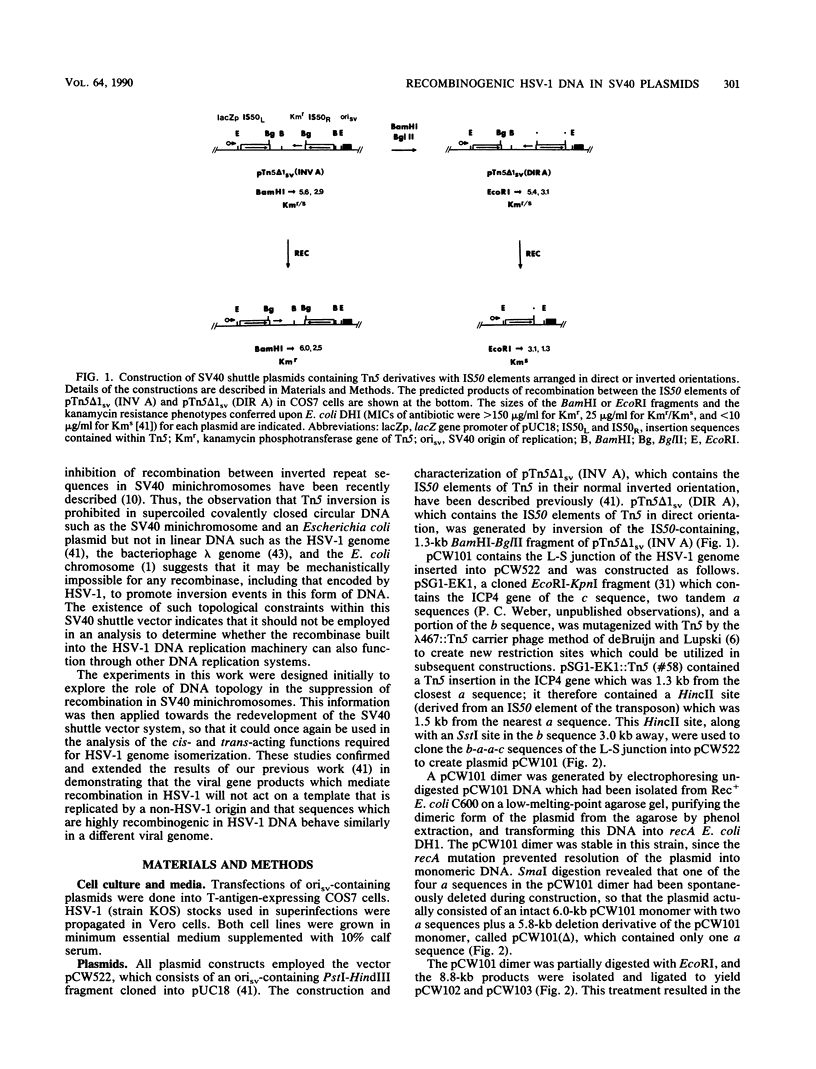
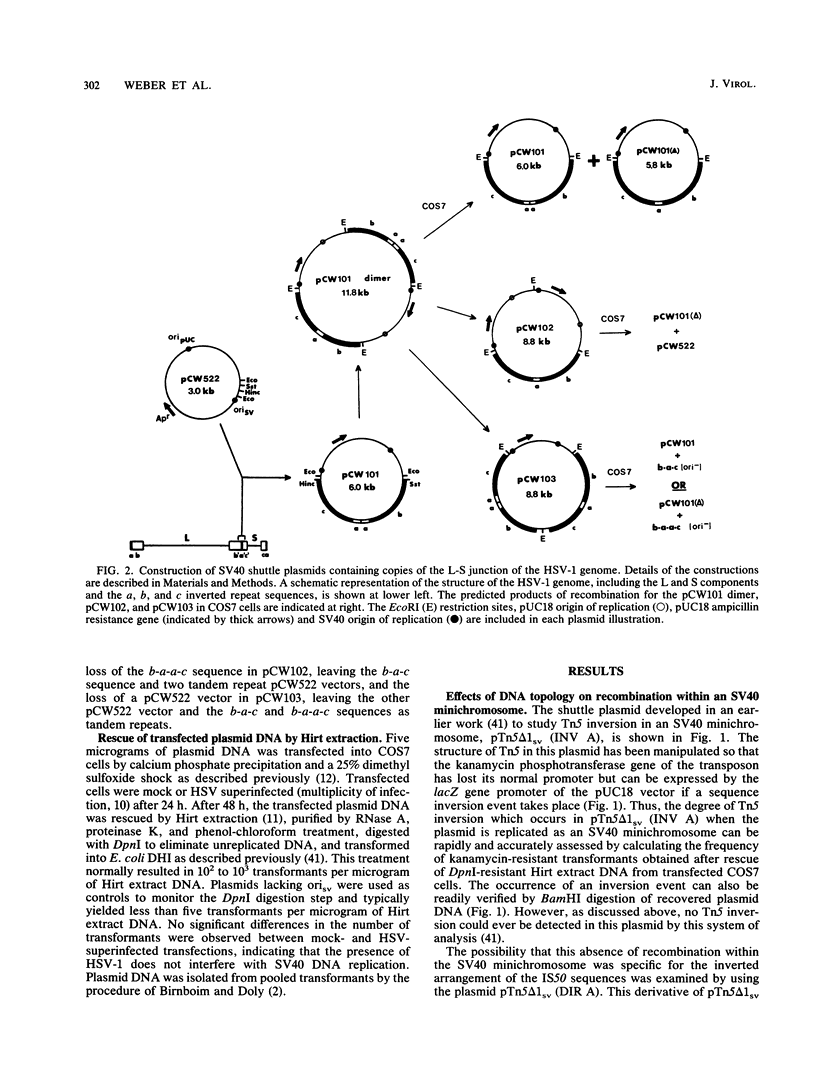
In a previous work, it was demonstrated that the bacterial transposon Tn5 is capable of undergoing sequence inversion via recombination between its duplicated IS50 elements when replicated by the herpes simplex virus type 1 (HSV-1) origin oris but not by the simian virus 40 (SV40) origin orisv. Further analysis of the latter phenomenon indicated that this lack of recombination was the result of topological constraints imposed by the SV40 minichromosome, such that recombination events could be readily detected in Tn5 derivatives in which the IS50 elements were arranged in a direct rather than inverted orientation. With this information, a second set of experiments were carried out to examine how the highly recombinogenic sequences which mediate the inversion of the long (L) and short (S) components of the HSV-1 genome behave in an SV40 minichromosome. Tandem copies of the L-S junction of the HSV-1 genome were observed to promote deletions in an SV40 shuttle plasmid at a frequency that was considerably greater than that of duplicated bacterial plasmid vector DNA. However, the presence of superinfecting HSV-1 did not enhance the frequency of these recombination events. These results support our previous findings that HSV-1 genome isomerization is mediated by a homologous recombination mechanism which is intimately associated with the act of viral DNA synthesis. Moreover, they demonstrate that the sequences which comprise the L-S junction appear to be inherently recombinogenic and, therefore, do not contain specific signals required for HSV-1 genome isomerization.
Full text
PDF
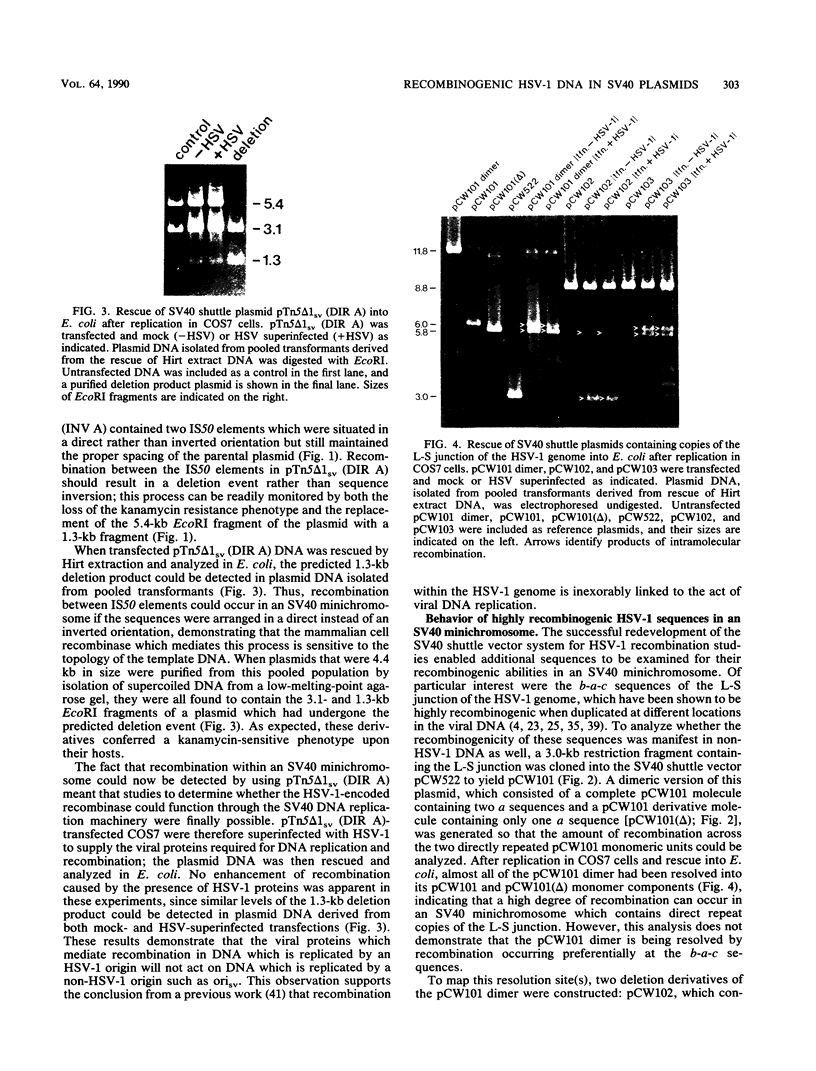



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg D. E. Control of gene expression by a mobile recombinational switch. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4880–4884. doi: 10.1073/pnas.77.8.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986 Jun 20;45(6):793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Gibson M. G., Spear P. G. Insertion mutants of herpes simplex virus have a duplication of the glycoprotein D gene and express two different forms of glycoprotein D. J Virol. 1983 Nov;48(2):396–404. doi: 10.1128/jvi.48.2.396-404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel S. S., Krysan P. J., Calos M. P., DuBridge R. B. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J Virol. 1988 Oct;62(10):3738–3746. doi: 10.1128/jvi.62.10.3738-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Homa F. L., Otal T. M., Glorioso J. C., Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases -34 to +124 relative to the 5' terminus of the mRNA. Mol Cell Biol. 1986 Nov;6(11):3652–3666. doi: 10.1128/mcb.6.11.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins F. J., Casadaban M. J., Roizman B. Application of the mini-Mu-phage for target-sequence-specific insertional mutagenesis of the herpes simplex virus genome. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4773–4777. doi: 10.1073/pnas.82.14.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins F. J., Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J Virol. 1986 Aug;59(2):494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. B., Glaccum M. B., Simon M. I. In vitro analysis of Hin-mediated site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:751–760. doi: 10.1101/sqb.1984.049.01.085. [DOI] [PubMed] [Google Scholar]
- Kennedy K. E., Iida S., Meyer J., Stålhammar-Carlemalm M., Hiestand-Nauer R., Arber W. Genome fusion mediated by the site specific DNA inversion system of bacteriophage P1. Mol Gen Genet. 1983;189(3):413–421. doi: 10.1007/BF00325903. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Symington L. S., Dyson P., Sherratt D. J. Transposon-encoded site-specific recombination: nature of the Tn3 DNA sequences which constitute the recombination site res. EMBO J. 1983;2(7):1055–1060. doi: 10.1002/j.1460-2075.1983.tb01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. T., Pogue-Geile K. L., Pereira L., Spear P. G. Expression of herpes simplex virus glycoprotein C from a DNA fragment inserted into the thymidine kinase gene of this virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6612–6616. doi: 10.1073/pnas.79.21.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Van de Putte P. Genetic switches by DNA inversions in prokaryotes. Biochim Biophys Acta. 1984 Jun 16;782(2):111–119. doi: 10.1016/0167-4781(84)90013-7. [DOI] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Tabares E., Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983 May;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Lee G. T., Spear P. G. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985 Feb;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Quinlan M. P., Knipe D. M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985 May;5(5):957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., Campbell M. E., Clements J. B. A tandemly reiterated DNA sequence in the long repeat region of herpes simplex virus type 1 found in close proximity to immediate-early mRNA 1. J Virol. 1984 Nov;52(2):715–718. doi: 10.1128/jvi.52.2.715-718.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott T. N., Simon M. I. Genetic analysis of the mechanism of the Salmonella phase variation site specific recombination system. Mol Gen Genet. 1982;188(2):313–321. doi: 10.1007/BF00332694. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Song K. Y., Chekuri L., Rauth S., Ehrlich S., Kucherlapati R. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985 Dec;5(12):3331–3336. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. K. Derivatives of ColE1 cer show altered topological specificity in site-specific recombination. EMBO J. 1989 Jan;8(1):309–315. doi: 10.1002/j.1460-2075.1989.tb03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Wohlrab F., McLean M. J., Wells R. D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987 May 5;262(13):6407–6416. [PubMed] [Google Scholar]
- Yagil E., Dower N. A., Chattoraj D., Stahl M., Pierson C., Stahl F. W. Chi mutation in a transposon and the orientation-dependence of Chi phenotype. Genetics. 1980 Sep;96(1):43–57. doi: 10.1093/genetics/96.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]