Abstract
The structure of truncated human apolipoprotein A-I (apo A-I), the major protein component of high density lipoprotein, has been determined at 4-Å resolution. The crystals comprise residues 44–243 (exon 4) of apo A-I, a fragment that binds to lipid similarly to intact apo A-I and that retains the lipid-bound conformation even in the absence of lipid. The molecule consists almost entirely of a pseudo-continuous, amphipathic α-helix that is punctuated by kinks at regularly spaced proline residues; it adopts a shape similar to a horseshoe of dimensions 125 × 80 × 40 Å. Four molecules in the asymmetric unit associate via their hydrophobic faces to form an antiparallel four-helix bundle with an elliptical ring shape. Based on this structure, we propose a model for the structure of apo A-I bound to high density lipoprotein.
Atherosclerosis is the underlying cause of most cardiovascular disease, the leading cause of death in affluent Western societies. It contributes to the progression of coronary artery disease and cerebrovascular insufficiency, which lead to ischaemic heart disease, myocardial infarction, and stroke. The results of several large clinical trials have demonstrated that elevated high density lipoprotein (HDL) levels correlate strongly with reduced risks of atherosclerosis and coronary artery disease (1–3). The importance of HDL to the prevention of atherosclerosis is consonant with its role as the mediator of reverse transport of cholesterol from peripheral tissues to the liver for catabolism (4, 5).
HDL is a heterogeneous collection of lipoprotein particles. Nascent HDL is a discoidal particle composed of protein, phosphatidylcholine, and small amounts of cholesterol. In contrast, mature HDL is a spherical particle that consists of half lipid (mostly phosphatidylcholine, cholesterol, cholesterol esters, and triglycerides) and half protein by weight. Apolipoprotein A-I (apo A-I) is the major protein in HDL. In addition to its structural role, apo A-I has a functional role in the reverse transport of cholesterol: it promotes the reversible efflux of cholesterol from peripheral cell plasma membranes into HDL; it activates lecithin-cholesterol acyl transferase, the enzyme that traps cholesterol within HDL by converting cholesterol to cholesterol esters (6); and it is responsible for the recognition of HDL by liver (and adrenal cell) HDL receptors (7–10). Of all the apolipoproteins, levels of apo A-I alone correlate with the protective effect of HDL against atherosclerosis (11, 12).
The structures of lipoproteins have been studied intensively for decades (13–15). Sequence analysis has shown that apo A-I and other apolipoproteins consist of repeated amphipathic α-helices, 11 or 2 × 11 amino acids in length, that provide the lipid-binding regions of these proteins (16). Detailed secondary (17) and tertiary (18) structural models of apo A-I have been proposed. Hydrodynamic methods, electron microscopy and small angle x-ray scattering showed that spherical HDL consists of an ≈84-Å diameter, low electron density core of neutral lipids and phospholipid acyl chains, surrounded by an ≈12-Å-thick, high electron density shell of phospholipid head groups and protein (19). Apo A-I is thought to form a mosaic structure on the surface of spherical HDL, with the amphipathic helices nestled between the phospholipid head groups. Discoidal HDL is composed of a phospholipid bilayer surrounded on the edge by apo A-I. Two models have been proposed for the conformation of apo A-I on the edge of the disc. In the “belt” model, apo A-I forms curved α-helices that wrap around the disc (helices perpendicular to the lipid acyl chains). In the “picket fence” model, apo A-I consists of short antiparallel α-helices separated by reverse turns (helices parallel to the lipid acyl chains; ref. 20).
We recently described a truncation mutant of human apo A-I, apo Δ(1–43)A-I, that contains residues 44–243 of native apo A-I (21). Apo Δ(1–43)A-I and native apo A-I exhibit similar lipid-binding properties, based on several lines of evidence: both associate exclusively with plasma HDL; both show identical exclusion pressures in egg phosphatidylcholine monolayers; both have similar free energies of binding to palmitoyl oleolyl phosphatidylcholine vesicles; and both proteins have similar α-helical contents when bound to lipid (21). Unlike apo A-I, no changes in secondary structure occur when apo Δ(1–43)A-I binds to lipid (21), and near-UV CD, 8-anilinonaphthalene sulfonate (ANS) binding, and thermodynamic stability demonstrate that lipid-free apo Δ(1–43)A-I adopts a tertiary structure that is distinctly different from that of lipid-free apo A-I. Furthermore, the proteolytic cleavage pattern of lipid-free apo Δ(1–43)A-I is essentially identical to that of lipid-bound apo A-I (D.P.R., L. M. Roberts, J. Lebowitz, J.A.E., and C.G.B., unpublished results). We suggested that these data are consistent with (i) a structural rearrangement of the C-terminal domain of apo A-I from a largely unfolded state to an α-helical, lipid binding-competent conformation in lipid-free apo Δ(1–43)A-I; and (ii) an overall structure for lipid-free apo Δ(1–43)A-I that is similar to lipid-bound apo A-I (ref. 21; D.P.R. et al., unpublished results).
To take advantage of this unique opportunity to study the putative lipid-bound structure of apo A-I using a more tractable, lipid-free protein, we undertook the crystallization and structure determination of apo Δ(1–43)A-I. The structure reveals that apo Δ(1–43)A-I adopts an unusual, curved α-helical conformation that has important implications for the structure and function of apo A-I when bound to HDL.
METHODS
Crystallization and Data Collection.
Apo Δ(1–43)A-I was overexpressed in Escherichia coli and purified by preparative reverse-phase HPLC as described (21). Greater than 97% of the protein was in the methionine-reduced form. Large bipyramidal crystals were obtained at 4°C from 1.2 M sodium citrate (pH 6.5–7.5). Precession photography showed the space group to be P212121, a = 97.47 Å, b = 113.87 Å, c = 196.19 Å. Crystal density measurements in mother liquors of varying density (23) indicated the presence of four molecules in the asymmetric unit (solvent content 72%). X-ray diffraction data were collected on a MAR image plate at 10°C using CuKα radiation (graphite monochromator), and processed with mosflm, scala, agrovata, and truncate (24). In the last resolution shell (4.22–4.00 Å), 25% of the observations have I/σI > 2, with an Rmerge of 0.340. Data from this last shell were therefore incorporated into the analysis (Table 1). The self-rotation function (ncode = 1; ref. 25) showed three noncrystallographic (pseudo)dyads at polar angles [ω,ϕ,κ] = [66,128,180] (rotates molecule A onto B, see below; 3.8σ); [90,45,180] (A/C; 7.2σ); and [25,–50,180] (A/D; 2.1σ) that suggested the presence of a 222-pseudosymmetrical tetramer in the asymmetric unit.
Table 1.
Summary of crystallographic analysis
| Diffraction data and phasing statistics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Data Set | Crystals | Resolution, Å | Unique reflections | Completeness, % | Redundancy | 〈I/σI〉 | Rmerge, % | Sites | Rderiv, % | Rc, % | Phasing power |
| Native | 2 | 30-4.0 | 16,089 | 85 (50) | 3.6 (1.3) | 3.7 (0.9) | 16.6 (73.2) | – | – | – | – |
| PIP39 | 2 | 24-4.2 | 13,760 | 84 (31) | 5.9 (2.2) | 3.8 (1.5) | 17.0 (46.0) | 7 | 37 | 62 | 1.32/0.96 |
| PIP4 | 1 | 75-4.5 | 11,678 | 89 (61) | 4.1 (1.8) | 2.9 (1.9) | 17.0 (34.7) | 6 | 24 | 64 | 1.23/0.88 |
| Averaging statistics | |||||
|---|---|---|---|---|---|
|
|
〈FOM〉 | 〈Δϕ〉 | FO/FCR, % | FO/FC CC, % | Map CC, % |
| Before averaging | 0.45 | – | 36.7 | 77.0 | 51.2 |
| After averaging | 0.75 | 58° | 10.6 | 98.6 | 90.7 |
| Refinement statistics | ||||||||
|---|---|---|---|---|---|---|---|---|
| Resolution, Å | Reflections | Rcryst, % | Rfree, % | Atoms | Bond length dev., rms | Bond angle dev., rms | Ramachandran outliers, residues | |
| F > 2σF | 27-4.0 | 15,543 | 38.2 (43.0) | 42.8 (46.2) | 3,294 | 0.010 Å | 1.66° | 31 of 402 |
Rmerge = Σ|I − 〈I〉|/ΣI, where I is the observed intensity and 〈I〉 is the average intensity of multiple observations of symmetry-related reflections. Numbers in parentheses refer to those data in the highest resolution shell (4.22–4.00 Å for the native data set).
Rderiv = Σ||FPH| − |FP||/Σ|FP|, where |FP| and |FPH| are the observed native and heavy-atom derivative structure factor amplitudes.
RC = Σ||FPH ± FP| − FH|/Σ|FPH ± FP|, where FH is the calculated heavy-atom structure factor amplitude; calculated for centric reflections only.
Phasing power = rms (|FH|/E), where E is the residual lack of closure error; listed for acentric/centric reflections.
FOM (mean figure of merit) = 〈ΣP(α)eiα/ΣP(α)〉, where α is the phase and P(α) is the phase probability distribution.
〈Δϕ〉 = rms phase change.
CC = [ΣFOFC − (ΣFOΣFC)/N]/{[ΣFO2 − (ΣFO)2/N][ΣFC2 − (ΣFC)2/N]}½, where |FO| and |FC| are the observed and calculated structure factor amplitudes, and N is the number of reflections.
R = Σ||FO| − |FC||/Σ|FO|; Rfree is calculated for a randomly-selected 5% of the reflections; Rcryst is calculated for the remaining 95% of the reflections used in refinement.
Phasing and Density Modification.
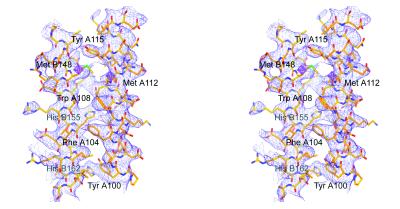
The structure was solved using MIRAS (multiple isomorphous replacement with anomalous scattering) phases derived from crystals soaked with 1 mM [Pt2I2en2](NO3)2 (PIP) for 4 or 39 h. Two sites were determined by inspection of the difference Patterson map Harker sections; difference Fourier syntheses revealed five additional sites. These sites were confirmed by a heavy-atom search (hassp; ref. 26). Choice of hand for the heavy-atom model was obvious from SIRAS (single isomorphous replacement with anomalous scattering) and MIRAS maps calculated using refined (heavy; ref. 27) PIP positions; these maps showed large solvent cavities surrounded by long tubes of density, consistent with α-helices, that curved throughout the unit cell. Solvent flattening (dm; ref. 24) with phase extension to 4.5-Å resolution resulted in a map that was remarkably clear and easily interpretable in terms of four curved, pseudo-continuous α-helices associated together in the shape of a ring. From a Cα trace built with o (28), it was apparent that two molecules of the tetramer had very similar but distinct shapes, and that these two molecules (A and B) were related to two other molecules (C and D) by application of the A/C noncrystallographic dyad. Iterative 2-fold noncrystallographic symmetry averaging (dm; mask around the A/B dimer; Table 1), using data sharpened by application of an artificial temperature factor (−70 Å2; ref. 29), dramatically improved the side chain density in the map. A portion of the final experimental electron density map is shown in Fig. 1.
Figure 1.
Stereoview of the solvent-flattened, noncrystallographic symmetry-averaged experimental electron density map, contoured at 1.25σ at 4-Å resolution, with the current model depicted. Helices A4 (right) and B6 (left) of apo Δ(1–43)A-I are shown, as are two Pt sites (magenta octahedra). The Trp, Tyr, Phe, Met, and His residues of these helices are labeled.
Model Building and Refinement.
A complete model was built into the sharpened, averaged map. Chain direction and register were unambiguously determined as described below. Positional refinement of the model was carried out against the sharpened data (27–4 Å; F > 2σF) using x-plor (30); 2-fold noncrystallographic symmetry was constrained throughout (data to parameter ratio, 1.6:1), and 5% of the data was reserved for calculation of the free R factor. Refinement of 24 group temperature factors (one for each secondary structural element) lowered the free R factor by 2.5%; these temperature factors appear to model varying levels of disorder/mobility along the length of the two molecules. The final model has an R factor of 38.2% (free R factor, 42.8%; Table 1). The high R factor is a result of weak data and a conservative refinement strategy. Both Rcryst and Rfree drop by 10% when data with F > 4σF are used (31). Our structure for apo Δ(1–43)A-I consists of two unique molecules, A and B, each comprising an overexpression vector-derived N-terminal methionine (residue −1) and residues 44–243 of apo A-I; 92% of the residues lie in the strictly allowed regions (32) of the Ramachandran plot. Molecules C and D are derived from A and B by application of the A/C noncrystallographic dyad. The experimental electron density map shows deviations from this noncrystallographic symmetry in the orientation of many side chains, especially at crystal packing contacts. We have not attempted to model these differences (33). Figures were prepared with o and ribbons (34). Solvent-accessible surface areas were calculated with grasp (ref. 35; 1.4-Å probe radius).
RESULTS
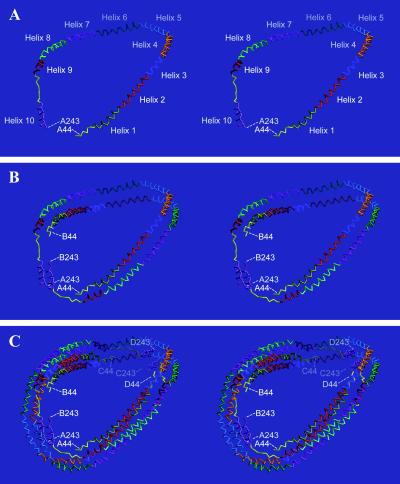
The structure of apo Δ(1–43)A-I determined at 4-Å resolution (Table 1 and Fig. 1) comprises four α-helical, horseshoe-shaped molecules that assemble in the crystal to form a tightly associated elliptical ring (see Figs. 2 and 4). Residues 44–243 of apo A-I are present in each molecule. The tetrameric ring can be divided into two pairs of molecules, A/B and C/D, that are related to one another by a noncrystallographic dyad. The two molecules within each dimer, which are arranged in an intimate, antiparallel fashion, possess very similar, but unique, conformations. The hydrophobic residues of each dimer form a strip that runs the length of the dimer. The two dimers bind one another in an antiparallel manner, thereby burying these hydrophobic strips, to form the tetramer. The unusual tertiary structure of apo Δ(1–43)A-I is composed of amino acids that possess normal geometry and phi/psi torsion angles (Table 1).
Figure 2.
Stereoviews of the apo Δ(1–43)A-I Cα trace. (a) The apo Δ(1–43)A-I monomer, molecule A. The N- and C-termini are labeled, as are each of the secondary structural elements described in the text. These elements are also colored differently. (b and c) The apo Δ(1–43)A-I A/B dimer, and the tetramer. The molecules are colored as in a to facilitate identification of the alignment of the extensive intermolecular interactions.
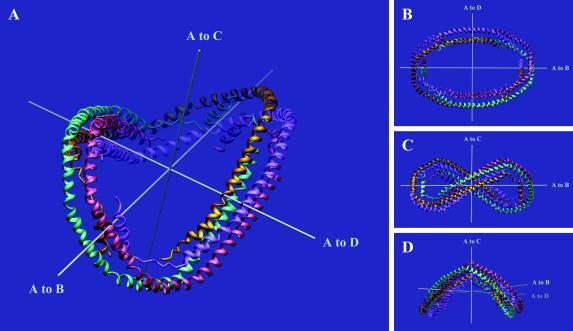
Figure 4.
ribbons (34) representation illustrating the elliptical and curved shape of the apo Δ(1–43)A-I tetramer. (a) The tetramer is shown with the three noncrystallographic (pseudo)dyads: molecule A is gold, molecule B is purple, molecule C is pink, and molecule D is green. (b) View down the A/C dyad. (c) As in b, rotated 90° around the horizontal axis—i.e., viewed down the A/D pseudo-dyad. (d) As in c, rotated ≈70° around the vertical axis.
Monomer Structure.
The apo Δ(1–43)A-I monomer adopts a sharply curved horseshoe shape (Fig. 2a). The molecule consists primarily of a pseudo-continuous, amphipathic α-helix that is punctuated by kinks at regularly spaced proline residues. A short α-helix at the extreme C-terminus is separated from the rest of the molecule by several residues that adopt an extended, nonhelical conformation. The curvature of the molecule places the N- and C-termini near one another (23 Å), despite the fact that the dimensions of the monomer are 125 × 80 × 40 Å.
Beginning at the N-terminus of apo Δ(1–43)A-I, residues 44–49 are random coil, whereas residues 50–65 form α-helix A1 (Fig. 2a; the sequence of apo A-I is shown in Fig. 3). These segments together constitute the first predicted helical repeat (17). The absence of residues 1–43 in apo Δ(1–43)A-I may cause residues 44–49 to be nonhelical. A slight kink at Pro-66 begins helix A2 (residues 66–87), which is essentially continuous with helix A3 (residues 88–98; residue 88 is a Lys). A more pronounced kink at Pro-99 initiates helix A4 (residues 99–120), a helix that both is unusually rich in aromatic residues and contains Met-112, one of two Pt ligands in the heavy-atom derivative. A second sharp kink at Pro-121 starts helix A5 (residues 121–142), and yet another kink at Pro-143 initiates helix A6 (residues 143–164), which contains the other Pt ligand, Met-148. Helix A7 (residues 165–186) also begins with a kink at Pro-165, but continues without interruption into helix A8 (residues 187–208; residue 187 is an Ala). A kink begins helix A9 (residues 209–219) at Pro-209, which is terminated by a 50° bend at Pro-220. The last 22-amino acid repeat of apo A-I, predicted to be helical, is actually composed of eight residues in an extended, nonhelical conformation (residues 220–227) followed by a 60° bend into helix A10 (residues 228–243). Regions of the monomer involved in crystal packing contacts are better-ordered than those that are not, resulting in group temperature factors that vary from 51 to 200 Å2. Residues 44–49 and 209–227 are the most mobile (although electron density is clearly present for these residues). The proline-induced kinks in apo Δ(1–43)A-I are similar to those found in other structures, except that the mean kink angle and its standard deviation (40 ± 17°; range 14–58°; Table 2) are larger than that reported (26 ± 5°; ref. 36). All of the helices except A10 are slightly curved between the kinks.
Figure 3.
Amino acid sequence of apo A-I. The secondary structural elements are indicated below the sequence, as is the N-terminal region that is not present in apo Δ(1–43)A-I. Proline residues are shown in italics. Residues in helices 2–7 located at positions a and d of the modified heptad repeat are shown in bold.
Table 2.
Interhelical angles
| Helix Junction | Angle, °
|
|
|---|---|---|
| Molecule A | Molecule B | |
| 1–2 | 15 | 14 |
| 2–3 | 19 | 8 |
| 3–4 | 47 | 51 |
| 4–5 | 47 | 57 |
| 5–6 | 58 | 42 |
| 6–7 | 48 | 54 |
| 7–8 | 12 | 18 |
| 8–9 | 17 | 29 |
Several facts demonstrate that the entire chain is traced correctly and that the direction and registration are also correct. (i) The experimental electron density map shows continuous density for the entire main chain. At no point were the connections between secondary structural elements ambiguous (in part a consequence of the linear nature of the molecule). (ii) Apo Δ(1–43)A-I contains only four methionines, at positions −1, 86, 112, and 148. Given this asymmetric distribution, it is not possible to reverse the directionality of our model and still keep methionines at the observed Pt binding sites. (iii) Helix 4 has four aromatic residues and a methionine, each of which has strong side chain electron density (Fig. 1). If our model were traced backwards, the sequence of helix 7, which has only one aromatic residue and no methionines, would occupy that region of electron density instead. (iv) Our model places all seven proline residues at kinks in the electron density; reversal of the polarity of the chain destroys this correspondence. The presence of prolines at the kinks in the electron density is also a sensitive indicator of the lack of out-of-register errors. (v) Essentially all of the aromatic residues, histidines, and methionines have strong side chain electron density. Many other residues (especially leucines) also have good side chain density. These five points are satisfied simultaneously for the two independent molecules in the asymmetric unit, and our structure results in chemically sensible packing of both noncrystallographically and crystallographically related molecules. Taken together, these facts indicate that our model is traced correctly. We expect that the overall structure and the relative alignment of the molecules with respect to one another will not be altered substantially as higher resolution data become available.
A helical hairpin model, in which the apo Δ(1–43)A-I monomer is folded back on itself in the middle of helix 5, would also be consistent with points ii–v noted above. This formal possibility is definitely excluded, however, by the unambiguous, unbroken main-chain electron density for helix 5 (i.e., helix 5 does not contain a reverse turn). No other helical hairpin models are consistent with the experimental electron density.
Dimer Structure.
Molecule A of apo Δ(1–43)A-I described above associates tightly with a similarly shaped molecule B to form an antiparallel dimer (Fig. 2b). The only part of a monomer not paired with its mate is residues 220–227. Molecules A and B are related by the A/B noncrystallographic pseudo-dyad (179.8°), which is parallel to the long axis of the horseshoe and runs between helices A5 and B5 at one end, and helices A10 and B10 at the other. Two consequences of this architecture are that the N terminus of one monomer is near residue 220 of its mate in the dimer, and that the C-terminal helices A10 and B10 are adjacent and antiparallel. Thus, the dimer is a closed elliptical ring, even though the monomer is an open horseshoe. This antiparallel arrangement causes specific regions of molecules A and B to be paired with one another (Fig. 2b): helices A10 and B10 are paired; A5/B5; A4/B6 and A6/B4; A3/B7 and A7/B3; A2/B7–B8 and A7–A8/B2; A1/B8–B9 and A8–A9/B1.
Alignment of molecules A and B by the A/B pseudo-dyad results in an rms deviation of 5.0 Å for 201 Cα atoms. When the corresponding helices from A and B are aligned individually, however, the rms deviations range from 0.5 to 1.3 Å. Thus, molecules A and B differ mainly in the angles of the proline-induced kinks between helices (Table 2).
Formation of the apo Δ(1–43)A-I dimer results in the loss of 4,120 Å2 of solvent-accessible surface area for each monomer, for a total buried area of 8,240 Å2 (21% of the total area of A and B). One-half of this buried area is due to the association of hydrophobic residues, and 30% is due to the association of charged residues. One face of the A/B dimer is predominantly hydrophilic; the monomers possess electrostatic complementarity, as evidenced by many inter-monomer Arg/Lys–Asp/Glu salt bridges. The other face of the dimer is strongly hydrophobic.
In addition to the tight, antiparallel A/B dimer described above, two other dimers are also present in the apo Δ(1–43)A-I crystal structure: the loose, parallel A/C dimer and the tight, antiparallel A/D dimer. Formation of the A/C and A/D dimers buries 1,600 and 6,980 Å2 of surface area, respectively. Both the A/B and the A/D dimers, but not the A/C dimer, possess a contiguous hydrophobic surface suitable for binding to lipid. It is notable that the A/B dimer can open from a ring to a horseshoe simply by breaking the short interaction between helices A10 and B10, whereas the A/D dimer can open only by unzipping one-half of the extensive A/D interface. The possible biological relevance of a facile conversion between topologically closed and topologically open forms of the A/B dimer is discussed below.
Tetramer Structure.
Two apo Δ(1–43)A-I dimers associate in an antiparallel fashion to form an elliptical tetrameric ring (Figs. 2c and 4), with outer dimensions of 135 × 90 Å, and an inner hole of 95 × 50 Å. The A/B and C/D dimers are related by the A/C noncrystallographic dyad that runs through the center of the ring (Fig. 4 a and b). The ring gently curves first one way then the other (Fig. 4 c and d); the thickness of the tetramer perpendicular to the axes of the ellipse is 60 Å. The tetramer consists of an up-down-up-down antiparallel four-helix bundle over about three-fourths of its circumference. The N and C termini of each molecule lie on the inside of the ring, whereas their mid-sections lie on the outside. Molecule A is related to molecule D (and B to C) by the A/D noncrystallographic pseudo-dyad (178.5°), which defines the short axis of the ellipse and passes between helices A2 and D8 on one side of the ring, and helices A8 and D2 on the other. The three noncrystallographic (pseudo)dyads intersect at the center of the ring (within 1 Å) and are mutually perpendicular (within 1°), giving the tetramer 222-pseudosymmetry (Fig. 4). This pseudosymmetry places the C-terminal helices A10/B10 of the A/B dimer next to the mid-point of the C/D dimer (helices C5/D5), at a crossing angle of 51°.
Formation of the tetramer buries 8,480 Å2 of surface area from each dimer, an amount comparable to the total area lost upon association of the monomers to form the A/B dimer. A total of 33,170 Å2 of surface area (42% of the area of the separated monomers) is lost upon tetramer formation. The exterior of the tetramer is hydrophilic, with a uniform electrostatic potential. The interior is composed mostly of leucine and valine side chains, which are sequestered in the core of the four-helix bundle that makes up most of the tetramer, and thus are not available to bind lipid. For this reason, we do not believe that the tetramer observed here is relevant to the lipid-bound structure of apo A-I. Rather, it seems likely that the tetramer forms in these crystals, which lack lipid, simply to shelter the lipid-binding hydrophobic face of the dimers from exposure to water.
DISCUSSION
Our crystal structure shows that residues 44–243 of apolipoprotein A-I, which correspond to exon 4 of the gene, adopt a continuously curved, amphipathic α-helical conformation (Figs. 2 and 4). Regularly spaced prolines insert kinks in the molecule that cause it to take on a horseshoe-like shape. A previous secondary structure prediction (17) of apo A-I is in good agreement with the apo Δ(1–43)A-I crystal structure, but an earlier tertiary structure prediction is not (18).
Modified Heptad Repeats.
On the basis of sequence homology, the 11-residue amphipathic α-helical motifs of apo A-I and other apolipoproteins can be segregated into two types, A and B (17, 37). Type A helices almost invariably begin with proline, whereas type B helices do not. In both types the pattern of hydrophobic residues is similar to the canonical heptad repeat of α-helical coiled-coil proteins: in a repeating seven amino acid motif (labeled by the letters a–g) hydrophobic residues are found at positions a and d. Whereas type B helices faithfully follow the canonical heptad repeat, type A helices exhibit a modified, interrupted heptad repeat. This is illustrated with the sequence of helices 4 and 5 (ABAB) of apo A-I (the heptad repeat, with the interrupting residue c′, is shown under the sequence): ![]()
The modified heptad repeat is continuous across adjacent helices when the c′ interruption is included. Also, proline residues are found only at position c of the heptad repeat. This pattern holds true for helices 2–7 of apo A-I (Fig. 3). Canonical heptad repeats are present on either side of this region; these canonical repeats are out of phase, however, with the modified repeats of helices 2–7. This phase-shift in the amino acid sequence is reflected by a structural transition in the A/B dimer, from side-by-side curved helices to straighter helices that twist around one another. Also, the dimer crosses from the outside to the inside of the tetrameric ring at this point.
Interruption of a canonical heptad repeat by the c′ residue rotates the orientation of hydrophobic residues following c′ by 100° relative to those preceding c′. This geometric consequence of the naturally occurring apolipoprotein amino acid sequences apparently has not been noticed previously (16). We observe just this rotation in the crystal structure of apo Δ(1–43)A-I. This effect, coupled with the proline-induced kinks, causes the direction of the hydrophobic face of the pseudo-continuous amphipathic α-helix in apo Δ(1–43)A-I to rotate every 11 residues. It is likely that this regular and frequent re-orientation of the hydrophobic face of apo A-I is required for the protein to wrap around curved lipid surfaces. Consistent with this notion, the two independent molecules in the crystal structure differ mainly in the interhelical angles at the proline-induced kinks (Table 2) and in the degree of bending between the kinks, near the c′ residue of the modified heptad repeat. These joints may give apo A-I the plasticity needed to accommodate its shape to the curvatures of lipid particles of different sizes. Finally, we note that many copies of the modified heptad repeat are present in apolipoprotein A-IV (17).
Biochemical Correlations.
The proteolytic cleavage sites observed for lipid-bound apo A-I are generally consistent with the crystal structure of apo Δ(1–43)A-I (38–41). The major sites cluster at the proline kinks, the kink between helices 2 and 3, and the sharp bend that initiates helix 10. Other sites occur in the poorly ordered N-terminal region and adjacent to the Gly-Gly dipeptide at the end of helix 7. Helices 1–3 and 7–9, which have high temperature factors, appear to be no more susceptible to proteolysis than the less-mobile helices 4–6 and 10.
The antiparallel association of two molecules of apo Δ(1–43)A-I mandates that some antibodies raised against apo A-I should have discontinuous epitopes—e.g., part of an epitope from helix 2, the rest from helices 7 or 8. Unfortunately, existing monoclonal antibody data (22, 42) reveal few epitopes at all in the C-terminal half of apo A-I; thus, we cannot state whether such widely discontinuous epitopes are indeed present.
Model for HDL.
The size and shape of apo Δ(1–43)A-I are ideal for wrapping around either discoidal or spherical HDL (diameters 80–120 Å; ref. 15). Relaxation of apo A-I from the elliptical shape seen here to a circular shape, more suitable for binding to HDL, could be accomplished simply by adjusting interhelical kinks and intrahelical bends, especially those kinks (≈15° in this structure; Table 2) between helices 1–3 and 7–9. We note, however, that the maturation of discoidal HDL into spherical HDL must involve, at some point, an ellipsoidal particle. The mobility of helices 1–3 and 7–9 in the apo Δ(1–43)A-I structure, coupled with their average susceptibility to proteolysis when apo A-I is bound to lipid (41), suggests that these two regions are well ordered and probably more curved, like helices 4–6, when apo A-I is bound to lipid.
HDL particles are often composed of two or four molecules of apo A-I. Therefore, we suggest that when pairs of apo A-I bind to HDL, they do so as the antiparallel A/B dimer observed here for apo Δ(1–43)A-I. Of the two dimers observed in this crystal structure that possess a contiguous lipid-binding surface, the A/B dimer has the tightest interaction and can also open from the topologically closed ring observed here to a topologically open horseshoe simply by breaking the short interaction between helices A10 and B10. Such a conversion may facilitate the assembly of HDL particles that contain more than two molecules of apo A-I.
We suggest also that apo A-I binds to discoidal HDL in the “belt” mode—i.e., the antiparallel A/B dimer of apo Δ(1–43)A-I wraps around the edge of the disc. Larger discoidal HDL particles comprised of four molecules of apo A-I can be formed from two topologically open A/B dimers interacting with one another; the interaction between open dimers would again be mediated by helix 10. Discoidal HDL particles comprised of three molecules of apo A-I require a doubled-back monomer and an open dimer.
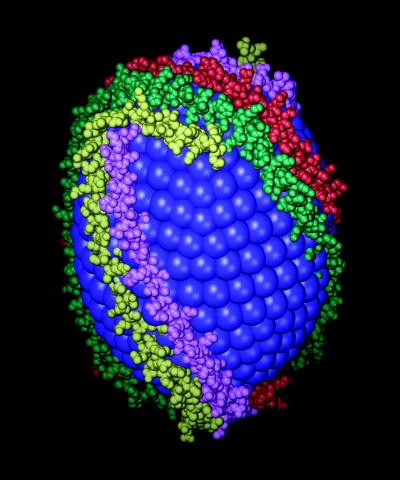
Finally, the shape and size of apo Δ(1–43)A-I suggest to us that apo A-I binds to spherical HDL in a similar manner. One possible model for spherical HDL comprised of four molecules of apo A-I is shown in Fig. 5. The HDL particle in this model, which was built with a minimum of distortion from the observed crystal structure, is slightly ellipsoidal. As noted above, ellipsoidal HDL is an obligate intermediate between discoidal and spherical HDL, and the model can be made spherical by slight adjustment of the interhelical angles.
Figure 5.
Hypothetical model of apo A-I bound to spherical HDL. Two dimers of apo Δ(1–43)A-I are shown as CPK models, colored as in Fig. 4. The lipid head groups are represented by blue balls. At the top of the model are helices A10, D5, C5, and B10 (left to right). Separation of helices A10 and B10 of the A/B dimer allows the C/D dimer to pass through the gap, over the top of the sphere.
What Is the True Lipid-Bound Conformation of Apo A-I?
Our structure of apo Δ(1–43)A-I is the first crystallographic evidence that supports any particular model (the “belt”) for how apo A-I binds to lipid. As described above, we are able to easily construct models for apo A-I bound to discoidal and spherical HDL based on the horseshoe-like shape of apo Δ(1–43)A-I. Because our structure contains no lipid, however, we must rely on the known biophysical properties of apo Δ(1–43)A-I, compared with intact apo A-I itself, in claiming that this structure of apo Δ(1–43)A-I represents the lipid-bound structure of apo A-I (21, 41). It is possible that the true lipid-bound structure is instead more akin to the “picket fence” model, or perhaps a zig-zag conformation intermediate between the belt and the picket fence models. Nevertheless, we believe that our crystal structure of apo Δ(1–43)A-I reveals, at a minimum, an unusual conformation that apo A-I can adopt, a conformation that may very well play a role in the function of apo A-I.
Our models for the structure of apo A-I bound to both discoidal and spherical HDL predict that the specific helix pairings characteristic of the A/B antiparallel dimer seen in the crystal structure of apo Δ(1–43)A-I are retained in apo A-I bound to HDL. This prediction is amenable to experimental verification by mutagenesis, crosslinking, fluorescence and spin label measurements.
Acknowledgments
We thank Margie Ray for assistance with production of apo Δ(1–43)A-I; Carolyn Cohen for suggesting PIP and encouragement; Lucile White, Tom Terwilliger, and David Rodgers for helpful discussions; and Stephen Harrison for insightful comments on the manuscript. The initial production of apo Δ(1–43)A-I was supported by National Institutes of Health Grant HL34343. Subsequent crystallization and characterization were supported by Southern Research Institute and the State of Alabama, respectively. D.W.B would like to dedicate this paper to the memory of his father, Dr. Nemat O. Borhani, whose many epidemiological studies over the past 40 years helped define and refine the causal connections between smoking, cholesterol and hypertension, and coronary artery disease and stroke.
ABBREVIATIONS
- Apo A-I
apolipoprotein A-I
- apo Δ(1–43)A-I
truncation mutant of apo A-I (residues 44–243)
- HDL
high density lipoprotein
- PIP
[Pt2I2en2](NO3)2
Note Added in Proof
X-ray diffraction data collected recently from a frozen apo Δ(1–43)A-I crystal at the Cornell High Energy Synchrotron Source (beamline A1, 100K, 30% glucose cryoprotectant) extended to 3.0-Å resolution.
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited with the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (accession no. 1av1), and are available prior to release upon e-mail request to D.W.B.
References
- 1.Miller G J, Miller N E. Lancet. 1975;i:16–19. doi: 10.1016/s0140-6736(75)92376-4. [DOI] [PubMed] [Google Scholar]
- 2.Miller N E, Førde O H, Thelle D S, Mjøs O D. Lancet. 1977;i:965–968. doi: 10.1016/s0140-6736(77)92274-7. [DOI] [PubMed] [Google Scholar]
- 3.Castelli W P, Garrison R J, Wilson P W F, Abbott R D, Kalousdian S, Kannel W B. J Am Med Assoc. 1986;256:2835–2838. [PubMed] [Google Scholar]
- 4.Miller N E, editor. High-Density Lipoproteins, Reverse Cholesterol Transport, and Coronary Heart Disease. Amsterdam: Elsevier; 1989. [Google Scholar]
- 5.Johnson W J, Mahlberg F H, Rothblat G H, Phillips M C. Biochim Biophys Acta. 1991;1085:273–298. doi: 10.1016/0005-2760(91)90132-2. [DOI] [PubMed] [Google Scholar]
- 6.Rothblat G H, Mahlberg F H, Johnson W J, Phillips M C. J Lipid Res. 1992;33:1091–1097. [PubMed] [Google Scholar]
- 7.Acton S, Rigotti A, Landschulz K T, Xu S, Hobbs H H, Krieger M. Science. 1993;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 8.Plump A S, Erickson S K, Weng W, Partin J S, Breslow J L, Williams D L. J Clin Invest. 1996;97:2660–2671. doi: 10.1172/JCI118716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozarsky K F, Donahee M H, Rigotti A, Iqbal S N, Edelman E R, Krieger M. Nature (London) 1997;387:414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 10.Morrison J R, McPherson G A, Fidge N H. J Biol Chem. 1992;267:13205–13209. [PubMed] [Google Scholar]
- 11.Schultz J R, Verstuyft J G, Gong E L, Nichols A V, Rubin E M. Nature (London) 1993;365:762–764. doi: 10.1038/365762a0. [DOI] [PubMed] [Google Scholar]
- 12.Warden C H, Hedrick C C, Qiao J-H, Castellani L W, Lusis A J. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- 13.Scanu A M. Biochim Biophys Acta. 1972;265:471–508. [Google Scholar]
- 14.Morrisett J D, Jackson R L, Gotto A M., Jr Biochim Biophys Acta. 1977;472:93–133. doi: 10.1016/0304-4157(77)90015-6. [DOI] [PubMed] [Google Scholar]
- 15.Brouillette C G, Anantharamaiah G M. Biochim Biophys Acta. 1995;1256:103–129. doi: 10.1016/0005-2760(95)00018-8. [DOI] [PubMed] [Google Scholar]
- 16.Segrest J P, Jones M K, De Loof H, Brouillette C G, Venkatachalapathi Y V, Anantharamaiah G M. J Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 17.Nolte R T, Atkinson D. Biophys J. 1992;63:1221–1239. doi: 10.1016/S0006-3495(92)81698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown E M, Kumosinski T F, Farrell H M., Jr ACS Symp Ser. 1994;576:100–112. [Google Scholar]
- 19.Atkinson D, Davis M A F, Leslie R B. Proc R Soc London B. 1974;186:165–180. doi: 10.1098/rspb.1974.0044. [DOI] [PubMed] [Google Scholar]
- 20.Brasseur R, De Meutter J, Vanloo B, Goormaghtigh E, Ruysschaert J M, Rosseneu M. Biochim Biophys Acta. 1990;1043:245–252. doi: 10.1016/0005-2760(90)90023-q. [DOI] [PubMed] [Google Scholar]
- 21.Rogers D P, Brouillette C G, Engler J A, Tendian S W, Roberts L, Mishra V K, Anantharamaiah G M, Lund-Katz S, Phillips M C, Ray M J. Biochemistry. 1997;36:288–300. doi: 10.1021/bi961876e. [DOI] [PubMed] [Google Scholar]
- 22.Bergeron J, Frank P G, Scales D, Meng Q-H, Castro G, Marcel Y L. J Biol Chem. 1995;270:27429–27438. doi: 10.1074/jbc.270.46.27429. [DOI] [PubMed] [Google Scholar]
- 23.Colman P M, Matthews B W. J Mol Biol. 1971;60:163–168. doi: 10.1016/0022-2836(71)90455-4. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 25.Tong L, Rossmann M G. Acta Crystallogr A. 1990;46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T C, Kim S-H, Eisenberg D. Acta Crystallogr A. 1987;43:1–5. [Google Scholar]
- 27.Terwilliger T C, Eisenberg D. Acta Crystallogr A. 1983;39:813–817. [Google Scholar]
- 28.Jones T A, Zou J Y, Cowan S W, Kjelgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Stehle T, Gamblin S J, Yan Y, Harrison S C. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 30.Brünger A T. x-plor: A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 31.Golmohammadi R, Fridborg K, Bundule M, Valegård K, Liljas L. Structure. 1996;4:543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 32.Kleywegt G J, Jones T A. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt G J, Jones T A. In: Making the Most of Your Model. Hunter W N, Thornton J M, Bailey S, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1995. pp. 11–24. [Google Scholar]
- 34.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 35.Nicholls A, Sharp K, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 36.Barlow D J, Thornton J M. J Mol Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 37.Li W-H, Tanimura M, Lou C-C, Datta S, Chan L. J Lipid Res. 1988;29:245–271. [PubMed] [Google Scholar]
- 38.Dalton M B, Swaney J B. J Biol Chem. 1993;268:19274–19283. [PubMed] [Google Scholar]
- 39.Lins L, Piron S, Conrath K, Vanloo B, Brasseur R, Rosseneu M, Baert J, Ruysschaert J M. Biochim Biophys Acta. 1993;1151:137–142. doi: 10.1016/0005-2736(93)90096-i. [DOI] [PubMed] [Google Scholar]
- 40.Ji Y, Jonas A. J Biol Chem. 1995;270:11290–11297. doi: 10.1074/jbc.270.19.11290. [DOI] [PubMed] [Google Scholar]
- 41.Roberts L M, Ray M J, Shih T-W, Hayden E, Reader M M, Brouillette C G. Biochemistry. 1997;36:7615–7624. doi: 10.1021/bi962952g. [DOI] [PubMed] [Google Scholar]
- 42.Marcel Y L, Provost P R, Koa H, Raffai E, Dac N V, Fruchart J-C, Rassart E. J Biol Chem. 1991;266:3644–3653. [PubMed] [Google Scholar]