Abstract
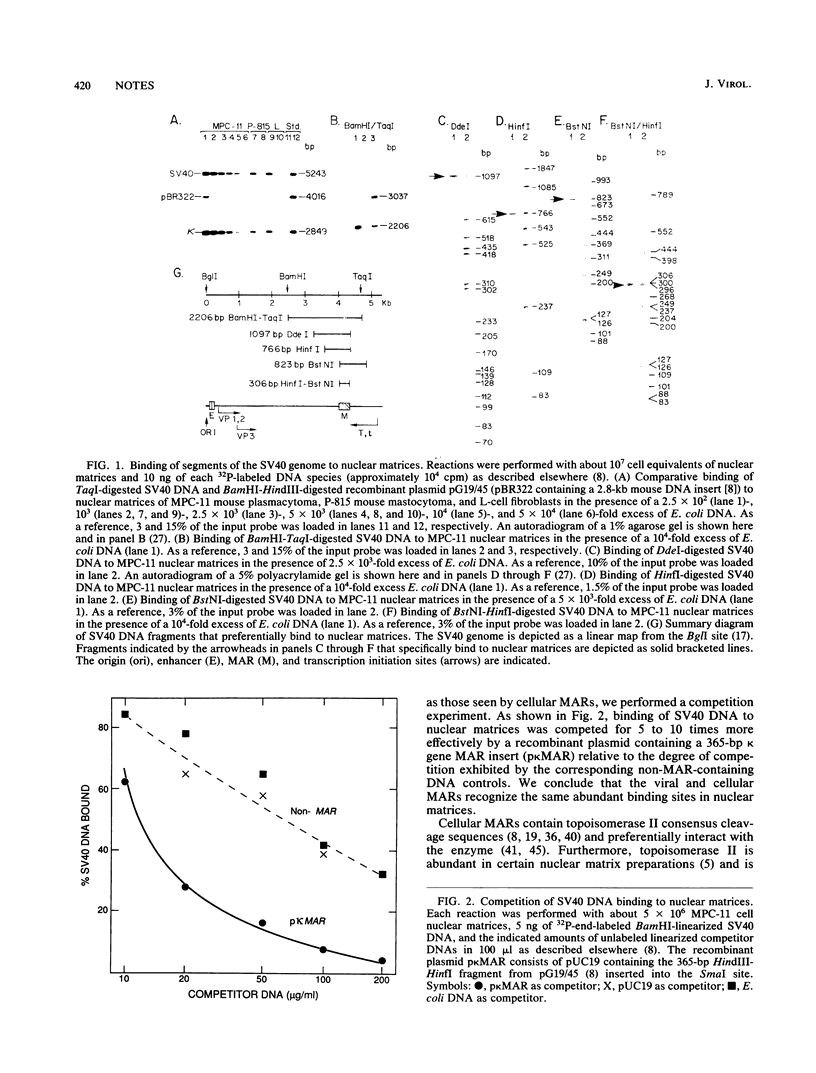
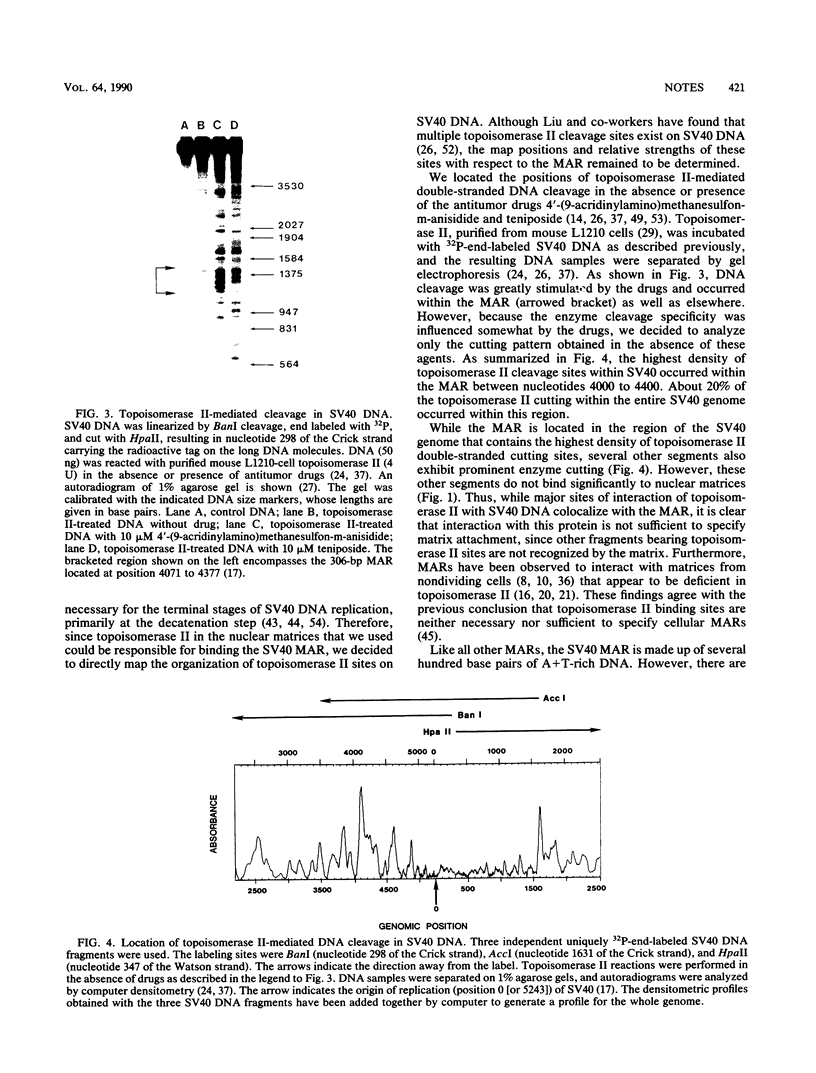
We demonstrate that the simian virus 40 genome contains a single MAR (matrix association region) that maps within a large T-antigen coding region (nucleotides 4071 to 4377). This region contains topoisomerase II cleavage sites, exhibits sequence similarity with cellular MARs, and recognizes the same evolutionarily conserved, abundant nuclear binding sites seen by cellular MARs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Berrios M., Osheroff N., Fisher P. A. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Maass K. Chromatin domain surrounding the human interferon-beta gene as defined by scaffold-attached regions. Biochemistry. 1988 Jun 28;27(13):4706–4711. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- Chen M., Lebowitz J., Salzman N. P. Hin D restriction mapping of upaired regions in simian virus 40 superhelical DNA I: considerations regarding structure-function relationships. J Virol. 1976 Apr;18(1):211–217. doi: 10.1128/jvi.18.1.211-217.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986 Aug 11;204(1):5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Yuen M. H., Garrard W. T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987 Apr 15;262(11):5394–5397. [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Conformational constraints in nuclear DNA. J Cell Sci. 1976 Nov;22(2):287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Covey L., Choi Y., Prives C. Association of simian virus 40 T antigen with the nuclear matrix of infected and transformed monkey cells. Mol Cell Biol. 1984 Jul;4(7):1384–1392. doi: 10.1128/mcb.4.7.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby M. K., Herrera R. E., Vosberg H. P., Nordheim A. DNA topoisomerase II cleaves at specific sites in the 5' flanking region of c-fos proto-oncogenes in vitro. EMBO J. 1986 Sep;5(9):2257–2265. doi: 10.1002/j.1460-2075.1986.tb04493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman R., Brutlag D. L. Expression of the Drosophila type II topoisomerase is developmentally regulated. Biochemistry. 1988 Jan 26;27(2):560–565. doi: 10.1021/bi00402a009. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M. M., Earnshaw W. C. Topoisomerase II: A specific marker for cell proliferation. J Cell Biol. 1986 Dec;103(6 Pt 2):2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M. M., Hittelman W. N., Earnshaw W. C. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Mirkovitch J., Laemmli U. K. Interaction of DNA with nuclear scaffolds in vitro. J Mol Biol. 1988 Mar 5;200(1):111–125. doi: 10.1016/0022-2836(88)90337-3. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaxel C., Kohn K. W., Pommier Y. Topoisomerase I interaction with SV40 DNA in the presence and absence of camptothecin. Nucleic Acids Res. 1988 Dec 9;16(23):11157–11170. doi: 10.1093/nar/16.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Chasin L. A. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J Mol Biol. 1987 Dec 20;198(4):677–692. doi: 10.1016/0022-2836(87)90209-9. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Rowe T. C., Yang L., Tewey K. M., Chen G. L. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem. 1983 Dec 25;258(24):15365–15370. [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Minford J., Pommier Y., Filipski J., Kohn K. W., Kerrigan D., Mattern M., Michaels S., Schwartz R., Zwelling L. A. Isolation of intercalator-dependent protein-linked DNA strand cleavage activity from cell nuclei and identification as topoisomerase II. Biochemistry. 1986 Jan 14;25(1):9–16. doi: 10.1021/bi00349a002. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S. M., Laemmli U. K. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988 Mar 5;200(1):101–109. doi: 10.1016/0022-2836(88)90336-1. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Spierer P., Laemmli U. K. Genes and loops in 320,000 base-pairs of the Drosophila melanogaster chromosome. J Mol Biol. 1986 Jul 20;190(2):255–258. doi: 10.1016/0022-2836(86)90296-2. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Location of the T4 gene 32 protein binding site on simian virus 40 DNA. J Virol. 1973 Dec;12(6):1631–1632. doi: 10.1128/jvi.12.6.1631-1632.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelkin B. D., Pardoll D. M., Vogelstein B. Localization of SV40 genes within supercoiled loop domains. Nucleic Acids Res. 1980 Dec 11;8(23):5623–5633. doi: 10.1093/nar/8.23.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Kerrigan D., Kohn K. Topological complexes between DNA and topoisomerase II and effects of polyamines. Biochemistry. 1989 Feb 7;28(3):995–1002. doi: 10.1021/bi00429a012. [DOI] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T., Udvardy A., Schedl P. Sequence dependence of Drosophila topoisomerase II in plasmid relaxation and DNA binding. J Mol Biol. 1987 Mar 20;194(2):219–229. doi: 10.1016/0022-2836(87)90370-6. [DOI] [PubMed] [Google Scholar]
- Shakhov A. N., Nedospasov S. A., Georgiev G. P. Deoxyribonuclease II as a probe to sequence-specific chromatin organization: preferential cleavage in the 72 bp modulator sequence of SV40 minichromosome. Nucleic Acids Res. 1982 Jul 10;10(13):3951–3965. doi: 10.1093/nar/10.13.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R. M., Powelson M. A., Strayer J. M. Swiveling and decatenation of replicating simian virus 40 genomes in vivo. Mol Cell Biol. 1988 Feb;8(2):515–521. doi: 10.1128/mcb.8.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R. M. Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol Cell Biol. 1986 Dec;6(12):4221–4227. doi: 10.1128/mcb.6.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry A. O., Blasquez V. C., Garrard W. T. Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5497–5501. doi: 10.1073/pnas.86.14.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staufenbiel M., Deppert W. Different structural systems of the nucleus are targets for SV40 large T antigen. Cell. 1983 May;33(1):173–181. doi: 10.1016/0092-8674(83)90346-x. [DOI] [PubMed] [Google Scholar]
- Sykes R. C., Lin D., Hwang S. J., Framson P. E., Chinault A. C. Yeast ARS function and nuclear matrix association coincide in a short sequence from the human HPRT locus. Mol Gen Genet. 1988 May;212(2):301–309. doi: 10.1007/BF00334700. [DOI] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Topoisomerase II cleavage in chromatin. J Mol Biol. 1986 Sep 20;191(2):231–246. doi: 10.1016/0022-2836(86)90260-3. [DOI] [PubMed] [Google Scholar]
- Verderame M. F., Kohtz D. S., Pollack R. E. 94,000- and 100,000-molecular-weight simian virus 40 T-antigens are associated with the nuclear matrix in transformed and revertant mouse cells. J Virol. 1983 May;46(2):575–583. doi: 10.1128/jvi.46.2.575-583.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Yang L., Rowe T. C., Liu L. F. Identification of DNA topoisomerase II as an intracellular target of antitumor epipodophyllotoxins in simian virus 40-infected monkey cells. Cancer Res. 1985 Nov;45(11 Pt 2):5872–5876. [PubMed] [Google Scholar]
- Yang L., Rowe T. C., Nelson E. M., Liu L. F. In vivo mapping of DNA topoisomerase II-specific cleavage sites on SV40 chromatin. Cell. 1985 May;41(1):127–132. doi: 10.1016/0092-8674(85)90067-4. [DOI] [PubMed] [Google Scholar]
- Yang L., Wold M. S., Li J. J., Kelly T. J., Liu L. F. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1987 Feb;84(4):950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]