Abstract
Covalent fusions between an mRNA and the peptide or protein that it encodes can be generated by in vitro translation of synthetic mRNAs that carry puromycin, a peptidyl acceptor antibiotic, at their 3′ end. The stable linkage between the informational (nucleic acid) and functional (peptide) domains of the resulting joint molecules allows a specific mRNA to be enriched from a complex mixture of mRNAs based on the properties of its encoded peptide. Fusions between a synthetic mRNA and its encoded myc epitope peptide have been enriched from a pool of random sequence mRNA-peptide fusions by immunoprecipitation. Covalent RNA-peptide fusions should provide an additional route to the in vitro selection and directed evolution of proteins.
In vitro selection experiments using RNA and DNA have shown that nucleic acid molecules with specific molecular recognition and catalytic properties can be isolated from complex pools of random sequences by repeated rounds of selection and amplification (for reviews see refs. 1–5). Directed in vitro evolution, in which mutagenic amplification is combined with continued selective pressure, has been widely used to select for nucleic acids with altered or improved binding or catalytic properties (e.g., refs. 6 and 7). Because proteins carry out a wider range of structural and catalytic roles in biology and are much more extensively used in diagnostic, therapeutic, and industrial applications, great interest has been generated in the development of methods for the in vitro selection and directed evolution of proteins. The main barrier to the development of effective methods for protein evolution has been the difficulty of recovering the information encoding a protein sequence after the protein has been translated. Until recently, most approaches to this problem have involved a step in which the DNA is transcribed and translated in vivo, and the resulting protein is expressed in such a way as to remain physically linked to the encoding nucleic acid, which then is recovered for amplification and further selection. Examples of such approaches include phage display (ref. 8 and recently reviewed in refs. 9 and 10), plasmid display (11), and completely in vivo genetic approaches (12–14). In general, such approaches are limited by the complexity of the sequence libraries that can be generated; for example, phage display libraries typically are limited by transfection efficiency to less than 109 independent members.
More recently, selection schemes based on the display of the nascent peptide chain on the surface of the ribosome have been developed (15–17). This approach has the advantage of being fully in vitro and potentially allowing larger libraries (>1012) to be explored; however, selections must be performed under conditions that preserve the integrity of the ribosome⋅mRNA⋅peptide ternary complex.
We sought to develop a simpler and more robust system in which an mRNA would become directly attached to the peptide or protein it encodes by a stable covalent linkage. We reasoned that because both the message and the adapters in protein synthesis are nucleic acids, it might be possible to generate an RNA that could act as both, i.e., a chimeric message-adapter (Fig. 1A). When translated, the adapter end would enter the ribosome and accept the nascent peptide (Fig. 1B) to generate a covalent mRNA-peptide fusion (Fig. 1C). In this way, the sequence information present in the peptide would be encoded in the covalently attached mRNA. This joint molecule then could act as a molecular Rosetta stone, allowing the unreadable information in the protein portion to be read and recovered via the attached mRNA.
Figure 1.
Proposed mechanism for RNA-peptide fusion formation on the ribosome. (A) The ribosome initiates protein synthesis on the mRNA and translocates toward the end of the template. (B) When the ribosome reaches the end of the RNA ORF, translation stalls at the RNA/DNA junction. The puromycin then enters the A site of the ribosome and accepts the nascent peptide. (C) Once formed the mRNA-peptide fusion is purified from the translation reaction by affinity chromatography.
The use of an mRNA with a charged tRNA linked to its 3′ end could, in principle, lead to the synthesis of the desired mRNA-peptide fusion molecules during translation; however, the resulting linkage would be an unstable aminoacyl ester. Puromycin, an antibiotic that mimics the aminoacyl end of tRNA, acts as a translation inhibitor by entering the ribosomal A site and accepting the nascent peptide as a result of the peptidyl transferase activity of the ribosome (18, 19). The resulting peptidyl-puromycin molecule contains a stable amide linkage between the peptide and the O-methyl tyrosine portion of the puromycin. The O-methyl tyrosine is, in turn, linked by a stable amide bond to the 3′-amino group of the modified adenosine portion of puromycin. Thus a synthetic mRNA with puromycin at its 3′ end would be expected to generate stable mRNA-peptide fusions.
MATERIALS AND METHODS
Synthesis of Controlled Pore Glass (CPG)-Puromycin.
Puromycin(HCl)2 was converted to the free base by dissolution in H2O, mixing with basic carbonate buffer, and extraction into chloroform. N-trifluoroacetyl puromycin was made by mixing the dried free base in 50/50 (vol/vol) dry pyridine (Fluka)/acetonitrile (Millipore) with an excess of trifluoroacetic anhydride (Fluka) for 1 hr at 25°C followed by workup with dilute ammonium hydroxide. N-trifluoroacetyl 5′-dimethoxytrityl (DMT) puromycin was made using DMT-Cl (Sigma) (20), and attached to an aminohexyl CPG (Sigma) support via the 2′OH using the standard protocol for attachment of DNA through its 3′OH (21) with the exception that the coupling step was carried out in the presence of ≈50 μmol activated puromycin per gram of CPG.
Synthesis of 3′ Puromycin Oligonucleotides and Templates.
CPG-puromycin was used as a solid support for automated synthesis of 30-P (5′-dA27dCdCP) and 43-P (the RNA sequence 5′-GGAGGACGAAAUG linked to 30-P) according to standard protocols for DNA and RNA synthesis (Millipore). 30-P and 43-P were deprotected in concentrated NH4OH (+ 25% vol/vol ETOH for 43-P) for 12 hr at 55°C and dried. 43-P was further deprotected in 1 M tetra butyl ammonium fluoride in tetrahydrofuran (Sigma) for 48 hr and desalted on a NAP-25 column (Pharmacia) before gel purification. The RNA portion of LP77 (5′-GGGAGGACGAAAUGGAACAGAAACUGAUCUCUGAAGAAGACCUGAAC) was produced by T7 transcription of partially single-stranded oligonucleotides, whereas the RNA portions of LP154 (5′-GGGACAAUUACUAUUUACAAUUACAAUGGCUGAAGAACAGAAACUGAUCUCUGAAGAAGACCUGCUGCGUAAACGUCGUGAACAGCUGAAACACAAACUGGAACAGCUGCGUAACUCUUGCGCU) and LP160 (5′-GGGACAAUUACUAUUUACAAUUACAAUG(NNS)27CAGCUGCGUAACUCUUGCGCU) were produced by transcription of dsDNA prepared by PCR (22). RNA samples were isolated on denaturing urea PAGE, electroeluted using an Elutrap (Schleicher & Schuell), and desalted on NAP-25 columns (Pharmacia). 30-P was 5′ phosphorylated by using polynucleotide kinase (New England Biolabs) in the presence of T4 DNA ligase buffer. LP77, LP154, and LP160 were produced by template-directed ligation of the RNA coding regions to 5′ phosphorylated 30-P (23) gel purified, electroeluted, and desalted as before.
Translation and Isolation of Templates.
Translations were performed in reticulocyte lysate (Novagen) according to the manufacturer’s specifications in 25 μl total volume. dT25 isolation of translated templates was performed by dilution of the lysate into isolation buffer (1.0 M NaCl/0.1 M Tris⋅Cl, pH 8.2/10 mM EDTA/1 mM DTT) containing >10-fold molar excess of dT25-biotin [synthesized on bioteg phosphoramidite columns (Glen Research, Sterling, Va)]-streptavidin agarose (Pierce) whose dT25 concentration was ≈10 μM (50/50 vol/vol slurry). The solid support then was washed with cold isolation buffer and isolated by centrifugation or filtration (Millipore ultrafree MC filters) followed by elution of the templates with 50- to 100-μl aliquots of 15 mM NaOH/1 mM EDTA. The base present in the eluent was quenched immediately after elution.
Isolation of translated templates based on disulfide bond formation was performed with thiopropyl Sepharose 6B (Pharmacia). Lysate was mixed 1:10 (vol/vol) with a 50/50 (vol/vol) slurry of thiopropyl Sepharose 6B in 1× TE, pH 8.2 (10 mM Tris⋅Cl/1 mM EDTA, pH 8.2), containing DNase-free RNase A (Boehringer Mannheim), and rotated for 1 hr at 4°C. The excess liquid was removed, and the mRNA-peptide eluted with two washes of isolation buffer containing 20 mM DTT. dT25- streptavidin agarose was added to this eluate and rotated for 1 hr at 4°C. The agarose was rinsed three times with cold isolation buffer and isolated via filtration, and the bound material eluted as above. Carrier tRNA was added, and the fusion product was ethanol-precipitated. The sample was resuspended in TE, pH 8.2 containing DNase-free RNase A to remove the RNA portion of the template.
Immunoprecipitation of Fusions.
Translation reactions were mixed with dilution buffer (10 mM Tris⋅Cl, pH 8.2/140 mM NaCl/1% vol/vol Triton X-100), protein G Sepharose (Sigma), and 1 μg anti-c-myc mAb 9E10 (Calbiochem) (24) and rotated for several hours at 4°C. The eluent was removed, the conjugate washed with dilution buffer, and DNase-free RNase A was added. The samples then were labeled with polynucleotide kinase by using [γ32P]ATP and separated by denaturing urea PAGE.
Selective Enrichment.
Translation reactions were performed on four mixtures of the myc template (LP154) and the pool template (LP160) with 1:0, 1:20, 1:200 and 1:2,000 mol ratios. The templates were isolated and eluted from dT25 agarose as described above to give 400 μl total volume. Forty microliters of slurry of thiopropyl Sepharose was added and rotated for 1 hr at 4°C. The solid support was washed three times with 1 ml 1× TE, pH 8.2 (10 mM Tris⋅Cl/1 mM EDTA, pH 8.2). One microliter of 1 M DTT was added to the solid (20–30 μl total volume), incubated for several hours, and washed four times with 20 μl H2O. Each sample was reverse-transcribed with Superscript reverse-transcriptase (United States Biochemical) for 1 hr (total volume ≈160 μl), and 10 μl of this “unselected” material then was reserved for PCR. The rest of the reaction was mixed with 1 ml of dilution buffer and 20 μl of Protein G/A conjugate (Calbiochem) and rotated for 1 hr at 4°C to preclear the sample. The eluent was isolated and mixed with 2 μg 9E10 antibody and 20 μl of conjugate and rotated for 2 hr at 4°C. The immunoprecipitate was isolated by centrifugation and washed three times with ice-cold dilution buffer and once with 10 mM Tris⋅Cl/100 mM NaCl. The selected material then was eluted with three washes of 4% HOAc and lyophilized to dryness.
PCR and Quantitation.
Concentrated NH4OH was added to the selected and unselected samples and incubated for 5 min at 55°, 70°, and 90°C followed by evaporation to dryness. Two hundred microliters of PCR reaction mixture containing 1× PCR buffer plus Mg2+ (Boehringer Mannheim); 1 μM of the 3′ primer 21.103 (5′-AGCGCAAGAGTTACGCAGCTG) and 5′ primer 42.108 (5′-TAATACGACTCACTATAGGGACAATTACTATTTACAATTACA); 200 μM of each dNTP; and 2 μl Taq polymerase was added. Sixteen cycles of PCR were performed on the unselected myc sequence, and 19 cycles were performed on all other selected and unselected samples. Aliquots of each sample then were added to PCR reaction mixtures as above containing 5′ 32P-labeled 21.103 primer, amplified with 4–7 cycles of PCR, and purified twice by using Wizard direct PCR purification kits (Promega). The resulting DNA was digested with AlwNI and assayed by 12% denaturing urea PAGE followed by quantitation by PhosphorImager (Molecular Dynamics).
RESULTS
Synthesis of mRNA-Peptide Fusions.
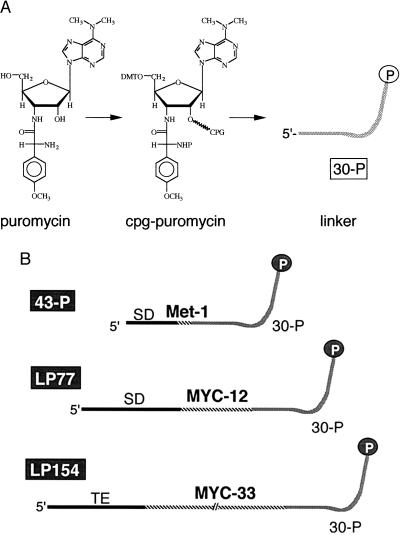
We began with the synthesis of an appropriately protected form of puromycin that could be coupled to a CPG solid support. The CPG-puromycin was used for the synthesis of an oligodeoxynucleotide linker (dA27dCdCP) with a 3′-terminal puromycin (Fig. 2A). We then ligated this linker to the 3′ end of three synthetic mRNA templates (Fig. 2B). The templates contained ORFs encoding one (the Met template, “43-P”), 12 (the short myc template, “LP77”), and 33 (the long myc template, “LP154”) amino acids. The ORF of each mRNA was joined directly to the dA27 portion of the linker, with the expectation that the DNA linker would act as a pause site for the ribosome, giving the 3′ puromycin time to enter the A site before dissociation of the nascent peptide chain. The dA27 sequence was chosen because poly dA is known to be a very poor template for polypeptide elongation on the ribosome (25). The Met and short myc templates begin with a Shine–Dalgarno ribosome binding sequence, whereas the long myc sequence contains a portion of the tobacco mosaic virus 5′ untranslated region sequence (26) with good initiation codon context (27) for efficient initiation of translation.
Figure 2.
Synthesis of translation templates containing 3′ puromycin. (A) Puromycin⋅(HCl)2 was protected and attached to a synthesis support to give CPG-puromycin, and the linker 30-P (dA27dCdCP) was made by automated synthesis (see Materials and Methods). (B) Translation templates used were: 43-P (Met template), LP77 (short myc template), and LP154 (long myc template) containing ORFs of 1, 12, and 33 codons, respectively, and no stop codon. The 5′ untranslated regions (UTRs) of 43-P and LP77 contain a Shine–Dalgarno sequence complementary to five bases of 16S rRNA (33) and spaced similarly to ribosomal protein sequences (34). The 5′ UTR of LP154 contains a 22-nt sequence derived from the tobacco mosaic virus 5′ UTR encompassing two ACAAAUUAC direct repeats (26). LP154 codes for the peptide used to raise the mAb 9E10 and LP77 codes for the recognition epitope EQKLISEEDL (24).
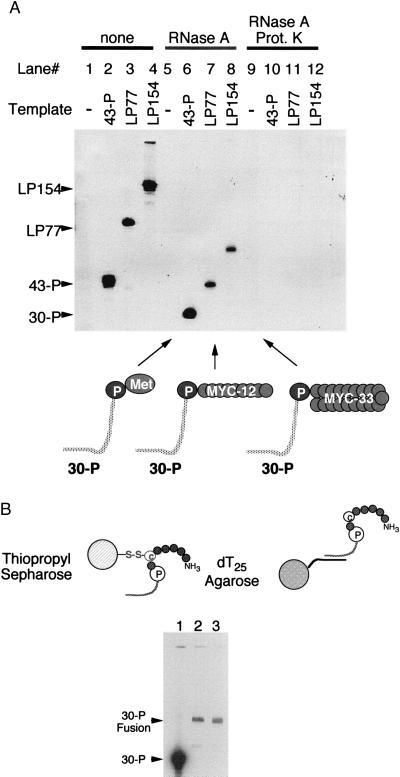
All three templates lead to the synthesis of RNA-peptide joint molecules when translated in vitro in a rabbit reticulocyte extract, as demonstrated by the incorporation of [35S]methionine into the template (Fig. 3A, lanes 1–4). Template labeling also occurs in bacterial (Met template) and wheat germ (all three templates) translation extracts and does not occur when the linker alone is used (data not shown). To investigate the nature of the attached peptide, we digested the isolated templates with RNase A to remove the variable RNA region and generate linker-peptide molecules differing only in the peptide moiety. The product of the one codon template migrated with the same mobility as the linker alone, consistent with the notion that a single amino acid (methionine) is attached to the 3′ end (Fig. 3A, lane 6). The linker fusions isolated from the short and long myc templates (Fig. 3A, lanes 7 and 8) each have significantly reduced mobility relative to 30-P, consistent with the notion that longer peptides are attached in each case. Digestion of the templates with Proteinase K and RNase A (Fig. 3A, lanes 9–12) removes all the 35S label, further supporting the notion that the decreased mobility is due to an attached peptide. The linker-peptide fusion molecules appear to be quite homogeneous by denaturing PAGE, suggesting that in most cases the ribosome translates to the end of the ORF and pauses, followed by transfer of the nascent chain to the 3′-terminal puromycin of the mRNA.
Figure 3.
Incorporation of [35S]Met into the Met (43-P), short myc (LP77), and long myc (LP154) translation templates. (A) Denaturing PAGE analysis of templates isolated from translation reactions. The gel shows templates isolated by dT25 affinity chromatography with no treatment (lanes 1–4), treatment with RNase A (lanes 5–8), and treatment with both RNase A and Proteinase K (lanes 9–12). The positions of unmodified 30-P, 43-P, LP77, and LP154 are indicated by arrows at the side of the gel. The schematic below indicates the proposed composition of the fragments in the RNase-treated lanes. (B) Sequential isolation of long myc linker fusion (30-P fusion) fragments by thiopropyl (TP) Sepharose and dT25 affinity chromatography. Lane 2 is fusion product purified on TP Sepharose followed by dT25, whereas lane 3 is material purified by dT25 only.
To confirm that the template labeling by [35S]Met was a consequence of translation, and more specifically resulted from the peptidyl transferase activity of the ribosome, we examined the effect of various inhibitors on the labeling reaction. The specific inhibitors of the eukaryotic peptidyl transferase anisomycin, gougerotin, and sparsomycin, (28) as well as the translocation inhibitors cycloheximide and emetine (28) all decrease RNA-peptide fusion formation by ≈95% with the long myc template in reticulocyte lysate translation extract. In addition, the specific inhibitors of the bacterial peptidyl transferase activity virginiamycin, gougerotin, and chloramphenicol (19) block 35S labeling of the Met template in an Escherichia coli in vitro translation reaction (data not shown).
Purification and Characterization of RNA-Peptide Fusions.
Purification of RNA-peptide joint molecules would be required for efficient selection experiments to avoid interference from mRNAs lacking an attached peptide and from free peptide. The joint molecules can be purified in a two-step procedure, by using oligonucleotide affinity chromatography to purify all RNAs that carry the linker sequence and activated thiopropyl Sepharose to purify all molecules that carry free sulfhydryl groups. The long myc-linker fusion has been purified by both oligonucleotide affinity and disulfide bond covalent affinity chromatography (Fig. 3B). Isolation of the fusion by disulfide bond formation provides independent confirmation that the ribosome is reading essentially to the end of the ORF on the message, because the cysteine is the penultimate residue of the peptide.
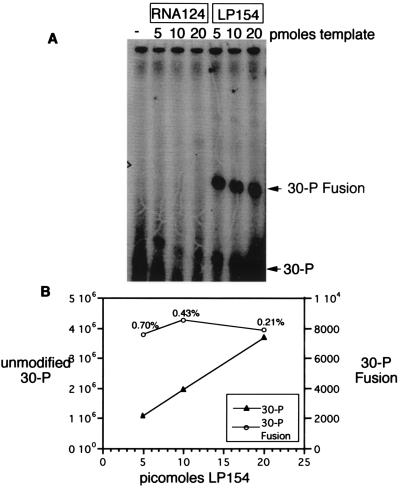
Further evidence for attachment of the desired peptide to the mRNA was obtained by immunoprecipitation of the long myc template fusion using a mAb that recognizes the myc epitope (Fig. 4A) (24). In control experiments, this antibody efficiently immunoprecipitates free peptide made by translation of the RNA portion of the long myc template (data not shown). When the long myc template (LP154) is translated, an RNA-peptide product is generated and immunoprecipitated, but no corresponding product is made when only the RNA portion of the template (RNA124) is translated. Quantitation of the ratio of unmodified linker to linker-myc peptide fusion shows that 0.2–0.7% of the input message is converted to fusion product. A higher fraction of the input RNA was converted to fusion product in the presence of a higher ribosome/template ratio; over the range of input mRNA concentrations that we tested approximately 0.8–1.0 × 1012 fusion molecules are made per ml of translation extract.
Figure 4.
Analysis and quantitation of fusion formation by immunoprecipitation. (A) Denaturing urea PAGE analysis of the 32P-labeled linker-peptide fusions isolated by immunoprecipitation-RNase A-kinase treatment of the translation reactions. Reactions containing no template or only the RNA portion of the long myc template (RNA124) show no fusion formation. Reactions containing the long myc template contain bands corresponding to long myc linker fusions (30-P fusion) indicated by the arrow. The amount of input template is indicated at the top. (B) Recovered unmodified linker 30-P (left y axis) is linearly proportional to input template (x axis), whereas linker-peptide fusion (right y axis) is constant, as determined by PhosphorImager counting.
For RNA-peptide fusions to be useful, the peptide attached to an mRNA must be the peptide encoded by that mRNA, i.e., the nascent peptide must not be transferred to the puromycin of some other mRNA. No indication of crosstransfer was seen when a linker (30-P) was coincubated with the long myc template in translation extract in ratios as high as 20:1 (data not shown), nor did the presence of free linker significantly decrease the amount of long myc fusion produced. Similarly, cotranslation of the short and long templates 43-P and LP154 produced only the fusion products seen when the templates are translated alone and none of intermediate mobility as would be expected for fusion of the short template with the long myc peptide (data not shown). Both of these results argue that fusion formation occurs primarily between a nascent peptide and mRNA bound to the same ribosome.
Selective Enrichment of an RNA-Peptide Fusion.
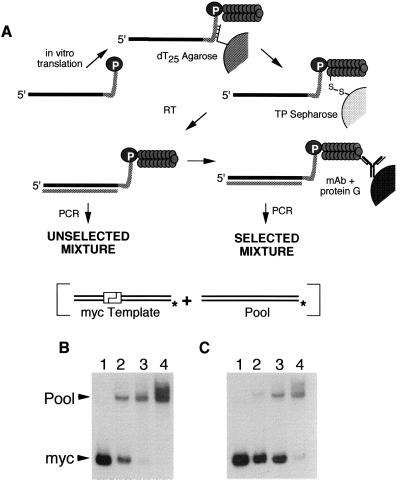
We have demonstrated the feasibility of using RNA-peptide fusions in selection and evolution experiments by enriching a particular RNA-peptide fusion from a complex pool of random sequence fusions on the basis of the encoded peptide. We prepared a series of dilutions of the long myc template (LP154) into a random sequence pool template (LP160), translated these mixtures, and purified the RNA-peptide fusion products by oligonucleotide and disulfide affinity chromatography. The myc-template fusions were selectively immunoprecipitated with anti-myc mAb (Fig. 5A). To measure the enrichment obtained in this selective step, aliquots of the mixture of RNA-peptide fusions from before and after the immunoprecipitation were reverse-transcribed and amplified by PCR in the presence of a radiolabeled primer. The amplified DNA was digested with a restriction endonuclease that cuts the myc template sequence but not the pool (Fig. 5 B and C). Quantitation of the ratio of cut and uncut DNA shows that the myc sequence was enriched by 20- to 40-fold relative to the random library by immunoprecipitation.
Figure 5.
Enrichment of myc dsDNA vs. pool dsDNA by in vitro selection. (A) Schematic of the selection protocol. Four mixtures of the myc and pool templates were translated in vitro and isolated on dT25 agarose followed by thiopropyl (TP) Sepharose to purify the template fusions from unmodified templates. The mRNA-peptide fusions then were reverse-transcribed to suppress any secondary or tertiary structure present in the templates. Aliquots of each mixture were removed before (B) and after (C) affinity selection, amplified by PCR in the presence of a labeled primer, and digested with a restriction enzyme that cleaves only the myc DNA. The input mixtures of templates were: lane 1, pure myc; lanes 2–4, 1:20, 1:200, and 1:2,000 myc/pool. The unselected material deviates from the input ratios because of preferential translation and reverse transcription of the myc template. The enrichment of the myc template during the selective step was calculated from the change in the pool/myc ratio before and after selection.
DISCUSSION
We propose a model for the mechanism of fusion formation in which translation initiates normally and elongation proceeds to the end of the ORF. When the ribosome reaches the DNA portion of the template, translation stalls. At this point, the complex can partition between two fates: dissociation of the nascent peptide or transfer of the nascent peptide to the puromycin at the 3′ end of the template. The efficiency of the transfer reaction is likely to be controlled by a number of factors that influence the stability of the stalled translation complex and the entry of the 3′-puromycin residue into the A site of the peptidyl transferase center. After the transfer reaction, the mRNA-peptide fusion probably remains complexed with the ribosome because the known release factors cannot hydrolyze the stable amide linkage between the RNA and peptide domains.
Both the classical model for elongation (29) and the intermediate states model (30) require that the A site be empty for puromycin entry into the peptidyl transferase center. For the puromycin to enter the empty A site, the linker either must loop around the outside of the ribosome or pass directly from the decoding site through the A site to the peptidyl transferase center. Our current data do not clearly distinguish between these alternatives because the shortest linker we have tested (21 nts) is still long enough to pass around the outside of the ribosome. In some models of ribosome structure (31), the mRNA is threaded through a channel that extends on either side of the decoding site, in which case unthreading of the linker from the channel would be required to allow the puromycin to reach the peptidyl transferase center through the A site.
Transfer of the nascent peptide to the puromycin appears to be slow relative to the elongation process as demonstrated by the homogeneity and length of the peptide attached to the linker (Fig. 3A). If the puromycin competed effectively with aminoacyl tRNAs during elongation, we would expect the linker-peptide fusions (e.g., Fig. 3A) present in the fusion products to be heterogeneous in size. Furthermore, the ribosome does not appear to read into the linker region as indicated by the similar gel mobility of the Met-template fusion to that of the unmodified linker. dA3n should code for (lysine)n, which certainly would decrease the mobility of the linker. The slow rate of unthreading of the mRNA may explain the slow rate of fusion formation relative to the rate of translocation. Preliminary results suggest that the amount of fusion product formed increases markedly after extended posttranslation incubation at low temperature, perhaps because of the increased time available for transfer of the nascent peptide to the puromycin.
The ability to synthesize covalent mRNA-peptide fusions by in vitro translation provides a different approach to the in vitro selection and directed evolution of peptides and proteins. This approach should have significant advantages over all approaches that require an in vivo step, because libraries of much greater complexity can be generated in vitro. This is a critical advantage for experiments in which a rare functional sequence is being selected from a completely random sequence initial library. Large libraries also provide an advantage in directed evolution experiments, in that sequence space can be explored to a greater depth around any given starting sequence. Current protocols are consistent with the generation of mRNA-peptide fusion libraries consisting of 1012–1013 independent members. We have generated a library of 1012 fusions, in which the peptide domain contains a random 27-aa sequence, in a 10-ml translation reaction. Improvements in the efficiency of the transfer reaction, coupled with a modest degree of scale-up, could increase the accessible library complexity to as much as 1015, which is comparable to the complexity of libraries that routinely are used for nucleic acid selection experiments. Further work will be needed to define the maximum length of peptide or protein that can be efficiently linked to mRNA in this approach. Short peptides of 12 and 33 amino acids seem to form fusions with comparable efficiency. Given that proteins >100 Kd can be generated in vitro in reticulocyte lysates (32) and that peptide chain elongation seems to be rapid compared with the transfer reaction, it is possible that much longer fusions will be generated with comparable efficiency.
Acknowledgments
This work was supported by a grant from Hoechst AG to Massachusetts General Hospital. R.W.R. was supported by fellowships from the Alfred P. Sloan Foundation for Studies in Molecular Evolution and the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: CPG, controlled pore glass.
References
- 1.Breaker R R. Chem Rev (Washington, DC) 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 2.Osborne S E, Ellington A D. Chem Rev (Washington, DC) 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 3.Williams K P, Bartel D P. Nucleic Acids Mol Biol. 1996;10:367–381. [Google Scholar]
- 4.Lorsch J R, Szostak J W. Acc Chem Res. 1996;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 5.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 6.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 7.Beaudry A A, Joyce G F. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]
- 8.Smith G P. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 9.Smith G P, Petrenko V A. Chem Rev (Washington, DC) 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 10.Harrison J L, Williams S C, Winter G, Nissim A. Methods Enzymol. 1996;267:83–109. doi: 10.1016/s0076-6879(96)67007-4. [DOI] [PubMed] [Google Scholar]
- 11.Schatz P J, Cull M G, Martin E L, Gates C M. Methods Enzymol. 1996;267:171–191. doi: 10.1016/s0076-6879(96)67012-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J-H, Dawes G, Stemmer W P C. Proc Natl Acad Sci USA. 1997;94:4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore J C, Arnold F H. Nat Biotechnol. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 14.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 15.Mattheakis L C, Bhatt R R, Dower W J. Proc Natl Acad Sci USA. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattheakis L C, Dias J M, Dower W J. Methods Enzymol. 1996;267:195–207. doi: 10.1016/s0076-6879(96)67013-x. [DOI] [PubMed] [Google Scholar]
- 17.Hanes J, Pluckthun A. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monro R E, Marcker K A. J Mol Biol. 1967;25:347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- 19.Monro R E, Vazquez D. J Mol Biol. 1967;28:161–165. doi: 10.1016/s0022-2836(67)80085-8. [DOI] [PubMed] [Google Scholar]
- 20.Jones R A. In: Oligonucleotide Synthesis: A Practical Approach. Gait M J, editor. Oxford: IRL; 1984. pp. 23–34. [Google Scholar]
- 21.Atkinson T, Smith M. In: Oligonucleotide Synthesis: A Practical Approach. Gait M J, editor. Oxford: IRL; 1984. pp. 35–81. [Google Scholar]
- 22.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 23.Moore M J, Sharp P A. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 24.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan A R, Wells R D, Khorana H G. J Mol Biol. 1967;26:477–497. doi: 10.1016/0022-2836(67)90316-6. [DOI] [PubMed] [Google Scholar]
- 26.Gallie D R, Sleat D E, Watts J W, Turner P C, Wilson T M A. Nucleic Acids Res. 1988;16:883–893. doi: 10.1093/nar/16.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez D. Inhibitors of Protein Biosynthesis. New York: Springer; 1979. [DOI] [PubMed] [Google Scholar]
- 29.Watson J D. Bull Soc Chim Biol. 1964;46:1399–1425. [PubMed] [Google Scholar]
- 30.Moazed D, Noller H F. Nature (London) 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 31.Frank J, Zhu J, Penczek P, Li Y, Srivasta S, Verschoor A, Radermascher M, Grassucci R, Lata R K, Grassucci R, Lata R K, Agrawal R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 32.Jackson R J, Hunt T. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 33.Steitz J A, Jakes K. Proc Natl Acad Sci USA. 1975;72:4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stormo G D, Schneider T D, Gold L M. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]