Abstract
During assembly, HLA class II molecules associate with the invariant chain. As the result, the peptide-binding groove is occupied by an invariant chain peptide termed CLIP (class-II-associated invariant chain peptide; sequence MRMATPLLM). By mass spectrometry, we have now characterized peptides that are naturally present in HLA-DQ2. This analysis revealed that 22 variants of Ii-derived peptides are associated with HLA-DQ2. Strikingly, the large majority of those do not contain the conventional CLIP sequence MRMATPLLM, but instead a peptide that partially overlaps with CLIP, sequence TPLLMQALPM. Peptide binding studies indicate that this alternative CLIP peptide has superior HLA-DQ2 binding properties compared to the conventional CLIP and that the minimal nine-amino-acid binding core consists of the sequence PLLMQALPM, findings that could be corroborated by molecular simulation. The alternative CLIP peptide was also found to be present in HLA-DQ2 molecules isolated from human thymus. Moreover, the alternative CLIP peptide was also found in association with HLA-DQ8. Together, these results indicate that HLA-DQ2 and HLA-DQ8 associate with an alternative CLIP sequence, a property that may relate to the strong association between HLA-DQ molecules and human autoimmune diseases.
Keywords: HLA class II, HLA-DQ2, CLIP, Alternative binding
Introduction
The MHC class II α and β subunits associate with an invariant chain (Ii) trimer shortly after biosynthesis in the endoplasmic reticulum, generating a stable complex (Roche et al. 1991). Part of the Ii-chain, commonly referred to as CLIP (class-II-associated invariant chain peptide), occupies the peptide-binding groove of MHC class II molecules, thus preventing peptide loading in the endoplasmatic reticulum (Bakke and Dobberstein 1990). Moreover, Ii facilitates the transport of the MHC class II molecules to the endosomal compartment where Ii is degraded by proteases. Subsequently, CLIP is released from the groove by the catalytic action of HLA-DM and exchanged for immunogenic peptides derived from proteins that have entered the endosomal/phagolysosomal system (Rocha and Neefjes 2008). Several autoimmune diseases, including type I diabetes and celiac disease, are HLA-associated.
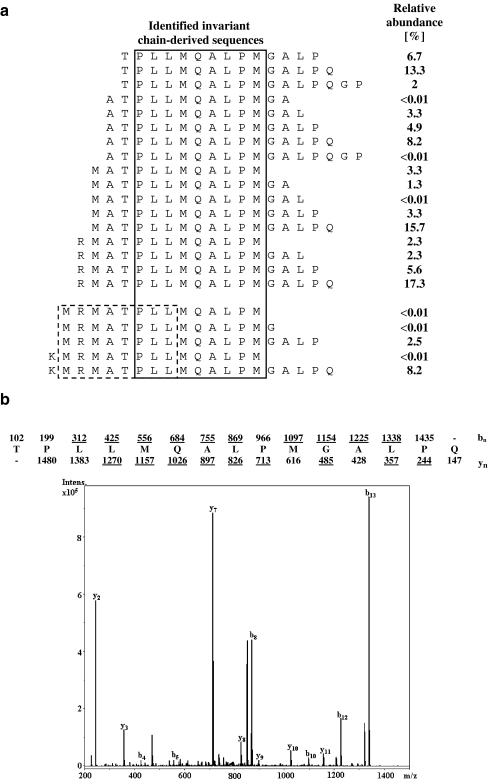
In a recent study, we have reported the characterization of a large number of naturally HLA-DQ2-bound peptides through mass spectrometry (Stepniak et al. 2008). In agreement with two previous studies, the most abundant peptide identified was the HLA-class-I-derived peptide IEQEGPEYW (van de Wal et al. 1996, Vartdal et al. 1996). Similarly, the second most abundant peptide was found to be invariant chain (li)-derived (van de Wal et al. 1996;Vartdal et al. 1996). We estimate that at least 10% of all HLA-DQ2 molecules is occupied by an invariant-chain-derived peptide. Twenty-two invariant-chain-derived peptides were identified, and a minimal core of ten amino acids was found to be present in all length variants [sequence TPLLMQALPM (CLIP 94–103); Fig. 1a]. Surprisingly, the sequence TPLLMQALPM was distinct from the CLIP peptide MRMATPLLM (CLIP 90–98) found to occupy the binding groove of all other human and murine class II molecules investigated (Chapman 1998). To determine the ratio between the CLIP and the alternative CLIP peptide in HLA-DQ2, we determined the relative abundance of the length variants of the Ii-derived peptides by mass spectrometry. This was done by integration of selected peaks in the mass chromatograms, done under the assumption that the relative response is similar for these species, which is appropriate since these species are length variants. The vast majority (approximately 90%) of the Ii-derived peptides contained the alternative CLIP sequence, while 10% contained the CLIP sequence (Fig. 1a).
Fig. 1.
Identification of an alternative CLIP peptide in HLA-DQ2. A Alignment of the length variants of the invariant-chain-derived peptides present in HLA-DQ2 expressed on EBV-transformed B cells. The solid line marks the predicted alternative CLIP sequence specific for HLA-DQ2; the dashed line marks the CLIP sequence found in other HLA class II molecules. B Tandem mass spectrum of one of DQ2-CLIP peptides. The amino acid sequence of the peptide is indicated on top of the spectrum, including the expected b- and y-fragment ions. Observed fragment ions are underlined and indicated in the spectrum. The correct identity of the peptide was proven by tandem mass spectrometry of the synthetic compound
To test if the alternative CLIP peptide is also present in HLA-DQ2 molecules expressed in the human thymus, we affinity purified HLA-DQ2 molecules from the thymus of two approximately 1.5-year-old HLA-DQ2-positive children. Subsequently, the bound peptides were eluted and characterized by mass spectrometry. A single Ii-derived peptide was identified which contained the CLIP sequence: TPLLMQALMGALPQ (CLIP 94–107; Fig. 1b). This peptide is identical to one of the most abundant peptides found in HLA-DQ2 molecules purified from an Epstein–Barr virus (EBV)-transformed cell line (Fig. 1a). Together, this indicates that also in the thymus, the alternative CLIP peptide is present in HLA-DQ2.
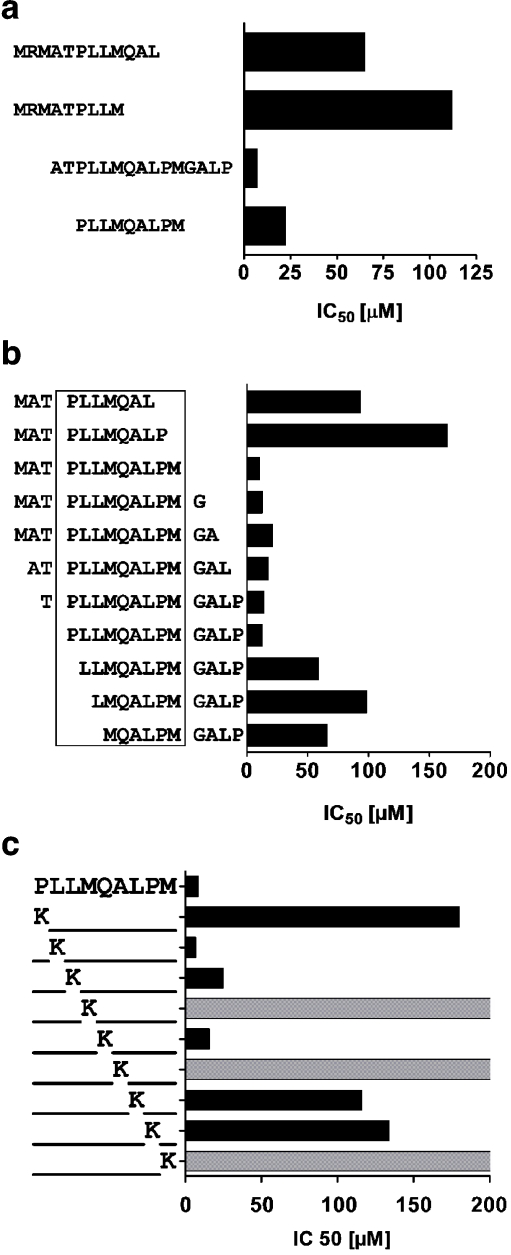
Two peptides, containing either CLIP (MRMATPLLMQAL; CLIP 90–101) or the alternative sequence (ATPLLMQALPMGALP; CLIP 93–107), were tested for their capacity to bind to HLA-DQ2 in a cell-free competition assay (Fig. 2a). The results showed a significantly lower IC50 value for the alternative CLIP sequence ATPLLMQALMGALP (CLIP 93–107), indicating a higher binding affinity. We further tested two nonamers containing CLIP (MRMATPLLM; CLIP 90–98) and the predicted binding frame of the alternative CLIP (PLLMQALPM: termed DQ2-CLIP hereafter; Fig. 2a) again showing a fourfold lower IC50 for DQ2-CLIP.
Fig. 2.
Identification of an alternative CLIP sequence. A Binding of two versions of the CLIP peptide to HLA-DQ2 and the binding of the predicted minimal core of DQ2-CLIP (PLLMQALPM). B Binding of a series of truncated versions of CLIP 90–105 to HLA-DQ2. C Binding of a series of CLIP 93–101 lysine substitution analogues. Gray columns indicate an IC50 above 200 μM representing weak or no binding. Each peptide was tested in at least two independent experiments; representative results are shown
To test the predicted binding frame of the DQ2-CLIP sequence, we next analyzed the binding of N- and C-terminally truncated length variants of CLIP 92–107 (MATPLLMQALPMGALP; Fig. 2b). Deletion of P95 and M103 was found to abrogate binding almost completely, indicating PLLMQALPM (CLIP 95–103) as the nine-amino-acid binding core.
To study the binding of DQ2-CLIP in more detail, we next determined which amino acids function as anchor residues in PLLMQALPM (CLIP 95–103). For this purpose, the impact of systematic substitution of amino acids in PLLMQALPM by lysine (K) was determined (Fig. 2c). Most dramatic effects could be seen for substitutions at positions p1, p4, p6, and p9, known to be important anchor positions where no positive charges are allowed. In addition, strong effects could also be seen at the p7 anchor position and the non-anchor position p8. This latter observation may reflect the preference of HLA-DQ2 for a proline at the p8 position (Stepniak et al. 2008).
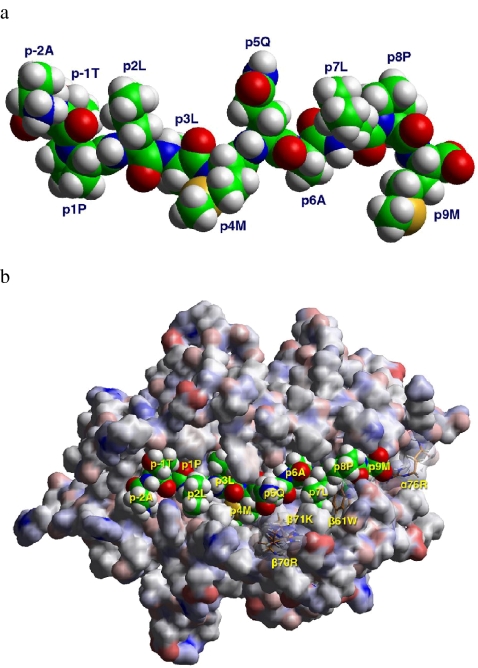
Additionally, modeling of HLA-DQ2 complexed with the peptide PLLMQALPM showed that it fits well in the groove of HLA-DQ2 (Fig. 3a and b). Moreover, it is predicted to bind better than the conventional CLIP peptide MRMATPLLM (unpublished data), a prediction that is confirmed by the observed superior binding of DQ2-CLIP (Fig. 2a).
Fig. 3.
Computer modeling of DQ2-CLIP in association with HLA-DQ2 based on the known DQ2-structure. A Side view of the DQ2-CLIP peptide (Ii 94–104) as it appears in complex with HLA-DQ2. B Shown is the TCR view of HLA-DQ2 (A1*0501/B1*0201) in complex with the DQ2-CLIP peptide (94–104 of Ii; core nonamer 96–104). The HLA-DQ2 molecule is shown in a van der Waals surface representation color coded according to the atomic charges (positive, blue; negative, red; neutral, gray); the surfaces for four residues (α76Arg, β61Trp, β70Arg, β71Lys) are transparent so that the respective residues can all be made visible. The peptide is shown in space filling mode (atomic color code: carbon, green; oxygen, red; nitrogen, blue; hydrogen, white; sulfur, yellow)
Since the spacing of the anchor residues in DQ2-CLIP is identical to that of the dominant HLA-class-I-derived peptide IEQEGPEYW (van de Wal et al. 1996; italicized amino acids are residues at anchor positions) as well as gluten-derived T cell stimulatory peptides (Vader et al. 2003), we conclude that the DQ2-CLIP peptide binds to HLA-DQ2 molecules in a similar binding register. In conclusion, the minimal core sequence within the DQ2-CLIP peptide that binds strongly to HLA-DQ2 is PLLMQALPM (residues 95–103) in which the italicized amino acids are residues at anchor positions.
Because celiac disease and type I diabetes are also associated with HLA-DQ8, we also analyzed naturally HLA-DQ8-bound peptides through mass spectrometry. The invariant chain was represented again by 22 peptides with a minimal core of ten amino acids present in all length variants, which is identical to that found in HLA-DQ2 [sequence TPLLMQALPM (CLIP 94–103); data not shown]. In adidtion, binding studies revealed that the peptide ATPLLMQALPMGALP (CLIP 93–107) binds slightly better than the peptide MRMATPLLMQAL (CLIP 90–101; data not shown). These results thus indicate that next to HLA-DQ2, HLA-DQ8 also appears to harbor an alternative CLIP peptide. Further studies will be required to determine the exact binding frame of this alternative CLIP peptide in HLA-DQ8.
It is now widely accepted that positive selection in the thymus is promiscuous (Ignatowicz et al. 1996), that is, a single self-peptide is able to mediate positive selection of large numbers of T cells with the ability to respond to various peptide antigens (Ignatowicz et al. 1997). It has also been demonstrated that in mice lacking DM molecules required for the dislocation of CLIP from MHC class II (Fukui et al. 1997; Martin et al. 1996), 15–50% of the normal number of CD4+ T cells are selected. Moreover, the T cell receptor repertoire in these mice was different from that in wild-type mice, suggesting that MHC class II–CLIP complexes can select a large and diverse repertoire of T cells. Given the dominant presence of DQ2-CLIP in HLA-DQ2, it is therefore possible that a substantial proportion of HLA-DQ2-restricted T cells in the periphery has been selected by HLA-DQ2–[DQ2-CLIP] complexes in the thymus. As HLA-DQ8 also appears to harbor an alternative CLIP peptide, this may similarly apply to T cell receptor selection in HLA-DQ8 individuals. It is tempting to speculate that this relates to the fact that HLA-DQ2 and HLA-DQ8 confer susceptibility for the common autoimmune diseases celiac disease and type I diabetes (Nepom and Kwok 1998; Todd et al. 1987).
Concluding remarks
In conclusion, we have observed that HLA-DQ2 molecules preferentially associate with an alternative form of the invariant-chain-derived CLIP peptide, DQ2-CLIP. The DQ2-CLIP has an at least four times higher binding affinity to HLA-DQ2 than the previously described classical CLIP sequence. Similarly, we observed that HLA-DQ8 harbors an alternative CLIP peptide. This property may relate to the strong association between these HLA-DQ molecules and human autoimmune diseases.
Acknowledgments
This research has been supported by a Marie Curie Fellowship of the European Community programme Drugs for Therapy under the contract number MRTN-CT-2004-512385 and the Celiac Disease Consortium, an Innovative Cluster approved by the Netherlands Genomics Initiative, and partially funded by the Dutch Government (BSIK03009). The authors thank B.O. Roep and J. van Bergen for critically reading the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- Ii
invariant chain
- CLIP
class-II-associated invariant chain peptide
References
- Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Chapman HA. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/S0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Ishimoto T, Utsuyama M, Gyotoku T, Koga T, Nakao K, Hirokawa K, Katsuki M, Sasazuki T. Positive and negative CD4+ thymocyte selection by a single MHC class II/peptide ligand affected by its expression level in the thymus. Immunity. 1997;6:401–410. doi: 10.1016/S1074-7613(00)80283-6. [DOI] [PubMed] [Google Scholar]
- Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/S0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- Ignatowicz L, Rees W, Pacholczyk R, Ignatowicz H, Kushnir E, Kappler J, Marrack P. T cells can be activated by peptides that are unrelated in sequence to their selecting peptide. Immunity. 1997;7:179–186. doi: 10.1016/S1074-7613(00)80521-X. [DOI] [PubMed] [Google Scholar]
- Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/S0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- Nepom GT, Kwok WW. Molecular basis for HLA-DQ associations with IDDM. Diabetes. 1998;47:1177–1184. doi: 10.2337/diabetes.47.8.1177. [DOI] [PubMed] [Google Scholar]
- Rocha N, Neefjes J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche PA, Marks MS, Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature. 1991;354:392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- Stepniak D, Wiesner M, de Ru AH, Moustakas AK, Drijfhout JW, Papadopoulos GK, van Veelen PA, Koning F. Large-scale characterization of natural ligands explains the unique gluten-binding properties of HLA-DQ2. J Immunol. 2008;180:3268–3278. doi: 10.4049/jimmunol.180.5.3268. [DOI] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Vader W, Stepniak D, Mearin ML, Thompson A, Spaenij L, Koning F. The HLA-DQ2 gene dose effect in Celiac Disease is directly related to the magnitude and breadth of gluten-specific T-cell respnoses. Proc Natl Acad Sci USA. 2003;100:12390–12395. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wal Y, Kooy YMC, Drijfhout JW, Amons R, Koning F. Peptide binding characteristics of the coeliac disease-associated DQ(alpha1*0501, beta1*0201) molecule. Immunogenetics. 1996;44:246–253. doi: 10.1007/BF02602553. [DOI] [PubMed] [Google Scholar]
- Vartdal F, Johansen BH, Friede T, Thorpe CJ, Stevanovic S, Eriksen JE, Sletten K, Thorsby E, Rammensee HG, Sollid LM. The peptide binding motif of the disease associated HLA-DQ (alpha 1* 0501, beta 1* 0201) molecule. Eur J Immunol. 1996;26:2764–2772. doi: 10.1002/eji.1830261132. [DOI] [PubMed] [Google Scholar]