Abstract
The major histocompatibility complex class I gene repertoire was investigated in a large panel of rhesus macaques of Chinese origin. As observed in Indian animals, subjects of Chinese derivation display Mamu-B gene copy number variation, and the sum of expressed genes varies among haplotypes. In addition, these genes display differential transcription levels. The majority of the Mamu-B alleles discovered during this investigation appear to be unique for the population studied. Only one particular Mamu-B haplotype is shared between Indian and Chinese animals, and it must have been present in the progenitor stock. Hence, the data highlight the fact that most allelic polymorphism, and most of the Mamu-B haplotypes themselves, are of relatively recent origin and were most likely generated after the separation of the Indian and Chinese rhesus macaque populations.
Keywords: Nonhuman primates, MHC, Macaques, Evolution
Introduction
The major histocompatibility complex (MHC) is a multigene family that plays a key role in initiating adaptive immune responses in vertebrates. Two main groups of genes and associated cell surface proteins are distinguished: class I molecules are involved in the binding and presentation of intracellular peptides, whereas class II gene products present processed extracellular antigen segments. An MHC class I or II molecule complexed with a peptide can interact with various types of receptors on distinct types of T and natural killer (NK) cells, which in turn may execute different kinds of effector functions. The main feature of the MHC is the abundant polymorphism of several of its genes, which may have a profound impact on disease susceptibility or resistance; it is also known to influence the outcome of organ transplantations. Moreover, the number of MHC class I and II genes may differ significantly between species (Kelley et al. 2005), as well as between individuals of a species (Robinson et al. 2003).
Due to its role in immune-related disorders, the MHC has been studied extensively, not only in humans (human leucocyte antigen (HLA) system) but also in non-human primates (Slierendregt et al. 1995; de Groot et al. 2002; O’Connor et al. 2003; Bontrop and Watkins 2005; Vierboom et al. 2005; Sauermann et al. 2008). In particular, the rhesus macaque (Macaca mulatta) is a commonly used animal model for the study of human diseases and vaccine development (Bontrop 2001). Orthologues of the HLA class I and II genes have been identified in rhesus monkeys and are named Mamu-A, Mamu-B, Mamu-DP, Mamu-DQ and Mamu-DR (Bontrop et al. 1995; Boyson et al. 1996; Doxiadis et al. 2001). Equivalents of non-classical class I genes, which are characterised by low levels of polymorphism and restricted tissue distributions, are also present in the rhesus macaque and are named Mamu-E and Mamu-F (Otting and Bontrop 1993; Boyson et al. 1995). In contrast to humans, the highly polymorphic classical class I A and B genes are multiplied in rhesus macaques (Daza-Vamenta et al. 2004; Kulski et al. 2004), whereas an equivalent of the HLA-C gene has not been observed (Vogel et al. 1999; Otting et al. 2005). Up until now, seven different Mamu-A genes, with differential transcription levels, have been defined. Combinations of two or three of these loci are present for every rhesus macaque chromosome that harbours the MHC region (Otting et al. 2007).
Orthologues of the HLA-B gene appear to have undergone several rounds of duplication in the rhesus macaque, as the sequence analysis of a complete rhesus macaque MHC region revealed a haplotype comprising 19 different Mamu-B-like genes (Daza-Vamenta et al. 2004; Kulski et al. 2004). Based on the promoter and exon sequences, it was concluded that 14 of these genes may have a protein-encoding capacity. Analyses of cDNA in pedigreed animals have shown, however, that only two or three loci per haplotype are transcribed at substantial levels (majors). In addition to these majors, minor alleles were also found, which are characterised by reduced transcription levels (Otting et al. 2005). Some minor B-like sequences appear to be alleles of an oligomorphic gene and may represent non-classical with a specialized function. An example is provided by Mamu-I (Urvater et al. 2000).
Most rhesus macaques used in research are of Indian origin, and most of the genetic data that have been gathered so far are based on animals from this subcontinent. In recent years, rhesus monkeys of Chinese origin have been introduced into the research, as well as cynomolgus (Macaca fascicularis) and pig-tailed macaques (Macaca nemestrina). cDNA analyses have suggested that these three species of macaque may share a similar organisation of the MHC class I A genes (Krebs et al. 2005; Pratt et al. 2006; Lafont et al. 2007; Otting et al. 2007). Phylogenetic comparison shows intermingling of the Mamu-A, Mafa-A and Mane-A alleles, which reflects the common ancestry of these species.
Considerable research has been performed on MHC class I B genes in the cynomolgus macaque, and about 80 Mafa-B alleles are known (Uda et al. 2005; Wiseman et al. 2007). Genomic sequencing of the MHC region in the cynomolgus macaque has revealed the presence of 12 Mafa-B-like genes on that particular haplotype (Watanabe et al. 2007). The investigation of Chinese rhesus macaques was started recently, and one report on 12 animals has documented the presence of 23 Mamu-B alleles (Karl et al. 2008). In the present study, an extensive panel of Chinese-origin animals was examined, with the aim of gathering more information on the makeup of the Mamu-B region. The lexicon of Mamu-B sequences has increased considerably, and with these sequences we intend to define the loci within the Mamu-B region, as has already been successfully performed for the Mamu-A and Mafa-A region (Otting et al. 2007). More information on distinct loci, and hence on configurations in the Mamu-B region, may help to establish a more consistent nomenclature system for the Mamu-B alleles as well as for B alleles in other macaque species.
Materials and methods
Animals and cell lines
The Biomedical Primate Research Centre harbours a self-sustaining colony of approximately 1,000 rhesus macaques, mainly of Indian origin. The animals have been pedigreed based on the segregation of serologically defined MHC allotypes and other markers defined by molecular techniques (Doxiadis et al. 2003, 2006, 2007; Penedo et al. 2005). In recent years, the colony has been supplemented with individuals of Chinese origin, the pedigree status of which was unknown. A panel of 48 Chinese animals was selected for the present study. For most of these subjects, B-cell lines are available.
cDNA, cloning and sequencing
RNA was isolated from PBMCs or B cells (Rneasy kit, Qiagen) and subjected to One-Step reverse-transcriptase polymerase chain reaction (RT-PCR), as recommended by the supplier (Qiagen or Promega). The primers 5′MBS: AATTCATGGCGCCCCGAACCCTCCTCCTGC and 3′MBS: CTAGACCACACAAGACAGTTGTCTCAG were used that anneal specifically to Mamu-B transcripts in macaques. The final elongation step was extended to 30 min to generate a 3′dA overhang. The RT-PCR products were cloned using the InsT/Aclone kit (Fermentas) or the PCR cloning kit (Qiagen). After transformation, 32 to 48 colonies were picked for plasmid isolation. Sequencing reactions were performed using the BigDye terminator cycle sequencing kit, and samples were run on an automated capillary sequencing system (Applied Biosystems Genetic Analyser 3100). The methods to determine high or low transcription levels have been published (Otting et al. 2005, 2007).
Phylogenetic analyses and nomenclature
Sequences were analysed using Sequence Navigator Software version 1.0.1 (Applied Biosystems) and MacVector™ version 9.5.2 (Oxford Molecular Group), followed by manual adjustments. Phylogenetic comparisons were also performed with the MacVector software. Neighbour-joining trees were constructed with the Kimura 2 parameter method, and bootstrap analyses were based on 1,000 replications.
Mamu-B alleles have been named according to published nomenclature proposals (Klein et al. 1990; Robinson et al. 2003; Ellis et al. 2006), and the alleles were submitted to the European Molecular Biology Laboratory database under accession numbers AM902528–AM902585.
Results and discussion
Differential transcription levels: majors and minors
In the cohort of 48 Chinese rhesus macaques, 80 distinct Mamu-B sequences were detected, of which 51 have not yet been reported. The alleles are listed, together with the relevant accession numbers, as well as reference animals (Table 1). In most animals, three or four Mamu-B alleles were detected, which are considered to represent majors, as determined by the number of clones; this means that one to three majors may be present per haplotype. In most animals, additional Mamu-B sequences were found with low transcription levels, which represent minors (Table 1). Only those minor sequences were named and listed of which at least three identical clones had been detected. As a consequence, one should realise that the number of Mamu-B alleles detected in this study represents the tip of the iceberg since many minors may not have been picked up. Differential transcription levels were earlier described for the Mamu-B alleles in Indian rhesus monkeys (Otting et al. 2005) and for genes in the Mamu-A and Mafa-A region (Otting et al. 2007). This phenomenon of varying transcription levels has been observed in other species as well (Birch et al. 2006; Wallny et al. 2006; Shaw et al. 2007).
Table 1.
Mamu-B sequences observed in a panel of Chinese-origin rhesus macaques
| Designation | Reference animals | Accession number |
|---|---|---|
| Majors | ||
| Mamu-B*010101 | Ri126, Ri165 | U42837 |
| Mamu-B*010102 | Ri165 | AM902529 |
| Mamu-B*0201 | Ri011 | U41833 |
| Mamu-B*0301 | Ri037, Ri159 | U41825 |
| Mamu-B*0401 | Ri037, Ri159 | U41826 |
| Mamu-B*0702 | Ri165, Ri191 | AJ556875 |
| Mamu-B*0704 | Ri011, Ri189 | AM902528 |
| Mamu-B*1001 | Ri137, Ri185 | AM902538 |
| Mamu-B*1301 | Ri011, Ri189 | AM902539 |
| Mamu-B*1401 | Ri184, Ri233 | AM902540 |
| Mamu-B*1501 | Ri079a | AM902541 |
| Mamu-B*1502 | Ri018 | AM902542 |
| Mamu-B*1601 | Ri018 | AM902543 |
| Mamu-B*1801 | Ri253a | AM902534 |
| Mamu-B*1903 | Ri126 | AM902535 |
| Mamu-B*1902 | Ri028, Ri146 | EF580169 |
| Mamu-B*2102 | Ri078 | AM902536 |
| Mamu-B*2103 | Ri290a | AM902537 |
| Mamu-B*2301 | Ri284 | AM902530 |
| Mamu-B*2401 | Ri028, Ri146 | AJ556881 |
| Mamu-B*2501 | Ri289a | AM902531 |
| Mamu-B*280201 | Ri253a | AM902532 |
| Mamu-B*280202 | Ri290a | AM902544 |
| Mamu-B*2803 | Ri078 | AM902545 |
| Mamu-B*3002 | Ri191 | AJ844597 |
| Mamu-B*300302 | Ri281, Ri284 | AM902546 |
| Mamu-B*300303 | Ri078 | AM902547 |
| Mamu-B*3004 | Ri137, Ri142 | AM902548 |
| Mamu-B*3201 | Ri289a | AM902549 |
| Mamu-B*3301 | Ri293 | AM902550 |
| Mamu-B*3401 | Ri197 | AM902551 |
| Mamu-B*3501 | Ri185 | AM902552 |
| Mamu-B*3602 | Ri185 | AJ556887 |
| Mamu-B*3701 | Ri185 | AJ556888 |
| Mamu-B*3901 | Ri002, Ri026 | AJ556890 |
| Mamu-B*4001 | Ri056 | AJ556891 |
| Mamu-B*4002 | Ri018 | EF362448/EF580150 |
| Mamu-B*4201 | Ri184 | AM902553 |
| Mamu-B*440102 | Ri002 | AM902555 |
| Mamu-B*4402 | Ri205 | AM902556 |
| Mamu-B*4403 | Ri056, Ri189 | AM902557 |
| Mamu-B*4404 | Ri302 | AM902558 |
| Mamu-B*4502 | Ri185 | AJ556896 |
| Mamu-B*4701 | Ri009 | AJ556898 |
| Mamu-B*4702 | Ri136, Ri228 | AJ556899 |
| Mamu-B*4703 | Ri090a | AM902560 |
| Mamu-B*4704 | Ri228, Ri260 | AM902561 |
| Mamu-B*6102 | Ri078 | AM902564 |
| Mamu-B*6601 | Ri197, Ri226 | AJ844597 |
| Mamu-B*6701 | Ri009, Ri018 | AJ844598 |
| Mamu-B*6702 | Ri028, Ri159 | AM902568 |
| Mamu-B*680101 | Ri226, Ri233 | AJ844599 |
| Mamu-B*680102 | Ri009, Ri018 | AM902569 |
| Mamu-B*6802 | Ri018, Ri284 | EF362453/EF219482 |
| Mamu-B*6903 | Ri026 | EF219479 |
| Mamu-B*6904 | Ri037 | AM902574 |
| Mamu-B*7501 | Ri026 | EF219478 |
| Mamu-B*7602 | Ri302 | EF112569 |
| Mamu-B*7702 | Ri002, Ri184 | AM902580 |
| Mamu-B*8301 | Ri281 | EF580161 |
| Mamu-B*8501 | Ri094a | EF580165 |
| Mamu-B*8502 | Ri056, Ri189 | AM902581 |
| Mamu-B*8602 | Ri287 ?? | AM902582 |
| Mamu-B*8603 | Ri205 | AM902583 |
| Mamu-B*8701 | Ri137, Ri142 | EF580170 |
| Mamu-B*9102 | Ri009 | AM902584 |
| Minors | ||
| Mamu-B*0703 | Ri228 | AJ556876 |
| Mamu-B*1703 | Ri165 | AM902533 |
| Mamu-B*2702 | Ri228 | AM902559 |
| Mamu-B*3801 | Ri009 | AJ556889 |
| Mamu-B*5601 | Ri028, Ri159 | AM902562 |
| Mamu-B*5901 | Ri233 | AM902563 |
| Mamu-B*6201 | Ri094a | AM902565 |
| Mamu-B*6502 | Ri281 | EF580163 |
| Mamu-B*6503 | Ri233 | AM902567 |
| Mamu-B*6803 | Ri137, Ri142 | AM902570 |
| Mamu-B*6804 | Ri253a | AM902571 |
| Mamu-B*7002 | Ri189, 226 | AM902575 |
| Mamu-B*7201 | Ri182a | AM902576 |
| Mamu-B*7301 | Ri078 | AM902578 |
Sequences in bold have been published previously. The names of some new alleles may have a lower lineage number than those already published because a series of vacant numbers was used for present designations. One or two reference animals are given for each allele.
aB-cell lines are not available
In studies performed on Indian rhesus macaques, the peptide binding motifs of several Mamu-A and Mamu-B gene products have been defined (Evans et al. 1999; O’Connor et al. 2003; Sette et al. 2005; Kaizu et al. 2007; Loffredo et al. 2007). Comparisons indicated that the highly expressed Mamu class I molecules seem to execute the classical antigen presentation function, whereas the minors may represent non-classical genes. Alleles with a lower level of transcription are those of the oligomorphic Mamu-I locus (Urvater et al. 2000). Mamu-I has the characteristics of a non-classical and was probably once recruited by a duplication from one of the Mamu-B loci. The Mamu-I gene appears to be present on most haplotypes and will not be discussed in further detail.
Alleles that display the characteristics of a major in one animal and a minor in another have not been encountered. However, almost identical alleles were found that exhibit significant transcription differences. For instance, Mamu-B*0702 is a major in some animals, whereas Mamu-B*0703, which differs by only one base pair, behaves as a minor (Table 1). The question arises as to how these differences in transcription took place. It is probable that the pool of paralogous Mamu-B genes has been generated by duplications (Kulski et al. 2004). Some of these paralogues may have acquired other unique mutations later, leading to distinct loci controlling unique lineages. A further level of complexity arose by unequal crossing over, which is often seen in multigene families. It is possible that during such processes apparently intact Mamu class I genes that belong to the same locus–lineage may have been placed in the context of promoters that render differential activity. Another possibility is that the genes and their promoters are tightly linked and that the promoter region itself was affected by mutations, leading to different transcription levels.
Sharing of Mamu-B alleles between populations
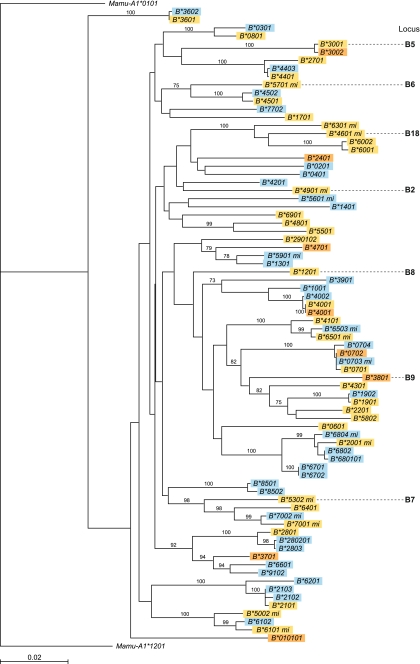
Phylogenetic analyses demonstrated that most of the alleles that are encountered in animals of Chinese or Indian origin share lineages. A phylogenetic tree, constructed of a selection of Mamu-B sequences (Fig. 1), illustrates, for instance, that members of the Mamu-B*07, Mamu-B*19, Mamu-B*21 and Mamu-B*68 lineages are present in both populations (B*2001 should be renamed into a B*68 allele). The difference between the alleles grouping within these lineage is explained by point mutations, although introns have not been sequenced to completely discard gene conversion mechanisms. Crossing over events may be responsible for the generation of new lineages. It seems that after the physical separation of the Indian and Chinese rhesus macaque populations; these animals generated unique Mamu-B allelic repertoires. Indeed, in our Chinese panel, eight alleles were detected: namely, Mamu-B*01, Mamu-B*0702, Mamu-B*2401, Mamu-B*3002, Mamu-B*3701, Mamu-B*3801, Mamu-B*4001 and Mamu-B*4701, which are present in Indian animals as well. However, three of these alleles are present on one haplotype and will be discussed in the next section. The sharing of alleles between populations seems to be independent of transcription levels. In the case of the Mamu-A locus, most of the alleles appeared to be population specific, and only one allele was shared between both groups (Otting et al. 2007; Karl et al. 2008). This would suggest that the Mamu-A alleles accumulate faster mutations than Mamu-B. This observation is in contrast to the situation encountered in humans and chimpanzees (Belich et al. 1992; Watkins et al. 1992; McAdam et al. 1994; McAdam et al. 1995; de Groot et al. 2000). It cannot be excluded that the population of Indian rhesus macaques has experienced a bottleneck (Hernandez et al. 2007). This phenomenon may explain the higher level of polymorphism within the population of Chinese rhesus monkeys.
Fig. 1.
Phylogenetic analysis of exons 2, 3 and 4 Mamu-B sequences obtained from Indian- and Chinese-origin rhesus macaques, as listed in Table 2. Indian-origin alleles are depicted in yellow; Chinese-origin alleles are shown in blue. The alleles depicted in brown are observed in both populations. The minor alleles have mi in their names. The locus numbers (in black) indicate the order in which transcribed alleles in haplotype B11a are present on the completely sequenced Mamu-B region (see also Table 3)
Mamu-B haplotypes
In the Indian population studied previously, it was possible to determine the combination of Mamu-B alleles that segregate on a chromosome because pedigree data are available (Otting et al. 2005). The pedigree status of the presently studied Chinese animals is unknown. However, haplotypes can be deduced on the basis that particular Mamu-B allele combinations are observed in at least three animals. As can be seen, the deduced haplotypes are numbered subsequently and listed (Table 2). The most common Indian haplotypes are named and listed according to their corresponding serotype. Only one allele combination, represented by haplotypes 1 and B26, is common to both populations. This haplotype (Mamu-B*01–B*0702–B*3002) is of specific interest, as the B26 specificity in Indian animals seems to control resistance to develop collagen-type-II-induced arthritis (CIA) in young Indian rhesus macaques (Bakker et al. 1992). The peptide binding specificity of the Mamu-B*01 gene product has been determined (Loffredo et al. 2005), so it will be of interest to assess whether Chinese animals, which express the Mamu-B*01 allotype, are resistant to the induction of CIA as well.
Table 2.
Deduced combinations of alleles on chromosomes (haplotypes)
| Haplotype | Majors | Minors |
|---|---|---|
| Chinese rhesus macaques | ||
| 1 | B*010101/2, B*0702, B*3002 | |
| 2 | B*1902, B*2401 | |
| 3 | B*3602, B*3701, B*4502 | |
| 4 | B*2102/3, B*2802/3, B*6102 | B*6804 (B*20-like) |
| 5 | B*0301, B*0401 | |
| 6 | B*6601, B*680101 | |
| 7 | B*6701/2 | B*5601 |
| 8 | B*4403, B*8501/2 | |
| 9 | B*4002, B*6802 | |
| 10 | B*1001, B*6802 | |
| 11 | B*0704, B*1301 | B*7002 |
| 12 | B*7702, B*4201 | |
| 13 | B*1401 | B*5901, B*6503 |
| 14 | B*0201 | |
| 15 | B*3901, B*0703 | |
| Indian rhesus macaques | ||
| B2 | B*3801, B*4701 | |
| B11a | B*1201, B*3001, B*3801 | B*4601, B*4901, B*5302, B*5701 |
| B11b | B*1201, B*3001, B*2201 a | B*4601, B*4901, B*5302, B*5701, B*7001 a |
| B13 | B*4101, B*4801, B*6401 | |
| B14 | B*0601, B*0801 | |
| B17 | B*1701, B*290102, B*6002 | B*6101 |
| B18 | B*5501, B*5802 | B*6301 |
| B20 | B*2101, B*2801 | B*2001 (B*68-like) |
| B24 | B*1901, B*2401 | |
| B25 | B*6901 | B*5002, B*6501 |
| B26 | B*010101, B*0701/2, B*3002 | |
| B29 | B*4001, B*4401 | |
| B32 | B*3601, B*3701, B*4501 | |
The Indian haplotypes are numbered according to associated serotypes. For the Chinese animals, reliable antisera are lacking, and the haplotypes are numbered successively. The B11a haplotype corresponds to the B region in the completely sequenced MHC.
a B*2201 and B*7001 sequences are not present on B11a.
The Chinese haplotypes 2, 3 and 4 are the equivalents of the B24, B32 and B20 haplotypes encountered in Indian monkeys, apart from a few base pair differences in some alleles. Two Mamu-B allele combinations, 5 and 15, were encountered only twice in our panel but were confirmed by other investigators (Boyson et al. 1996; Karl et al. 2008). A linked allele was not observed for Mamu-B*0201 (haplotype 14). In summary, the data show that in both macaque populations the number of majors transcribed per haplotype varies from one to three (Table 2). Although 14 functional Mamu-B-like loci may be present on the chromosome, only up to three loci are transcribed at considerable levels. Since an equivalent of HLA-C has never been found in the rhesus macaque, the overall number of classical class I gene products on the cell membrane in this animal seems to be comparable to the situation in humans.
Apart from the stable allele combinations, observed in both populations of macaques, majors are present that segregate with alleles from different lineages. The Mamu-B*6802 allele, for example, is found in combination with two majors: namely, Mamu-B*1001 and Mamu-B*4002, which are most likely different paralogous structures. Furthermore, the combination Mamu-B*4001–Mamu-B*4401 is frequent in Indian animals, whereas in Chinese animals members of the B*40 and B*44 lineages are not observed on the same chromosome (Table 2). The gene content of Mamu-B haplotypes may have been rearranged by recombination-like processes. The arrangement of the different Mamu-B genes is more prone to reshuffling than those of the Mamu-A region, which may be due in part to the high gene copy number variation in the Mamu-B region. This reshuffling of genes, in combination with the varying transcription levels, represents an alternative method for bearing polymorphisms within a population.
An attempt to define Mamu-B loci
One of the main consequences of the reshuffling of Mamu-B genes is the tremendous difficulty in assigning particular sequences to defined B genes or loci, as has been done for the Mamu-A and the Mafa-A regions. We have tried to group Mamu-B alleles into loci by comparing them to the only published genomic rhesus macaque MHC region. The Mamu-B region published by Daza-Vamenta and co-workers matches our B11a serotype in Indian animals, and the major and minor alleles are listed in Table 3. Chinese and Indian animals share Mamu-B lineages, as is indicated by the intermingling of alleles in the phylogenetic tree (Fig. 1). The major and minor alleles of the B11 haplotype are indicated by a locus number that reflects their position on the sequenced B region. In the tree, clades are present that do not contain any of the loci present on the sequenced haplotype, for example, those with the Mamu-B*21 and Mamu-B*28 alleles. The phylogenetic analyses were also performed including the non-transcribed loci; however, clustering with B*21 and B*28 was again not observed (data not shown). Moreover, the sequenced haplotype appears to have a locus, represented by the major Mamu-B*1201, for which no orthologues are present in any other haplotype in our Chinese panel.
Table 3.
Comparison of genomic sequences and cDNA transcripts for the Mamu-B11 region configuration
| Locus–gene | Allele | Transcription level |
|---|---|---|
| Mamu-B2 | Mamu-B*4901 | Minor |
| Mamu-B3 | Mamu-I | Minor |
| Mamu-B5 | Mamu-B*3001 | Major |
| Mamu-B6 | Mamu-B*5701 | Minor |
| Mamu-B7 | Mamu-B*5302 | Minor |
| Mamu-B8 | Mamu-B*1201 | Major |
| Mamu-B9 | Mamu-B*3801 | Major |
| Mamu-B18 | Mamu-B*4601 | Minor |
In the first column, the Mamu-B loci as they appear on the completely sequenced rhesus MHC region are listed, in the next column which transcripts are observed as well as their transcriptions levels. For the other loci (B1, B4, etc.), no transcripts are detected with cDNA sequencing.
This observation underlines the fact that the number and combination of Mamu-B genes present per chromosome may differ dramatically between haplotypes, leading to various region configurations. At this stage, we do not understand whether any of the majors in different region configurations share paralogous or orthologous relationships. As discussed earlier, orthology or paralogy may be obscured by unequal crossing over events. Therefore, it is impossible at present to set up a nomenclature system for Mamu-B similar to that implemented for Mamu-A (Otting et al. 2007). Thus far, the Mamu-B alleles have been numbered mostly based on the order of detection. It is possible that the Mamu-B region is as plastic as the kinase inhibitory region in humans. Should this be the case, a nomenclature system for B alleles, which is based solely on lineage numbers, may provide a solution. However, a definitive choice with regards to nomenclature can only be made when additional data become available on the genomic organisation of more Mamu-B regions as well as on the B region of other macaques species.
Acknowledgement
The authors wish to thank Donna Devine for editing the manuscript and Henk van Westbroek for preparing the figure.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
This study was supported in part by the National Institutes of Health, project 1-R24-RR16038-01 (Catalog of federal assistance 93.306).
References
- Bakker NP, van Erck MG, Otting N, Lardy NM, Noort RC, t Hart BA, Jonker M, Bontrop RE. Resistance to collagen-induced arthritis in a nonhuman primate species maps to the major histocompatibility complex class I region. J Exp Med. 1992;175:933–937. doi: 10.1084/jem.175.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belich MP, Madrigal JA, Hildebrand WH, Zemmour J, Williams RC, Luz R, et al. Unusual HLA-B alleles in two tribes of Brazilian Indians. Nature. 1992;357:326–329. doi: 10.1038/357326a0. [DOI] [PubMed] [Google Scholar]
- Birch J, Murphy L, MacHugh ND, Ellis SA. Generation and maintenance of diversity in the cattle MHC class I region. Immunogenetics. 2006;58:670–679. doi: 10.1007/s00251-006-0137-y. [DOI] [PubMed] [Google Scholar]
- Bontrop RE. Non-human primates: essential partners in biomedical research. Immunol Rev. 2001;183:5–9. doi: 10.1034/j.1600-065x.2001.1830101.x. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Otting N, Slierendregt BL, Lanchbury JS. Evolution of major histocompatibility complex polymorphisms and T-cell receptor diversity in primates. Immunol Rev. 1995;143:33–62. doi: 10.1111/j.1600-065X.1995.tb00669.x. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Watkins DI. MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol. 2005;26:227–233. doi: 10.1016/j.it.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM, et al. The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]
- Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, et al. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Arguello R, Watkins DI, Doxiadis GG, Madrigal JA, et al. Major histocompatibility complex class I diversity in a West African chimpanzee population: implications for HIV research. Immunogenetics. 2000;51:398–409. doi: 10.1007/s002510050638. [DOI] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS, Heeney JL, van Rood JJ, et al. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci U S A. 2002;99:11748–11753. doi: 10.1073/pnas.182420799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, de Groot NG, Bontrop RE. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol Rev. 2001;183:76–85. doi: 10.1034/j.1600-065x.2001.1830106.x. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, De Groot NG, De Groot N, Rouweler AJ, Noort R, et al. Evolutionary stability of MHC class II haplotypes in diverse rhesus macaque populations. Immunogenetics. 2003;55:540–551. doi: 10.1007/s00251-003-0590-9. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, et al. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, Claas FH, Doxiadis II, van Rood JJ, Bontrop RE. A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc Natl Acad Sci U S A. 2007;104:8907–8912. doi: 10.1073/pnas.0702964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, et al. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- Evans DT, O’Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, et al. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- Kaizu M, Borchardt GJ, Glidden CE, Fisk DL, Loffredo JT, Watkins DI, et al. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, et al. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics. 2008;60:37–46. doi: 10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, et al. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol. 2004;21:2079–2091. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- Lafont BA, McGraw CM, Stukes SA, Buckler-White A, Plishka RJ, Byrum RA, et al. The locus encoding an oligomorphic family of MHC-A alleles (Mane-A*06/Mamu-A*05) is present at high frequency in several macaque species. Immunogenetics. 2007;59:211–223. doi: 10.1007/s00251-007-0190-1. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, et al. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–5997. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, et al. A uniquely high level of recombination at the HLA-B locus. Proc Natl Acad Sci USA. 1994;91:5893–5897. doi: 10.1073/pnas.91.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, et al. Chimpanzee MHC class I A locus alleles are related to only one of the six families of human A locus alleles. J Immunol. 1995;154:6421–6429. [PubMed] [Google Scholar]
- O’Connor DH, Mothe BR, Weinfurter JT, Fuenger S, Rehrauer WM, Jing P, et al. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol. 2003;77:9029–9040. doi: 10.1128/JVI.77.16.9029-9040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, Bontrop RE. Characterization of the rhesus macaque (Macaca mulatta) equivalent of HLA-F. Immunogenetics. 1993;38:141–145. doi: 10.1007/BF00190901. [DOI] [PubMed] [Google Scholar]
- Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, et al. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A. 2005;102:1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, et al. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics. 2005;57:198–209. doi: 10.1007/s00251-005-0787-1. [DOI] [PubMed] [Google Scholar]
- Pratt BF, O’Connor DH, Lafont BA, Mankowski JL, Fernandez CS, Triastuti R, et al. MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics. 2006;58:995–1001. doi: 10.1007/s00251-006-0164-8. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauermann U, Siddiqui R, Suh YS, Platzer M, Leuchte N, Meyer H, et al. Mhc class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)-infected rhesus macaques. Genes Immun. 2008;9:69–80. doi: 10.1038/sj.gene.6364448. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Bui HH, del Guercio MF, Alexander J, Loffredo J, et al. Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics. 2005;57:53–68. doi: 10.1007/s00251-004-0749-z. [DOI] [PubMed] [Google Scholar]
- Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, Riegert P, et al. Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol. 2007;178:5744–5752. doi: 10.4049/jimmunol.178.9.5744. [DOI] [PubMed] [Google Scholar]
- Slierendregt BL, Hall M, t Hart B, Otting N, Anholts J, Verduin W, Claas F, Jonker M, Lanchbury JS, Bontrop RE. Identification of an Mhc-DPB1 allele involved in susceptibility to experimental autoimmune encephalomyelitis in rhesus macaques. Int Immunol. 1995;7:1671–1679. doi: 10.1093/intimm/7.10.1671. [DOI] [PubMed] [Google Scholar]
- Uda A, Tanabayashi K, Fujita O, Hotta A, Terao K, Yamada A. Identification of the MHC class I B locus in cynomolgus monkeys. Immunogenetics. 2005;57:189–197. doi: 10.1007/s00251-005-0782-6. [DOI] [PubMed] [Google Scholar]
- Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarczyk MS, et al. Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol. 2000;164:1386–1398. doi: 10.4049/jimmunol.164.3.1386. [DOI] [PubMed] [Google Scholar]
- Vierboom MP, Jonker M, Bontrop RE, t Hart B. Modeling human arthritic diseases in nonhuman primates. Arthritis Res Ther. 2005;7:145–154. doi: 10.1186/ar1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel TU, Evans DT, Urvater JA, O’Connor DH, Hughes AL, Watkins DI. Major histocompatibility complex class I genes in primates: co-evolution with pathogens. Immunol Rev. 1999;167:327–337. doi: 10.1111/j.1600-065X.1999.tb01402.x. [DOI] [PubMed] [Google Scholar]
- Wallny HJ, Avila D, Hunt LG, Powell TJ, Riegert P, Salomonsen J, et al. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci U S A. 2006;103:1434–1439. doi: 10.1073/pnas.0507386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, et al. A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics. 2007;89:402–412. doi: 10.1016/j.ygeno.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Watkins DI, McAdam SN, Liu X, Strang CR, Milford EL, Levine CG, et al. New recombinant HLA-B alleles in a tribe of South American Amerindians indicate rapid evolution of MHC class I loci. Nature. 1992;357:329–33. doi: 10.1038/357329a0. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, et al. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]