Abstract
Gain-of-function NOTCH1 mutations are found in 50%–70% of human T cell acute lymphoblastic leukemia/lymphoma (T-ALL) cases. Gain-of-function NOTCH1 alleles that initiate strong downstream signals induce leukemia in mice, but it is unknown whether the gain-of-function NOTCH1 mutations most commonly found in individuals with T-ALL generate downstream signals of sufficient strength to induce leukemia. We addressed this question by expressing human gain-of-function NOTCH1 alleles of varying strength in mouse hematopoietic precursors. Uncommon gain-of-function NOTCH1 alleles that initiated strong downstream signals drove ectopic T cell development and induced leukemia efficiently. In contrast, although gain-of-function alleles that initiated only weak downstream signals also induced ectopic T cell development, these more common alleles failed to efficiently initiate leukemia development. However, weak gain-of-function NOTCH1 alleles accelerated the onset of leukemia initiated by constitutively active K-ras and gave rise to tumors that were sensitive to Notch signaling pathway inhibition. These data show that induction of leukemia requires doses of Notch1 greater than those needed for T cell development and that most NOTCH1 mutations found in T-ALL cells do not generate signals of sufficient strength to initiate leukemia development. Furthermore, low, nonleukemogenic levels of Notch1 can complement other leukemogenic events, such as activation of K-ras. Even when Notch1 participates secondarily, the resulting tumors show “addiction” to Notch, providing a further rationale for evaluating Notch signaling pathway inhibitors in leukemia.
Introduction
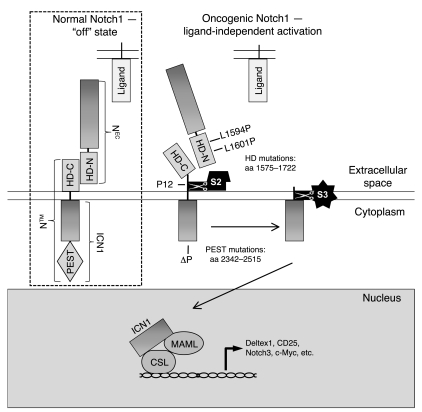
Notch1 belongs to a unique family of type I transmembrane receptors that regulate differentiation, proliferation, and survival in a dose- and context-dependent fashion (reviewed in ref. 1). During transit to the cell surface, Notch1 is cleaved by a furin-like protease at site S1 just external to the transmembrane domain, yielding a mature heterodimeric receptor that consists of noncovalently associated extracellular (NEC) and transmembrane (NTM) subunits (2). NEC consists of 36 iterated EGF-like repeats that bind Notch ligands; 3 iterated Lin-12/Notch repeats (LNRs); and a conserved approximately 100-amino-acid region that lies just N-terminal of the furin cleavage site. This last region binds the approximately 70-amino-acid extracellular portion of NTM that lies between the furin cleavage site and the transmembrane domain. Together, the 2 regions flanking the furin cleavage site (designated HD-N and HD-C, respectively) constitute the Notch heterodimerization domain (HD), which maintains a stable association between the NEC and NTM subunits when the receptor is in the off state (Figure 1) (3). The intracellular portion of NTM includes 7 ankyrin-like repeats, a strong transcriptional activation domain, and a C-terminal PEST sequence that regulates protein stability.
Figure 1. Schematic of normal and oncogenic Notch1 signaling.
In the absence of ligand, Notch is locked in an “off” state, which blocks S2 cleavage. However, mutations that disrupt the integrity of the HD domain (e.g., L1594P, L1601P, and P12) permit ligand-independent S2 cleavage, leading to formation of ICN, which translocates to the nucleus, where it engages CSL and mastermind-like (MAML) to form a transcriptionally active complex. PEST mutations (represented by ΔP) improve protein stability by removing negative regulatory elements in the C terminus that shorten the half-life of ICN. Transcriptional activation in the hematopoietic system (previously described for a construct containing only ICN) leads to the Notch GOF phenotype that includes ectopic BM T cell development, circulating CD4+CD8+ (DP) T cells, induction of T cell leukemia, suppression of B lymphoid development, and suppression of myeloid development.
Ligand binding initiates a series of additional enzymatic cleavages that activate Notch1 signaling (reviewed in ref. 1). An ADAM metalloprotease first cleaves the HD-C portion of the NTM subunit at site S2, leaving a short-lived membrane-bound intermediate (NTM*) (4). A multiprotein complex with γ-secretase activity then cleaves NTM* within its transmembrane domain at site S3, releasing the intracellular domain of Notch1 (ICN1), which translocates to the nucleus (5). Here, ICN1 activates transcription by associating with the DNA-binding transcription factor CSL (RBPJ) (6–9), creating a dual-binding surface that recruits coactivators of the mastermind-like family (MAML1–3) (10). ICN1 is short lived due to the presence of multiple degron motifs within its C-terminal PEST domain that mark it for proteosomal destruction (11–19).
Ligand-independent activation of Notch leads to gain-of-function (GOF) phenotypes in organisms ranging from Caenorhabditis elegans to humans (1). In mice, retroviral expression of ICN1 in BM progenitors drives thymus-independent T cell development at the expense of B cell development and rapidly and efficiently induces T cell acute lymphoblastic leukemia/lymphoma (T-ALL) (20, 21). In terms of the levels of ICN1 that are produced, this model is reminiscent of human T-ALLs that are associated with a t(7;9) chromosomal translocation that juxtaposes 3ι regions of NOTCH1 and TCRB gene enhancer/promoter elements. The resulting chimeric TCRB/NOTCH1 fusion gene drives high-level expression of truncated, constitutively active, ICN1-like polypeptides (22). However, translocations involving Notch1 in human T-ALL are rare. Much more commonly, human T-ALLs harbor acquired GOF point mutations, which occur predominantly within the extracellular HD and the C-terminal PEST domains (23). Such mutations are found in more than 50% of human T-ALLs and may occur either alone or in combination in cis. HD mutations, which include amino acid substitutions and short in-frame insertions and deletions, most often destabilize the HD domain and cause ligand-independent S2 cleavage (24, 25), whereas PEST domain mutations introduce premature stop codons or cause frameshifts that lead to the loss of C-terminal degron sequences (11, 17–19, 23). These types of Notch1 mutations occur in all of the major molecular subtypes of human T-ALL subtypes and many T-ALL–prone genetic backgrounds in mice (reviewed in ref. 1), suggesting a central role for aberrant Notch1 signaling in T-ALL that is not easily phenocopied by other oncogenes.
The outcome of Notch signaling is highly dose dependent in many developmental contexts, such as hematopoietic progenitors (26–30), making it probable that dosage has an impact on pathophysiologic processes, such as leukemogenesis, as well. Notch1 receptors bearing PEST deletions alone have little intrinsic signaling activity, while receptors bearing various HD mutations generate between 1% and 10% of the activity of constructs encoding ICN1 as judged by reporter gene assays (ref. 23 and our unpublished observations). Even constructs bearing dual HD/PEST mutations (a combination that is found in about 15%–20% of pediatric T-ALLs) have levels of activity well below those of constructs encoding ICN1. Mouse modeling of Notch-associated T-ALL has thus far been limited predominantly to enforced expression of ICN1 from strong promoters, which causes the rapid induction of T-ALL. The levels of ICN1 activity generated in such models may approximate those found in rare human T-ALLs associated with the t(7;9) chromosomal translocation but may not be relevant to human cases associated with more common, weaker GOF Notch1 mutations.
It is thus an open question whether the levels of ICN1 activity generated by the mutations found in human T-ALL are sufficient to initiate T-ALL, or whether Notch1 GOF mutations are generally complementary events in this cancer. The distinction may be clinically relevant, as it has been suggested that initiating lesions that uniformly participate in the establishment of the transformed state (e.g., the Bcr-Abl fusion kinase in chronic myeloid leukemia) are the most promising therapeutic targets. To investigate this issue, we scored a panel of Notch1 mutations in a robust BM transplantation (BMT) assay that permits evaluation of various forms of Notch with respect to effects on hematopoiesis, T-ALL induction, and oncogenic complementation. Our data suggest that the typical Notch1 mutations found in human T-ALL generate insufficient ICN1 to initiate leukemia efficiently but nevertheless interact with an activated form of K-ras to accelerate T-ALL development. Importantly, T-ALLs that develop in the context of K-ras activation retain a dependency on Notch signals for growth and survival, indicating that an initiating role for Notch is not essential for the development of Notch pathway “addiction.”
Results
Selection of Notch1 GOF mutations.
Four Notch1 mutations, 3 involving the HD and 1 involving the PEST domain, were selected for detailed study (Table 1). Two of the HD mutations, L1594P and L1601P, are relatively common mutations in human T-ALL that destabilize the HD (3). The third HD mutation is a 13-amino-acid insertion identified in the cell line P12-Ichikawa (P12). This insertion creates a “deprotected” S2 cleavage site that is susceptible to ligand-independent proteolysis (3). The single PEST domain mutation selected is a frameshift identified in the ALL-SIL T-ALL cell line, in which amino acids 2,473–2,555 are deleted (ΔP). The missing region contains multiple residues implicated in ICN1 degradation, including WSSSSP (amino acids 2,521–2,526) (11), a sequence targeted by Fbw7-E3 ligase complexes (amino acids 2,510–2,516) (18, 19), and serine residues that are putatively targeted by CDK8 phosphorylation for degradation (S2514, S2517, and S2538; ref. 17).
Table 1 .
Notch mutations selected for mouse modeling studies
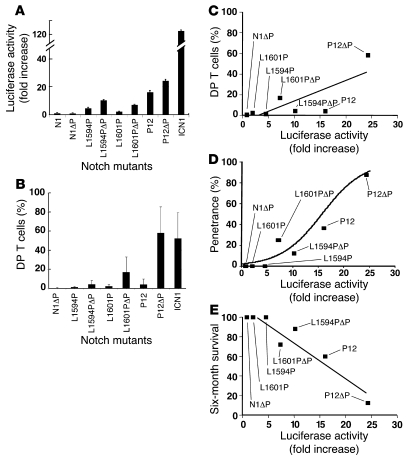
Consistent with previous reports (23, 24), each of the 3 HD mutations increased the activation of a Notch-dependent luciferase reporter gene, with P12 having the greatest potency (Figure 2A). ΔP alone had no effect on Notch1 signaling, but when present in cis with each of the 3 Notch1 HD mutations, it caused further increases in Notch1 activation (Figure 2A). The level of Notch1 activation caused by each of these constructs was significantly lower than that generated by ICN1.
Figure 2. Relative strength of Notch1 receptors bearing T-ALL–associated mutations.
(A) U2OS cells were transfected in triplicate with empty pcDNA3 plasmid or pcDNA3 plasmids encoding various mutated forms of human Notch1 (10 ng/well), the pGL2-CSLx4-luciferase reporter plasmid (250 ng/well), and a Renilla luciferase internal control plasmid (5 ng/well). Firefly luciferase activity was normalized to Renilla luciferase activity in cell lysates prepared 44–48 hours after transfection and then normalized to empty pcDNA3 vector activity, which was arbitrarily set to 1. N1 is normal full-length human Notch1. (B) Forms of Notch1 bearing T-ALL–associated mutations induce DP T cell development with varying degrees of strength. Lethally irradiated mice were reconstituted with 5-FU–treated donor BM cells transduced with various Notch1 alleles. The percentages of DP T cells in the GFP-positive compartment at 6 weeks after transplantation are charted with respect to each Notch mutant. See Figure 3 for representative profiles. (C) Mean percentage of DP T cells at 6 weeks after BMT in the GFP+ compartment plotted against reporter activity for each mutant. Linear regression performed at the individual mouse level calculated r = 0.5633 and P < 0.0001. (D) Mean percentage of leukemic penetrance at 1 year plotted against the reporter activity for each mutant. Logistical regression calculated P < 0.001 and odds ratio, 1.28 (95% CI, 1.15–1.43). (E) Survival rate (percentage leukemia-free at 6 months) plotted against the reporter activity for each mutant. Error bars represent single SDs of the mean.
Leukemia-associated Notch1 mutations induce ectopic T cell development and perturb hematopoiesis in mice in a dose-dependent manner. Our panel of mutated Notch1 cDNAs (Table 1) was cloned into the MigR1 vector, which expresses genes of interest and GFP from a bicistronic RNA containing an internal ribosomal entry site (31). cDNAs in MigR1 were packaged into ecotropic retroviruses and transduced into adult murine BM progenitors. Reconstituted animals were bled 6 weeks after BMT to assess the presence of CD4+CD8+ double-positive (DP) T cells in the peripheral blood, an aberrant cell population created by Notch1-induced ectopic T cell development in the BM (20) (Figure 2B and Figure 3). At this time point, T-ALL had not yet developed but all hematopoietic lineages had been reconstituted. L1594P, L1601P, and P12 induced DP T cell development. ΔP alone did not promote DP T cell development but augmented DP T cell development when present in cis with a HD mutation. The P12ΔP mutation generated as many DP T cells as ICN1, suggesting that its strength is sufficient to cause maximal GOF in this assay. Statistical analysis revealed a significant correlation between the effect of these mutations on ectopic T cell development and their relative signal strength in reporter gene assays (Figure 2C; r = 0.56, P < 0.0001).
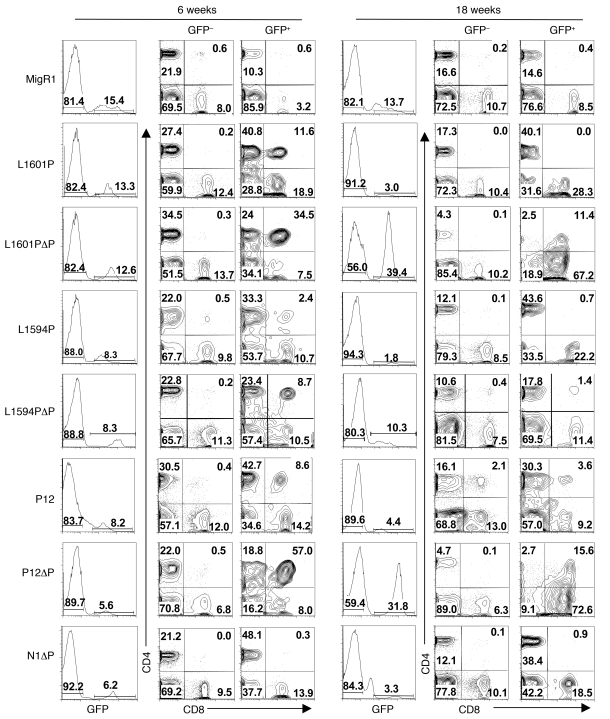
Figure 3. Differential effects of Notch1 receptors of varying strength to induce ectopic T cell development and T cell leukemia.
Lethally irradiated mice were reconstituted with 5-FU–treated donor BM cells transduced with various Notch1 alleles or GFP alone (MigR1). GFP percentages in the blood are shown at early (6 weeks) and late (18 weeks) time points after BMT. CD4/CD8 profiles are shown for both GFP+ and internal control GFP– populations. Note that GFP percentages and DP T cell generation fell at 18 weeks except in leukemic mice; shown here are examples of leukemic L1601PΔP and P12ΔP mice. Representative profiles are shown. Numbers within scatter plots refer to percentages of live gated cells. Experiments were performed at least twice.
By 18 weeks after BMT, the percentage of both GFP-expressing cells and DP T cells declined in nonleukemic mice, whereas these parameters increased further in mice that developed T-ALL (Figure 3). GFP+ cells in nonleukemic mice disappeared from the peripheral blood by 24 weeks after BMT (data not shown). This phenotype is similar to that observed with another relatively weak GOF form of Notch1, ΔEGFΔLNR, which bears a deletion that “deprotects” the S2 cleavage site in a fashion similar to the HD mutations (3, 11). The disappearance of GFP+ cells in these mice is causally related to an increase in Notch signaling, since GFP+ cells persist indefinitely in mice reconstituted with progenitors transduced with empty MigR1 virus (data not shown). Preliminary analysis suggests a renewal defect in a multipotential stem cell compartment that manifests in the periphery at late time points (data not shown). Thus, analysis at the early, 6-week time point provided a window into the effects of Notch GOF on hematopoiesis uncomplicated by effects on precursor stem cells or by leukemia that confounds analyses at later time points.
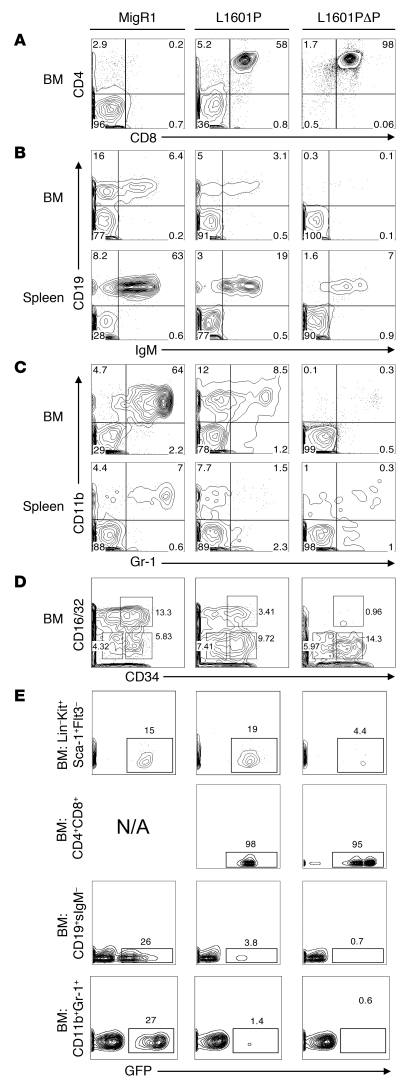
Additional studies were performed with L1601P and L1601PΔP cDNAs to more completely assess the effects of weak and moderately strong Notch1 GOF alleles on hematopoiesis at the 6-week time point. Strong Notch1 GOF alleles, such as ICN1, promote T lineage commitment at the expense of B lineage commitment (20) and myeloid development (32). At 6 weeks after BMT, flow cytometric analysis of BM and spleen cells showed that L1601P promoted DP T cell development (Figure 4A), which was also reflected in the peripheral blood (Figure 3). L1601P also inhibited B cell development and mature B cell production (Figure 4B) and inhibited myeloid development and mature granulocyte production (Figure 4C). Each of these effects was enhanced by addition of the ΔP mutation in cis. The observed block in myelopoiesis with both forms of Notch1 occurred after the common myeloid progenitor (CMP) stage, prior to transition to the granulocyte-macrophage progenitor stage (GMP) (Figure 4D). The enhanced T cell generation at the expense of B lymphoid and myeloid cell development was confirmed by the finding that the percent GFP reconstitution was higher in the DP T cell compartment and lower in the B lymphoid and myeloid compartments relative to the HSC compartment (Figure 4E). These data indicate that even weak GOF HD domain mutations have the capacity to alter lymphopoiesis and myelopoiesis and that the degree of alteration correlates with signal strength.
Figure 4. Effect of Notch1 receptors bearing mutations of varying strength on myeloid and lymphoid development at 6 weeks after BMT.
Lethally irradiated mice were reconstituted with 5-FU–treated donor BM cells transduced with empty MigR1 (negative control), L1601P, or L1601PΔP. Mice were sacrificed 6 weeks after BMT, and the GFP+ BM and spleen compartments were analyzed by flow cytometry for CD4+CD8+ T cells (A); precursor, immature, and mature B cells (B); Gr-1+CD11b+ granulocytes (C); and the Lin–Sca-1–Kit+ subpopulation (D), which is subdivided into megakaryocyte-erythroid progenitors (MEPs) (CD16/32+/–CD34–), granulocyte-macrophage progenitors (GMPs) (CD16/32+CD34+), and common myeloid progenitors (CMPs) (CD16/32+/–CD34+). (E) Comparison of the percentage of GFP expression in different BM hematopoietic subsets (HSCs, Lin–Kit+Sca-1+Flt3–; DP T cells; pro/pre-B cells, CD19+surface IgM– [CD19+sIgM–]; and granulocytes, CD11b+Gr-1+) for each mouse is shown. N/A, not applicable. Side scatter is plotted on the y axis. Numbers within scatter plots refer to the percentages of live gated cells, except in D, where the numbers refer to the percentages of Lin–Sca-1–Kit+ cells, and E, where the numbers refer to the percentages of cells in the indicated hematopoietic subsets that were positive for GFP expression. Representative plots are shown. Experiments were performed twice.
Leukemia-associated Notch1 mutations induce murine T-ALL in a dose-dependent manner.
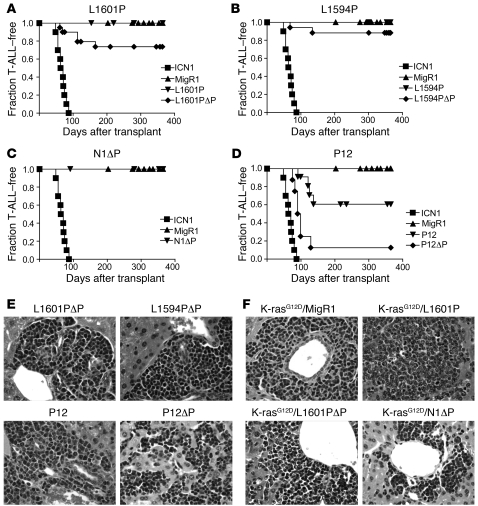
Strong Notch1 GOF alleles, such as ICN1, uniformly and efficiently induce T-ALL when expressed in BM progenitors (21, 33). In contrast, Notch1 cDNAs bearing our test panel of mutations showed a more limited ability to induce T-ALL that again correlated with signal strength (Figure 5). None of the L1601P, L1594P, or Notch 1 with ΔP (N1ΔP) mice developed T-ALL in 2 or more cohorts of at least 8 animals each (Figure 5, A–C). One N1ΔP mouse in 2 cohorts of 16 transplanted mice developed T-ALL at approximately 12 weeks after transplantation. However, examination of this tumor by Southern blot analysis showed a rearrangement in the provirus that is predicted to lead to the production of ICN1ΔP-like polypeptides (data not shown); as a result, this animal was removed from the N1ΔP cohort. The strongest HD mutation, P12, induced T-ALL with a penetrance of approximately 36% (Figure 5D). Combining ΔP in cis with each of the HD mutations increased leukemogenesis. Specifically, combining ΔP with L1601P or L1594P raised the incidence of T-ALL from 0% to 25% and 12.5%, respectively, while combining ΔP with P12 increased the incidence of T-ALL from 36% to 88%. None of the mutations tested induced T-ALL as rapidly or with as high a penetrance as ICN1, which caused T-ALL in all mice. Based on logistic regression analysis, leukemic penetrance at 1 year was significantly associated with the reporter activity of each mutation (Figure 2D; odds ratio, 1.28; 95% CI, 1.15–1.43; P < 0.001). Fold increase in stimulation in the reporter assays was inversely associated with survival time in the accelerated failure time regression model (Figure 2E; P < 0.0001). Thus, in our panel of mutated receptors, there was a significant association between leukemia induction (penetrance or survival rate) and Notch1 signal strength in reporter gene assays.
Figure 5. Notch1 GOF allele strength and T-ALL induction.
Lethally irradiated mice were reconstituted with 5-FU–treated donor BM cells that were transduced with the indicated Notch1 alleles. Kaplan-Meier graphs show the fraction of mice without T-ALL as a function of time. Only tumors having intact proviral integrants were included in the analysis. MigR1 and ICN1 mice were negative and positive controls, respectively, that were compared with L1601P and L1601PΔP (A); L1594P and L1594PΔP (B); N1ΔP (C); and P12 and P12ΔP (D). Individual cohorts contained 6–12 mice transduced with each allele, and each experiment was performed at least twice. Representative tissue sections showing Notch-associated T-ALLs infiltrating the liver (H&E stain) are shown in E and F. Original magnification, ×400.
The presentation and characteristics of the T-ALLs resembled those previously described for T-ALLs induced with ICN1 (21, 31). Proviral integrants (except for the outlier N1ΔP tumor already mentioned) were of the expected size (data not shown) and were 1–3 in number (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI35090DS1). Physical findings included splenomegaly (Supplemental Table 1) and lymphadenopathy. All tumors exhibited immunophenotypic and immunohistochemical features compatible with an immature T cell phenotype (Figure 5E and data not shown). Tumors were readily serially transferred to secondary recipients, and Western blot analyses revealed the presence of Notch1 polypeptides of the expected size (data not shown).
The nonleukemogenic HD mutant L1601P poorly induces c-Myc transcription.
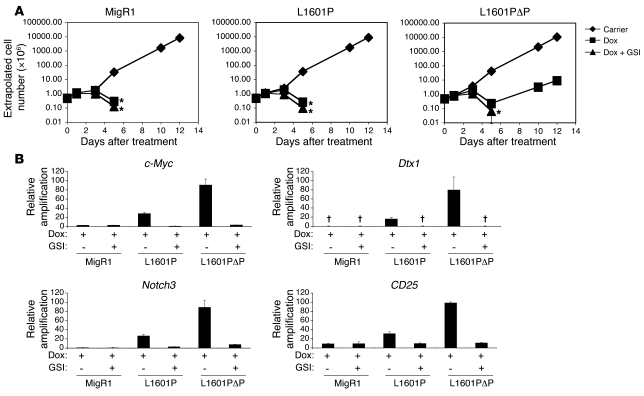
The results obtained with mutated forms of Notch1 of varying strength suggest that there is a threshold level of signal intensity required for efficient induction of T-ALL. Recent observations identified c-Myc as a direct downstream target of ICN1 that plays an important role in Notch-dependent T-ALL induction and growth maintenance (34–36). For example, enforced expression of c-Myc can induce T-ALL (37–39) and rescue multiple Notch1-dependent T-ALL cell lines from Notch pathway inhibition (34, 35, 40). To explore the relationship among Notch dosage, c-Myc expression, and T-ALL cell growth, we compared the effects of the nonleukemogenic L1601P mutant and the modestly leukemogenic L1601PΔP mutant in 8946 cells. This cell line is derived from a murine T-ALL induced with a human c-Myc transgene under the control of the doxycycline-dependent transactivator. 8946 cells undergo apoptosis upon doxycycline withdrawal but can be rescued from death by transduction of strong GOF forms of Notch1, such as ICN1, which upregulate the expression of endogenous murine c-Myc (34). We observed that L1601PΔP rescued 8946 cells treated with doxycycline, whereas empty MigR1 and L1601P did not (Figure 6A). The rescue by L1601PΔP was prevented by treatment with a γ-secretase inhibitor (GSI), implicating Notch1 signaling in this activity. The observed functional difference correlated with the ability to induce the expression of endogenous murine c-Myc, which was most strongly upregulated by L1601PΔP, only weakly upregulated by L1601P, and unaffected by MigR1 (Figure 6B). Similarly, L1601PΔP induced higher levels of expression of other Notch1 target genes, such as Dtx1, Notch3, and CD25, relative to L1601P (Figure 6B). Thus, these data suggest that relatively strong doses of Notch1 are required to achieve sufficient activation of certain targets downstream of Notch1, such as c-Myc, that appear to play a critical role in T-ALL oncogenesis
Figure 6. Differential ability of L1601P and L1601PΔP to induce c-Myc expression and to rescue the growth of c-Myc–dependent 8946 cells.
8946 cells were transduced with empty MigR1 (negative control), L1601P, or L1601PΔP. Two days later, sorted GFP+ cells were treated with DMSO carrier, doxycycline (Dox; 20 ng/ml), or doxycycline and 1 μM JC19 (GSI) in triplicate. (A) Cell numbers were measured (×106 cells/ml) using a logarithmic scale and were extrapolated over time. Conditions in which the cell number dropped below 0.1 × 106 cells/ml failed to yield any viable cells over the 12 days of culture. *No live cells detected after this time. (B) Eighteen hours after treatment, RNA was harvested and assayed for murine c-Myc, Dtx1, Notch3, and CD25 expression with real-time PCR. Target gene amplification normalized to Hprt amplification is shown relative to the signal generated by doxycycline-treated cells transduced with L1601PΔP. †No transcript detected. Error bars represent single SDs of the mean.
The leukemogenicity of the oncogene K-rasG12D is enhanced cooperatively by the nonleukemogenic allele L1601P.
In T-ALL–prone mice, weak GOF Notch1 mutations, such as deletions removing the PEST domain, are common secondary events (1, 41–43), suggesting that the dose of Notch1 that is required for oncogenic cooperativity is lower than that needed for tumor initiation. To study this possibility, L1601P, N1ΔP, and L1601PΔP were transduced into hematopoietic progenitor cells engineered to express a constitutively active form of K-ras in T lineage progenitors. In addition to long-standing developmental genetic evidence of complex crosstalk between Notch and Ras (reviewed in ref. 44), insertional mutagenesis screens have also identified the Notch and Ras signaling pathways as likely collaborators in the induction of T-ALL (45), providing a rationale for the use of this model. Furthermore, NRAS or KRAS mutations occur in about 15% of human ALL samples (46–48), and tumor samples from Notch mutant mice were found to have active Ras/MAPK signaling (Supplemental Figure 2A and Supplemental Figure 3).
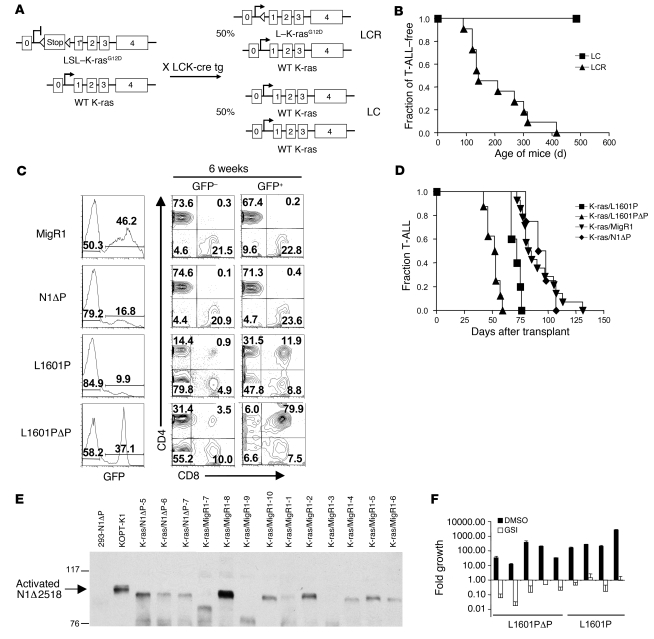
The mouse strain used, LSL–K-rasG12D, has a K-rasG12D allele knocked into the endogenous K-ras locus downstream of a floxed stop cassette (49) (Figure 7A). In the presence of Cre recombinase, the stop cassette is removed, allowing expression of the constitutively active K-rasG12D mutation. Heterozygous LSL–K-rasG12D mice were mated with homozygous mice expressing Cre recombinase under the control of the proximal Lck promoter (LCK-Cre mice) to generate offspring expressing either K-rasG12D (LCR mice) or normal K-ras (LC mice) in the T lineage. LCR mice develop T-ALLs at a median of approximately 180 days after birth (Figure 7B) that resembled those produced by retrovirally expressed or transgenic H-ras (41, 50, 51). BM progenitors from K-rasG12D–expressing LCR mice were transduced with MigR1, L1601P, N1ΔP, or L1601PΔP and then transferred to lethally irradiated LC littermates. At 6 weeks after BMT, flow cytometric analyses of peripheral blood showed that L1601P- and L1601PΔP-transduced progenitors generated circulating GFP+ DP T cells, whereas MigR1 and N1ΔP-transduced progenitors did not (Figure 7C); similar results were obtained in the wild type (non–K-ras) background (Figure 3). The L1601P and L1601PΔP alleles significantly shortened the median time to leukemic presentation (from ~84 days to ~72 and ~53 days, respectively), as compared with empty MigR1 virus (Figure 7D and Supplemental Table 1). In addition, at presentation, the spleens of the L1601P and L1601PΔP mice were larger and the white blood cell counts were higher than in MigR1 control or N1ΔP mice (Supplemental Table 1). All affected mice displayed splenomegaly (Supplemental Table 1), lymphadenopathy, and widespread organ infiltration (data not shown). The tumors showed intact proviral integrations (Supplemental Figure 4A); were monoclonal or oligoclonal (Supplemental Figure 4B); expressed Notch1 polypeptides of the expected sizes (data not shown); and were composed of immature T cells, based on pathology (Figure 5F) and flow cytometry (Supplemental Figure 5). In all tumors, the stop cassette was excised (Supplemental Figure 6) and ERK was phosphorylated (Supplemental Figure 2B), confirming Ras activation.
Figure 7. The nonleukemogenic HD mutant L1601P accelerates the development of T-ALL induced by the K-rasG12D oncogene.
(A) Heterozygous LSL–K-rasG12D mice were bred to homozygous LCK-Cre transgenic mice to generate mice expressing K-rasG12D in the T cell lineage (LCR) and littermate control mice not expressing K-rasG12D (LC). (B) Kaplan-Meier graph showing that LCR mice spontaneously develop T-ALL with 100% penetrance and a median latency of approximately 180 days, while LC control mice do not. (C) Lethally irradiated LC recipient mice were reconstituted with 5-FU–treated donor LCR BM cells after transduction with empty MigR1 (negative control), L1601P, L1601PΔP, or N1ΔP. Mice were bled 6 weeks after transduction and evaluated for circulating DP cells. Numbers within scatter plots refer to the percentages of live gated cells. (D) Kaplan-Meier graph shows the fraction of mice developing T-ALL over time. K-ras/L1601P: P = 0.005, K-ras/L1601PΔP: P < 0.0001, K-ras/N1ΔP: P = 0.802 versus K-ras/MigR1. (E) Western blot for activated Notch1 antibody (anti-V1744) shows activated, truncated forms of Notch1 in both K-ras/N1ΔP mice and K-ras/MigR1 mice. Extract from 293T cells transiently transfected with N1ΔP is shown as a negative control. Extract from the T-ALL cell line KOPT-K1, which contains both an HD mutation and a PEST deletion (Δ2518), is shown as a positive control. (F) Primary cells cultured from tumors that developed in mice reconstituted with K-ras/L1601P or K-ras/L1601PΔP BM cells were treated with 1 μM JC19 (GSI) or DMSO carrier in triplicate wells and measured for fold cell growth (relative to initial cell number) after 6 days. Cell counts were extrapolated. In experiments involving mice, at least 5 mice were present in each test group, and each experiment was performed twice. Error bars represent single SDs of the mean.
In contrast, transduction of N1ΔP did not significantly shorten the mean time to leukemic presentation, despite the expression of N1ΔP (as judged by Western blotting [data not shown] and immunohistochemistry [Supplemental Figure 7]) and the presence of low levels of ICN1ΔP in K-rasG12D/N1ΔP tumor cells (as assessed by Western blotting; Figure 7E). Flow cytometric analyses showed that all N1ΔP tumors were GFP+, while only a subset of tumors in MigR1 animals expressed GFP (data not shown), which may indicate that cells expressing N1ΔP have a weak selective advantage. Of note, even high-level expression of N1ΔP in 293T cells did not lead to detectable Notch activation (Figure 7E), suggesting that extrinsic (ligand-mediated) or intrinsic (HD mutation) mechanisms are activating N1 in the N1ΔP tumors. To rule out acquired mutations within the MigR1 proviruses as a mechanism for the generation of ICN1ΔP in K-rasG12D/N1ΔP tumor cells, we cloned 32 independent amplicons from 4 N1ΔP tumors spanning the proviral regions encoding exons 25–28 (which encode the LNRs, HD regions, and transmembrane domain). Bidirectional sequencing of 8 clones from each tumor did not reveal mutations (data not shown). Since these tumors contained at most 3 proviruses, the odds that a provirus will not be represented at least once in these reads is at most (2/3)8, or 3.9%. These data suggest that extrinsic mechanisms (such as Notch ligands) mediate Notch activation in the N1ΔP tumors; however, the possibility of an effect of mutations outside of the exon 25–28 region cannot be excluded.
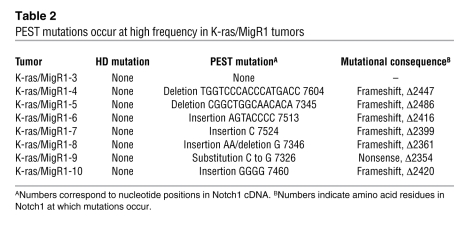
The ability of weak GOF forms of Notch1 to cooperate with K-rasG12D suggested that tumors arising after MigR1 transduction might also have acquired spontaneous activating mutations involving murine Notch1. This was initially evaluated by Western blot analysis using an antibody specific for the neoepitope at the N terminus of ICN1, which showed that a high fraction of K-rasG12D/MigR1 tumors contained C-terminally truncated forms of ICN1 (Figure 7E). DNA sequencing confirmed the presence of heterozygous frameshift or stop codon mutations involving Notch1 exon 34 (which encodes the PEST sequence) in 7 of 8 (88%) tumors analyzed (Table 2). Each of these mutations is predicted to cause the expression of abnormally stable truncated forms of ICN1 that lack conserved negative regulatory motifs, including the Fbw7 degron, the WSSSSP motif, and CDK8 phosphorylation sites. Thus, activated K-ras joins the list of murine T-ALL models in which Notch1 mutations have a complementary role.
Table 2 .
PEST mutations occur at high frequency in K-ras/MigR1 tumors
Interactions between the Notch and Ras pathways have long been recognized in multiple developmental and cellular contexts, including oncogenesis, but the mechanism that underlies their cooperation in cancer remains poorly defined. To explore potential mechanisms, we developed a T-ALL cell line–based assay that models the cooperativity between Notch and Ras observed in the K-rasG12D/Notch T-ALLs. 8946 T-ALL cells were cotransduced with both a Notch-expressing retroviral vector (which coexpressed GFP as a surrogate marker) and a K-rasG12D–expressing retroviral vector (which coexpressed a truncated version of the nerve growth factor receptor [NGFR] as the surrogate marker) and then treated with doxycycline for 6 days to withdraw expression of transgenic human c-Myc. The addition of K-rasG12D enhanced the rescue of the MigR1-, L1601P-, or L1601PΔP-transduced cells relative to control treatment (Supplemental Figure 8A), thus confirming that this system models Ras/Notch cooperativity. One report suggests that the Ras pathway activates the Notch pathway by increasing Delta-1 and presenilin-1 expression, which presumably facilitates S3 cleavage (52). To test the possibility that the K-rasG12D allele augments Notch1 activation, we explored the effect of K-rasG12D on the signal strength of L1601P and L1601PΔP in 8946 cells. We found that the K-rasG12D allele did not influence the level of upregulation of Notch target genes such as c-Myc or Dtx1 (Supplemental Figure 8B), whereas activation of the Ras/MAPK pathway was seen, as expected (Supplemental Figure 8C). Thus, we found no evidence for “supra-activation” of the Notch pathway in the presence of K-rasG12D.
A more refined interpretation of the role of Notch1 dosage in leukemogenesis requires assessment of the relative levels of Notch1 expression driven by the MigR1 retroviral promoter and the endogenous Notch1 promoter. We first approached this question by performing immunohistochemistry with a previously characterized antibody against the intracellular domain of Notch1 that works well in paraffin-embedded tissues (18). Although there was substantial variation among tumors (which is to be expected, given that each harbors different proviral insertions), on average, Notch1 staining was stronger in tumors in which expression was retrovirally driven (Supplemental Figures 7, 9, and 10) than in K-rasG12D/MigR1 tumors (Supplemental Figures 7 and 9). We also noted that the levels of activated Notch1 (ICN1) in K-rasG12D/MigR1 tumors and K-rasG12D/N1ΔP tumors were roughly comparable (Figure 7E), despite the fact that N1ΔP is the weakest Notch1 allele scored in our assay. Finally, we compared the levels of retroviral N1ΔP expression and endogenous Notch1 expression directly in extracts prepared from K-ras/N1ΔP tumors (Supplemental Figure 11). The ΔP deletion is sufficiently large to distinguish the 2 different NTM subunits created by furin cleavage at site S1, NTMΔP (derived from the provirus) and NTM (derived from the endogenous Notch1 alleles). Moreover, since the ectodomains of these 2 proteins are identical and N1ΔP is a very weak allele (and therefore the least likely of any allele tested to influence endogenous Notch1 expression), retroviral N1ΔP is likely to provide the best comparison with endogenous Notch1 expression. Western blotting revealed that the levels of NTMΔP were consistently several-fold higher than the levels of NTM (Supplemental Figure 11). Together, these findings suggest that retroviral expression is unlikely to underestimate (and may overestimate) the leukemogenic potential of the various Notch1 alleles tested to date.
T cell tumors initiated by K-ras and accelerated by the nonleukemogenic L1601P allele acquire a dependence on continued Notch1 signaling. Current therapeutic strategies are focused on targeting initiating oncoproteins based on the belief that tumors are most likely to be addicted to signaling alterations that occur early in tumor development. To test this idea in the K-rasG12D–initiated T-ALL model, we treated a panel of 5 independent K-rasG12D/L1601PΔP tumors and 4 independent K-rasG12D/L1601P tumors ex vivo with DMSO carrier, the PI3K inhibitor LY294002, or the MAPK inhibitor U0126. Both of these inhibitors, which target different nodes of the signaling pathways downstream of activated K-ras, abrogated tumor growth (Supplemental Figure 11), indicating that these tumors (as expected) depend on activated K-ras. Since these inhibitors may have off-target effects, we cannot rule out the possibility that these Ras-initiated tumors may partly depend on additional pathways. Because HD mutations such as L1601P lack the strength to initiate T-ALL oncogenesis in mice, it was unclear whether the K-ras/L1601P tumors would also develop a dependency on Notch1 signals for growth. To address this, tumor cells isolated from the K-ras/L1601PΔP mice and K-ras/L1601P mice were treated in short-term cultures with GSI or DMSO carrier (Figure 7F). GSI abrogated the growth of both types of tumors, thus confirming Notch addiction even in tumors where Notch1 is playing a collaborative oncogenic role.
Discussion
The finding that more than 50% of human T-ALLs are associated with Notch1 mutations has focused attention on Notch signaling as a central mediator of T-ALL pathogenesis. Although certain strong GOF Notch1 constructs, such as ICN1, initiate T-ALL in murine models efficiently, it was unclear whether the Notch1 GOF mutations that are commonly found in human T-ALL would behave similarly. This is because the diverse GOF Notch1 alleles found in human T-ALL (with rare exceptions) generate substantially lower levels of activated Notch1 than constructs previously evaluated in mouse models. The experiments described here show that, unlike ICN1, neither a set of 3 HD mutations nor a PEST mutation originally identified in human T-ALLs induced leukemia efficiently when expressed alone in murine BM progenitors. Pairing each HD mutation with the same PEST mutation in cis increased signaling activity synergistically to a level that was sufficient to induce T-ALL, but less rapidly and with a lower efficiency than ICN1. Importantly, the transforming activities of these alleles were compared using a BMT assay that allows us to precisely titer our viruses, which improves accuracy and reproducibility. Since the levels of Notch1 driven by the retroviral promoter are comparable to or greater than the levels of Notch1 driven by the endogenous promoter in K-ras–initiated T-ALLs, it is unlikely that our assay has underestimated the leukemia-initiating capability of these alleles.
In contrast to the inefficient induction of T-ALL, each HD mutation generated signals of sufficient strength to drive ectopic T cell development and inhibit myeloid and B cell development. Taken together, these data suggest the existence of at least 2 Notch signaling thresholds (Figure 8): a lower, “physiological” threshold for influencing hematopoiesis and a higher, “supraphysiological” threshold for induction of T-ALL. The precise molecular basis for these thresholds is unclear, but it seems likely that low doses of Notch1 fail to drive expression of one or more critical downstream targets to levels needed for efficient T-ALL induction. The important targets could be normal Notch1-regulated genes in T cell progenitors and/or genes that are only activated in the context of supraphysiological doses of Notch1. Although we are not yet able to distinguish between these possibilities, our data provide some early leads. Addition of a PEST mutation to the L1601P HD mutation increases the expression of c-Myc, a conserved direct target gene that may be activated by Notch1 in a context-specific manner (34, 35, 40, 53, 54). Notably, enforced expression of c-Myc induces T-ALL (37–39) and rescues multiple Notch1-dependent T-ALL cell lines from Notch1 pathway inhibition (34, 35, 40), suggesting that it is a particularly important determinant of Notch1-mediated oncogenic activity. Since c-Myc is induced by L1601PΔP, but not L1601P, the signaling threshold for c-Myc induction by Notch1 appears to be relatively high. Whether other leukemogenic targets are differentially induced by L1601P and L1601PΔP is unclear.
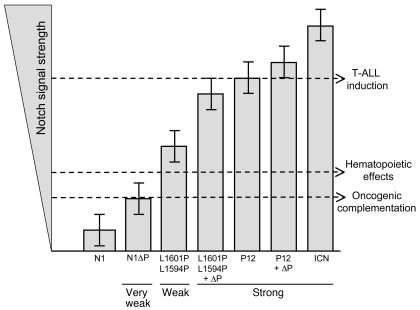
Figure 8. Proposed dose-dependent model for Notch involvement in hematopoiesis and T-ALL.
The lowest threshold is required for Notch1 to collaborate with initiating oncogenes, such as E2A-PBX1 (55), c-Myc (38), Bcr-Abl (56), and K-rasG12D. A middle threshold is required for Notch signaling to turn on targets that influence hematopoiesis. A third, supraphysiological threshold is required for Notch signaling to turn on targets sufficiently to initiate T-ALL. These thresholds divide Notch mutations into 3 major categories: (a) “very weak” alleles, such as N1ΔP, that collaborate with T-ALL initiators but fail to influence hematopoiesis or initiate T-ALL; (b) “weak” alleles that influence hematopoiesis and collaborate with T-ALL initiators but fail to initiate T-ALL; and (c) “strong” alleles that influence hematopoiesis and initiate T-ALL. Error bars signify the variability of expression of each mutant based on the random element inherent in retroviral integration and genetic background.
Assuming that human and murine BM progenitors are similar in their sensitivity to Notch1 signals, our results suggest that the majority of single Notch1 mutations associated with human T-ALL are unlikely to initiate T-ALL development. Such an activity may be limited to rare, exceptionally strong Notch1 mutations, such as the truncated Notch1 alleles created by the t(7;9) chromosomal translocation (21, 31) and the unusual P12 insertional HD mutation. Weaker GOF mutations that are more typical of human T-ALL, such as L1594P and L1601P (Figure 1 and ref. 24), initiate T-ALL inefficiently or not at all, even when combined in cis with a PEST mutation.
In contrast, doses of ICN1 that are insufficient to initiate T-ALL, such as those generated by L1594P and L1601P, appear to accelerate the induction of T-ALL by strong oncogenes, such as K-rasG12D. In fact, even very weak Notch1 alleles, such as N1ΔP, appear to generate a mild selective advantage when expressed in the context of activated K-ras. In line with our observations, other groups have found that N1ΔP alleles also complement T-ALL initiated by transgenic E2a/Pbx (55), c-Myc (38), and Bcr-Abl (56). These observations suggest that the dose of Notch that is required for complementation is substantially lower than that needed for T-ALL induction, and perhaps even lower than that needed to produce effects on hematopoiesis (Figure 8).
The timing of Notch1 mutations relative to other events that collaborate in the genesis of human T-ALL remains to be fully determined. With the exception of the t(7;9) translocation (57), which likely occurs during TCRβ rearrangement, the mutations in human T-ALL could occur in committed T cell progenitors following the acquisition of other oncogenic events or as an early event in a multipotent hematopoietic progenitor cell prior to T cell commitment. In support of the former scenario, Mansour et al. have reported that human T-ALLs sometimes have Notch1 mutations in only a minority (less than 10%) of blasts and may differ in their Notch1 mutational status at presentation and relapse (58), suggesting that, at least on occasion, Notch1 mutations are late events during the multistep genesis and progression of human T-ALL. Alternatively, in line with the results from our model, Eguchi-Ishimae et al. have shown that Notch1 HD mutations can sometimes be detected in peripheral blood cells at birth, prior to the acquisition of other oncogenic events and well in advance of the clinical appearance of childhood T-ALL (59). One novel, but relatively simplistic, possible explanation for disease acceleration in our model is that weak GOF Notch1 mutations divert BM progenitor cells toward the T cell fate and thereby expand the pool of T cell progenitors that are at risk for transformation by K-rasG12D. However, the dependence of T-ALLs expressing weak GOF Notch1 alleles on Notch signaling for continued growth and the strong selective pressure for acquisition of similar Notch1 mutations in sporadic human T-ALL and T-ALL–prone murine models indicate that even weak Notch signals have additional ongoing roles in the development and progression of T-ALL beyond effects on the size of at-risk pools. Primary among these oncogenic effects are upregulation of c-Myc (34, 35, 40), activation of mTOR (60), and increased flux through the PI3K/Akt signaling axis (61), all of which have been implicated in the sustained growth of T-ALL cells by Notch1.
The high prevalence of Notch1 mutations in T-ALL has provided a rationale for clinical trials of GSI in relapsed/refractory T-ALL. Our data suggest that even when Notch acts in a complementary fashion, tumors can acquire addiction to Notch. However, addiction may not be an “all-or-none” phenomenon. For example, the human T-ALL cell lines that are most sensitive to GSI either possess dual HD domain and PEST domain mutations lying in cis, a combination that produces a relatively high level of Notch1 activation (23), or have unusual t(7;9) translocations (36). It will be of interest to determine in preclinical models whether the relative level of ICN1 in Notch-dependent tumors is predictive of responsiveness to Notch pathway inhibitors. Finally, it is hoped that a more detailed understanding of the crosstalk that allows Notch to collaborate in T-ALL with many other oncogenes, such as K-ras, will allow for development of rational combinations of Notch pathway inhibitors and other targeted therapies.
Methods
Mice.
Four- to 8-week-old C57BL/6 mice were obtained from Taconic. LSL–K-rasG12D f/+ mice were a gift from David Tuveson (Cancer Research UK Cambridge Research Institute, Cambridge, United Kingdom) and were generated as described previously (49). Protocols for PCR-based genotyping and detection of Cre-mediated excision of loxP sites were performed as described previously (49). To activate K-rasG12D in the T lineage, LSL–K-rasG12D f/+ mice were crossed to Lck-Cre+/+ transgenic mice (Taconic). LSL–K-rasG12D–positive offspring, designated LCR mice, were used as donors for BMT studies, while littermate LSL–K-rasG12D–negative offspring, designated LC mice, were used as recipients. Donors were 4–12 weeks old and were divided into donor pools that were age matched for each experimental condition (11).
Constructs and retroviruses.
The control GFP-expressing MSCV-IRES-GFP construct (MigR1) has been described previously (62). High-titer retroviral supernatant was produced using transient transfection of 293T cells and assessed for GFP titer by plating on 3T3 fibroblasts (20). Construction and cloning of the Notch1 mutants found in human T-ALL were performed as described previously (23).
Antibodies.
Antibodies from BD Biosciences — Pharmingen or eBioscience were as follows: CD4 (RM4-5), CD8 (clone 53-6.7), CD19 (1D3), Ter119, TCRβ (H57-597), CD3ε (14502C11), c-Kit (2B8), Sca-1/Ly-6A/E (E13-161.7), NK1.1 (PK136), B220 (RA3-B2), CD11b (M1/70), Gr-1 (RB6-8C5), and CD11c (HL3). Biotinylated antibodies were revealed with Streptavidin-Pacific Blue (Molecular Probes; Invitrogen) or Streptavidin-PerCP (BD Biosciences — Pharmingen). Lineage-positive cells were identified with anti–Gr-1, -Ter119, -B220, -CD19, -CD11c, -CD8, -CD4, -CD3ε, -TCRβ, and -NK1.1.
Flow cytometry.
Cells were stained on ice in PBS containing 2% FBS, 10 mM HEPES, and 0.02% NaN3 after blocking with rat and mouse IgG (Sigma-Aldrich) and 24G2 cell supernatant. The 24G2 block was omitted when staining for CMP, megakaryocyte-erythroid progenitor (MEP), and GMP markers. Acquisition was performed on a FACSCalibur (BD) or LSRII (BD). Dead cells and doublets were excluded based on FSC-W and SSC-W characteristics and DAPI staining. Data were analyzed with FlowJo (Tree Star) software. Flow sorting was performed on either a FACS-DIVA (BD) or Mo-Flo (Dako).
BM transduction and transplantation.
Retroviral transduction of BM cells and transfer into lethally irradiated recipients was performed as described previously (20, 31). Briefly, BM cells were collected from 6- to 12-week-old mice 4 days after intravenous administration of fluorouracil (5-FU) (250 mg/kg). The cells were cultured overnight in the presence of IL-3 (6 ng/ml), IL-6 (5–10 ng/ml), and SCF (100 ng/ml). The cells were then washed, resuspended in retroviral supernatant that had been normalized based on GFP titer, placed in the same cytokine cocktail containing polybrene (4 μg/ml), and centrifuged at 1,290 g for 90 minutes. A second round of spinoculation was performed the following day. After washing with PBS, at least 5 × 105 cells were injected intravenously into lethally irradiated (9 Gy) recipients. Mice were maintained on antibiotics in drinking water 2 weeks after BMT. Mice were bled every 2–3 weeks to monitor blood counts and evaluate the presence of circulating immature T cell progenitors by flow cytometry. All experiments were performed in accordance with NIH guidelines for the care and use of animals under an animal protocol approved by the University of Pennsylvania Animal Care and Use Committee.
Cell cultures.
Primary tumor cells extracted from leukemic mice were grown in RPMI 1640 (Invitrogen) supplemented with 10% FBS (GIBCO; Invitrogen), 2 mM l-glutamine, 2-mercaptoethanol (0.0005% [v/v], Sigma-Aldrich), and antibiotics. 293T cells were maintained in DMEM (Invitrogen) with the same supplements except 2-mercaptoethanol. Cells were grown at 37°C under 5% CO2. 8946 is a murine T-ALL cell line derived from a tumor created with a tetracycline-dependent human c-Myc transgene that was kindly provided by Dean Felsher (Stanford University, Palo Alto, California, USA) (37). Retroviral transduction of these cells, sorting, and analysis in the presence of the GSI JC-19 (1 μM) and doxycycline (20 ng/ml) were performed as described previously (34). JC-19 was kindly provided by Yueming Li (Memorial Sloan-Kettering Cancer Center, New York, New York, USA).
Reporter assays.
HD domain and PEST domain (ΔP) mutations were introduced alone and in combination by site-directed mutagenesis (Stratagene) into Notch1 cDNAs, which were subcloned into the vectors pcDNA3 and MigR1. Luciferase reporter gene assays were performed as described previously (31). Briefly, U2OS cells plated in a 24-well format were transfected in triplicate using Lipofectamine (Invitrogen) plus 10 ng of pcDNA3 plasmid, 250 ng of pGL2-CSLx4-luciferase reporter plasmid, and 5 ng of pRL-TK Renilla luciferase internal control plasmid. Firefly and Renilla luciferase activities were measured in cell lysates prepared 44–48 hours after transfection with the Promega Dual-Luciferase kit per the manufacturer’s instructions, using a Turner BioSystems Dual Luminometer.
Western blotting.
Western blots of extracts prepared from the spleens of recipient mice were stained with a rabbit polyclonal antibody raised against the intracellular domain of human Notch1 as described previously (21). Cleaved Notch1 was detected using the antibody recognizing the V1744 epitope (Cell Signaling Technology Inc.).
DNA sequencing.
Sequencing for murine HD and PEST mutations in primary mouse tumor samples was performed as described previously (42).
Quantitative real-time PCR.
Total RNA was prepared using TRIzol (Invitrogen). Random-primed total RNAs (2 μg) were reverse transcribed with SuperScript II (Invitrogen). Mouse Dtx1 (Mm00492297_m1), Notch3 (Mm00435270_m1), CD25 (Mm00434261_m1), and c-Myc (Mm00487803_m1) expression was validated using primer/probe sets from TaqMan Gene Expression Assays (Applied Biosystems). The Hprt forward primer sequence was 5′-CTCCTCAGACCGCTTTTTGC-3′, and the Hprt reverse primer sequence was 5′-TAACCTGGTTCATCATCGCTAATC-3′. The Hprt probe sequence was 5′-VIC-CCGTCATGCCGACCCGCAG-TAMRA-3′. Transcripts were amplified with either TaqMan Universal PCR Master Mix or SYBR Green PCR Master Mix (Applied Biosystems) on the ABI Prism 7900 sequence detection system (Applied Biosystems).
Statistics.
Linear regression analysis was used to evaluate the relationship between DP T cell production at 6 weeks and the reporter activity of the mutations. Survival curves were computed using the Kaplan-Meier procedure, and these curves were used to determine the 6-month survival rates shown in Figure 2E. Logistic regression analysis was used to evaluate the relationship between penetrance and the relative potency of various Notch1 alleles judged by activity in reporter gene assays (fold stimulation). An accelerated failure time model was used to evaluate the association between the survival times and the mutation types. Comparison of survival curves was performed using log-rank (χ2) analysis provided through the Prism 4.03 software package (GraphPad Software). Logistic regression and survival analyses were performed using Proc Logistic, Proc Lifetest, and Proc Lifereg, version 9.1.3 of the SAS System for PC (SAS Institute Inc.). A P value of less than 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Hong Sai for excellent technical assistance, Suman Sood of the University of Pennsylvania Center for Clinical Epidemiology and Biostatistics for excellent statistical support, and Cinquetta Bush and colleagues in the University Laboratory Animal Resource (ULAR) Core for outstanding animal care. We are grateful to Chelsea Combs, Dean Felsher, David Tuveson, and Yueming Li for providing invaluable reagents. We are also grateful to the following cores at the University of Pennsylvania: the ULAR Core, the Abramson Cancer Center Flow Cytometry and Biostatistics Cores, and the AFCRI Core. We also thank the Hematopathology Core of the Dana Farber/Harvard Cancer Center. This study was supported by grants from the NIH and the Leukemia and Lymphoma Society SCORE Program to W.S. Pear and J.C. Aster. M.Y. Chiang was supported by a career development award from the National Cancer Institute (5K08 CA120544-02) and a training grant from the NIH/NIDDK (T32-DK007780-07).
Footnotes
Nonstandard abbreviations used: BMT, BM transplantation; DP, double-positive; 5-FU, fluorouracil; GOF, gain-of-function; GSI, γ-secretase inhibitor; HD, heterodimerization domain; ICN1, intracellular domain of Notch1; LNR, Lin-12/Notch repeat; N1ΔP, Notch1 with ΔP; NEC, Notch1 extracellular subunit; NGFR, truncated nerve growth factor receptor; NTM, Notch1 transmembrane subunit; ΔP, deletion spanning amino acids 2,473–2,555; P12, 13-amino-acid insertion identified in the cell line P12-Ichikawa; T-ALL, T cell acute lymphoblastic leukemia/lymphoma.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3181–3194 (2008). doi:10.1172/JCI35090
References
- 1.Aster J.C., Pear W.S., Blacklow S.C. Notch signaling in leukemia. Annu. Rev. Pathol. 2007;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logeat F., et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Irizarry C., et al. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brou C., et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell. 2000;5:207–216. doi: 10.1016/S1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 5.Fortini M.E. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat. Rev. Mol. Cell Biol. 2002;3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh J.J., et al. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen S., Kodoyianni V., Bosenberg M., Friedman L., Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 8.Schweisguth F., Posakony J.W. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-O. [DOI] [PubMed] [Google Scholar]
- 9.Hamaguchi Y., et al. Cloning and characterization of a protein binding to the J kappa recombination signal sequence of immunoglobulin genes. Adv. Exp. Med. Biol. 1991;292:177–186. doi: 10.1007/978-1-4684-5943-2_20. [DOI] [PubMed] [Google Scholar]
- 10.Wu L., Sun T., Kobayashi K., Gao P., Griffin J.D. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell. Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang M.Y., et al. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol. Cell. Biol. 2006;26:6261–6271. doi: 10.1128/MCB.02478-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. The notch intracellular domain is ubiquitinated and negatively regulated by the mammalian sel-10 homolog. J. Biol. Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 13.Wu G., et al. SEL-10 Is an inhibitor of Notch signaling that targets Notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard E.J., Wu G., Kitajewski J., Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta-Rossi N., et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 16.Tetzlaff M.T., et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fryer C.J., White J.B., Jones K.A. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 18.O’Neil J., et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson B.J., et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pui J.C., et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/S1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 21.Pear W.S., et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellison L.W., et al. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 23.Weng A.P., et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 24.Malecki M.J., et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon W.R., et al. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 26.Schweisguth F. Suppressor of Hairless is required for signal reception during lateral inhibition in the Drosophila pupal notum. Development. 1995;121:1875–1884. doi: 10.1242/dev.121.6.1875. [DOI] [PubMed] [Google Scholar]
- 27.Robey E., Fowlkes B.J. The alpha beta versus gamma delta T-cell lineage choice. Curr. Opin. Immunol. 1998;10:181–187. doi: 10.1016/S0952-7915(98)80247-1. [DOI] [PubMed] [Google Scholar]
- 28.Delaney C., Varnum-Finney B., Aoyama K., Brashem-Stein C., Bernstein I.D. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106:2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellstrom M., et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 30.Krebs L.T., et al. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aster J.C., et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 2000;20:7505–7515. doi: 10.1128/MCB.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Pooter R.F., et al. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. . J. Immunol. 2006;176:5267–5275. doi: 10.4049/jimmunol.176.9.5267. [DOI] [PubMed] [Google Scholar]
- 33.Aster J.C., et al. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 34.Weng A.P., et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma V.M., et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palomero T., et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 37.Felsher D.W., Bishop J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 1999;4:199–207. doi: 10.1016/S1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 38.Girand L., et al. Frequent proviral insertional mutagenesis of NOTCH1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and NOTCH1 for oncogenesis. . Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 39.Langenau D.M., et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 40.Palomero T., et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumortier A., et al. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 2006;26:209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y.W., Nichols R.A., Letterio J.J., Aplan P.D. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Neil J., et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundaram M.V. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- 45.Dupuy A.J., et al. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 46.von Lintig F.C., Huvar I., Law P., Diccianni M.B., Yu A.L., Boss G.R. Ras activation in normal white blood cells and childhood acute lymphoblastic leukemia. Clin. Cancer Res. 2000;6:1804–1810. [PubMed] [Google Scholar]
- 47.Perentesis J.P., et al. RAS oncogene mutations and outcome of therapy for childhood acute lymphoblastic leukemia. Leukemia. 2004;18:685–692. doi: 10.1038/sj.leu.2403272. [DOI] [PubMed] [Google Scholar]
- 48.Kawamura M., et al. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leuk. Res. 1999;23:115–126. doi: 10.1016/S0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]
- 49.Tuveson D.A., et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/S1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 50.Hawley R.G., Fong A.Z., Ngan B.Y., Hawley T.S. Hematopoietic transforming potential of activated ras in chimeric mice. Oncogene. 1995;11:1113–1123. [PubMed] [Google Scholar]
- 51.Haupt Y., Harris A.W., Adams J.M. Retroviral infection accelerates T lymphomagenesis in E mu-N-ras transgenic mice by activating c-myc or N-myc. Oncogene. 1992;7:981–986. [PubMed] [Google Scholar]
- 52.Weijzen S., et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 53.Klinakis A., et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krejci A., Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldman B.J., Hampton T., Cleary M.L. A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A-PBX1 transgenic mice. Blood. 2000;96:1906–1913. [PubMed] [Google Scholar]
- 56.Mizuno T., et al. Overexpression/enhanced kinase activity of BCR/ABL and altered expression of Notch1 induced acute leukemia in p210BCR/ABL transgenic mice. Oncogene. 2008;27:3465–3474. doi: 10.1038/sj.onc.1211007. [DOI] [PubMed] [Google Scholar]
- 57.Ellisen L.W., et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 58.Mansour M.R., et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 2007;13:6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- 59.Eguchi-Ishimae M., Eguchi M., Kempski H., Greaves M. NOTCH1 mutation can be an early, prenatal genetic event in T-ALL. Blood. 2008;111:376–378. doi: 10.1182/blood-2007-02-074690. [DOI] [PubMed] [Google Scholar]
- 60.Palomero T., et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan S.M., Weng A.P., Tibshirani R., Aster J.C., Utz P.J. Notch signals positively regulate activity of the mTOR pathway in T cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weng A.P., et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.