Abstract
The hairpin ribozyme requires functional groups from Ade38 to achieve efficient bond cleavage or ligation. To identify molecular features that contribute to catalysis, structures of position 38 base variants 2,6-diaminopurine (DAP), 2-aminopurine (AP), cytosine (Cyt), and guanine (Gua) were determined between 2.2 and 2.8 Å resolution. For each variant, two substrate modifications were compared: (1) a 2′-O-methyl-substituent at Ade-1 was used in lieu of the nucleophile to mimic the precatalytic state, and (2) a 3′-deoxy-2′,5′-phosphodiester linkage between Ade-1 and Gua+1 was used to mimic a reaction-intermediate conformation. While the global fold of each variant remained intact, the results revealed the importance of Ade38 N1 and N6 groups. Absence of N6 resulting from AP38 coincided with failure to localize the precatalytic scissile phosphate. Cyt38 severely impaired catalysis in a prior study, and its structures here indicated an anti base conformation that sequesters the imino moiety from the scissile bond. Gua38 was shown to be even more deleterious to activity. Although the precatalytic structure was nominally affected, the reaction-intermediate conformation indicated a severe electrostatic clash between the Gua38 keto oxygen and the pro-Rp oxygen of the scissile bond. Overall, position 38 modifications solved in the presence of 2′-OMe Ade-1 deviated from in-line geometry, whereas variants with a 2′,5′ linkage exhibited S-turn destabilization, as well as base conformational changes from syn to anti. These findings demonstrate the importance of the Ade38 Watson–Crick face in attaining a reaction-intermediate state and the sensitivity of the RNA fold to restructuring when electrostatic and shape features fail to complement.
Keywords: hairpin ribozyme, hepatitis-δ-virus ribozyme, phosphoryl-transfer reaction, nucleobase substitution, reaction-intermediate analog
INTRODUCTION
The hairpin ribozyme is a small RNA enzyme whose family members catalyze a reversible, site-specific phosphodiester bond-cleavage reaction within a complementary RNA substrate (McKay and Wedekind 1999; Doherty and Doudna 2001). Cleavage occurs when the 2′-hydroxyl group at Ade-1 attacks the Gua+1 phosphorus (Fig. 1A; Buzayan et al. 1986). The trigonal-bipyramidal intermediate is resolved by formation of a cyclic-2′,3′-phosphodiester and free 5′-OH (Long and Uhlenbeck 1993). This chemical reaction is essential to the replication of several plant viroids (Symons 1992), as well as the human hepatitis δ virus (Perrotta and Been 1992), the mitochondrial Varkud satellite from Neurospora (Lilley 2004), and the glmS metabolite-sensing riboswitch encountered among gram-positive bacteria (Winkler et al. 2004). Larger, more complex ribozymes pervade biological systems, such as the ribosome (Noller et al. 1992; Nissen et al. 2000), self-splicing introns (Cech et al. 1981; Grabowski et al. 1981; Ferat and Michel 1993), and RNase P (Guerrier-Takada et al. 1983). However, the diminutive sizes of small ribozymes make them tractable systems to investigate the general principles of RNA catalysis using molecular structure and functional approaches (Doudna and Lorsch 2005; Bevilacqua and Yajima 2006).
FIGURE 1.
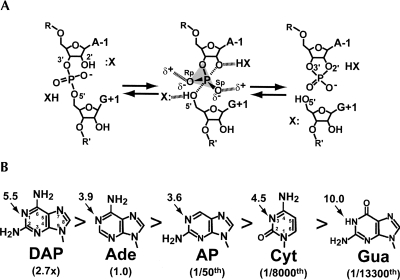
Schematic diagrams of the hairpin ribozyme chemical reaction and variants at position 38. (A) The chemical reaction of the hairpin ribozyme where X: and XH are proton shuttling groups. The reaction proceeds via a trigonal-bipyramidal transition state. (B) The base structures, cleavage activity relative to WT, and solution pKa values of base imino groups (arrows). Enzymatic rate values were recorded at 25°C at pH 7.5 in the context of a 4WJ construct (Kuzmin et al. 2005). Solution pKa values were derived by the 5′-monophosphorylated nucleoside (Ward et al. 1969; Saenger 1984).
Previous investigations of the hairpin ribozyme demonstrated the importance of bases Gua8 and Ade38. These residues were poised in crystal structures to participate directly in catalysis (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002). At a minimum, these residues contribute to electrostatic stabilization of the transition state (Ryder et al. 2001; Lebruska et al. 2002; Rupert et al. 2002; Kuzmin et al. 2004, 2005). Additional evidence suggests that catalysis could proceed through general acid/base chemistry based upon nucleobase substitutions at Gua8 (Pinard et al. 2001; Wilson et al. 2006), as well as simulations of pH rate profiles that accounted for titrations at both Gua8 and Ade38 nucleobases (Bevilacqua 2003). Alkylation of Gua8 via incidental contact with a 2′-bromoacetamide group added at Ade-1 in lieu of the nucleophile suggested the close proximity of these residues in solution (Thomas and Perrin 2006).
The function of Gua8 as a general base catalyst has been probed by investigations of abasic hairpin ribozymes in which the nucleobase is ablated, leaving only a 2′-deoxyribose. Whereas abasic modifications at Ade9 and Ade10 within the compact core led to a reduction in hairpin ribozyme cleavage activity of <10-fold (Lebruska et al. 2002), the 2′-deoxy abasic substitution at Gua8 exhibited 850-fold reduction in cleavage at pH 7.5 (Kuzmin et al. 2004). By contrast, an abasic modification at Ade38 resulted in a 5100-fold activity loss under comparable conditions (Kuzmin et al. 2005). Rescue experiments in which exogenous nucleobases were added in trans to abasic variants suggested that the key functional group restored at position 38 was an amidine moiety (i.e., an exocyclic amine alpha to a ring nitrogen), which contributed electrostatic stabilization to the transition state (Kuzmin et al. 2005). This investigation also posited water as a participant in cleavage by protonation of the 5′-leaving group. Crystallographic evidence for water at the active site of the hairpin ribozyme was presented recently for minimal variants representative of the precatalytic and reaction-intermediate states, although well-defined waters were not observed to interact with the O5′-moiety of Gua+1 (Salter et al. 2006; Torelli et al. 2007).
To investigate the importance of various functional groups at position 38, a series of “mutants” was constructed and analyzed for cleavage activity (Fig. 1B). With the exception of DAP38, which was threefold more reactive than wild type (WT), each variant showed reduced activity relative to Ade38 spanning four orders of magnitude (Kuzmin et al. 2005). The trend in reactivity did not appear to be the result of changes from an N1 to N3 moiety or to be correlated strictly with the substituted group's imino pKa. Moreover, no effort was made to relate the substituted nucleobase to its propensity to favor the syn conformation, which is preferred by Ade38 in the native state (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Alam et al. 2005; Salter et al. 2006; MacElrevey et al. 2007; Torelli et al. 2007, 2008). Although it is tempting to accept such modifications as pure indicators of chemical functionality, previous studies indicated that single nucleobase changes could evoke large, unforeseen conformational alterations, thus obfuscating the direct assignment of catalytic determinants (Alam et al. 2005; Salter et al. 2006).
The goal of this investigation was to correlate prior functional observations of position 38 hairpin ribozyme variants with detailed structural information. Four position 38 modifications (Fig. 1B) were examined in the context of a minimal, hinged hairpin ribozyme rendered into two distinct active-site configurations. A “precatalytic” conformation with near in-line geometry was produced by incorporating a 2′-OMe modification at Ade-1, thereby inactivating the nucleophile as reported for the WT ribozyme (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002), as well as several Gua8 variants (Salter et al. 2006). A “reaction-intermediate” conformation was generated by use of an inert 2′,5′ linkage in place of the scissile bond. This linkage restrains the nonbridging oxygens of the scissile phosphate to interact with the exocyclic amines of Gua8, Ade9, and Ade38, as observed for an oxo-vanadium transition-state mimic (Torelli et al. 2007). The significance of the imino and exocyclic moieties at position 38 is discussed in light of an inductive activation interaction with the pro-Rp oxygen of the scissile bond. Similarities are apparent when considering conformational nucleobase variations at position γCyt75(76) of the hepatitis δ virus ribozyme and the general-acid role attributed to this position. Overall, the localized structural rearrangements reported herein highlight the flexible nature of the hairpin ribozyme and provide a molecular perspective that accounts for activity losses by means that might not otherwise be considered.
RESULTS
Structural quality of hairpin ribozyme position 38 variants
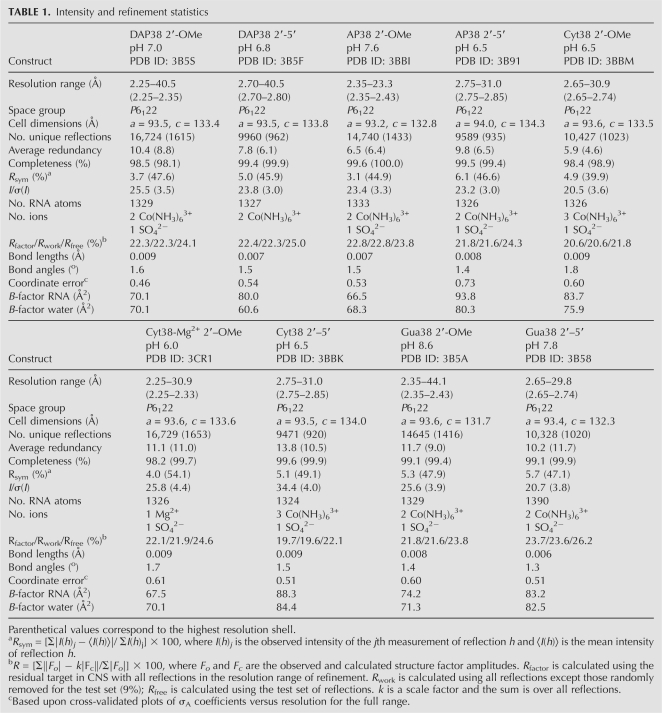
The results of this investigation provide insight into how position 38 nucleobase substitutions influence the active-site architecture, which has implications for determining the spatial orientation of key reactive groups on the reaction coordinate. This work was conducted in the context of a minimal, hinged hairpin ribozyme construct reported previously (MacElrevey et al. 2007; Torelli et al. 2007, 2008; Fig. 2). The Ade38 substitutions interrogated were DAP, AP, Cyt, and Gua (Fig. 1B). Each variant was crystallized in the context of two active-site configurations mimicking precatalytic (2′-OMe Ade-1) and reaction-intermediate (2′,5′) states. The structures discussed herein were solved and refined to resolutions between 2.25 and 2.75 Å (Table 1). The quality of each structure is indicated by the fit of the model to representative σA-weighted simulated-annealing-omit electron-density maps calculated independently for the respective position 38 base and nucleotides Ade-1 to Gua+1 (Fig. 3). Each structure exhibited reasonable refinement statistics including a final R factor ≤ 23.7%, with working and free R-values ≤23.6% and ≤26.2%, respectively (Table 1). The average R factor and R free values for all nine structures were 21.8% and 24.0%, respectively. These values are comparable to the average obtained from another small ribozyme whose structure was determined eight times (Klein et al. 2007) in a similar resolution range (R factor 21.3% and R free 23.9%). In general, the RNA atoms for each structure were well ordered in electron density maps, although the quality was noticeably poorer for the nucleotides of H1, as observed previously (Alam et al. 2005). Electron density maps with coefficients 2mFo-DFc were contiguous for the RNA of all structures with the exception of the S-turn of the Gua38 2′,5′ structure (discussed below). In the latter case, two alternate conformations were modeled for residues 37–39 according to features in σA-weighted simulated-annealing-omit electron-density maps. Occupancy was modeled at 0.5 for each conformation as described in the Materials and Methods.
FIGURE 2.
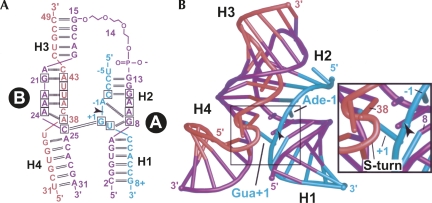
Hairpin ribozyme constructs and global structure of this investigation. (A) Secondary structure of the minimal, hinged hairpin ribozyme. The substrate strand is green, the S-turn strand is red, and the interdomain linker strand is purple. This color scheme is preserved throughout, except where noted. Helix-loop-helix domains are labeled “A” and “B” with flanking helices H1–H4. An arrowhead denotes the cleavage site, here and elsewhere, and was modified by a 2′-OMe Ade-1 or 2′,5′ phosphodiester linkage between Ade-1 and Gua+1 to prevent cleavage. Conserved residues are boxed. Watson–Crick hydrogen-bond pairs are indicated by double lines; heavy dashed lines indicate single hydrogen bonds. (B) Ribbon diagram of the minimal hinged hairpin ribozyme. (Inset) Close-up view of the active site emphasizing the spatial relationship of the scissile bond relative to Ade38.
TABLE 1.
Intensity and refinement statistics
FIGURE 3.
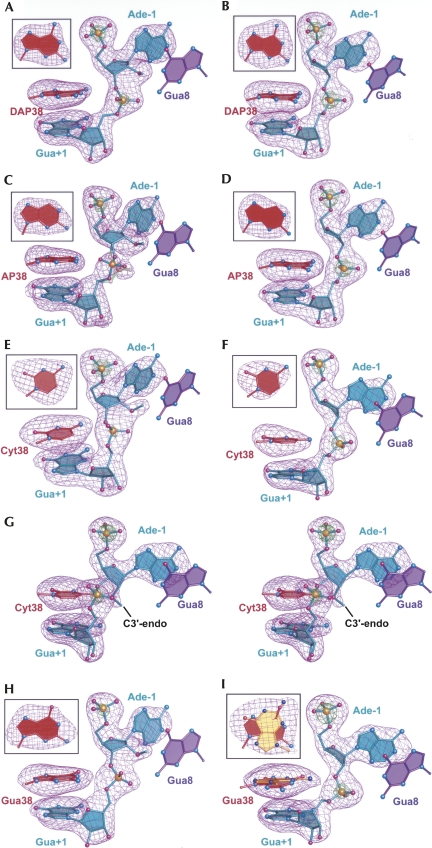
Representative σA-weighted simulated-annealing-omit electron-density maps in which the nucleobase at position 38, Ade-1, Gua+1, and atom O3′ of Cyt-2 were omitted from the phase calculation. The purple mesh represents mFo-DFc maps contoured at 4σ; the green mesh is contoured at 12σ. Models are shown as ball-and-stick representations. (Insets) Omit electron density for nucleobase 38 rotated ∼90° for clarity. (A) DAP38 with a 2′-OMe Ade-1. (B) DAP38 with a 2′,5′ scissile bond. (C) AP38 with a 2′-OMe Ade-1. This variant exhibited two conformations with bonds depicted in teal (40% occupancy) and light green (60% occupancy). (D) AP38 with a 2′,5′ scissile bond. (E) Cyt38 with a 2′-OMe Ade-1. (F) Cyt38 with a 2′,5′ scissile bond. (G) Stereoview depicting the 2′-OMe-modified Cyt38 structure solved in the presence of Mg2+. (H) Gua38 with a 2′-OMe Ade-1. (I) Gua38 with a 2′,5′ scissile bond. This variant exhibited two conformations of equal occupancy (0.5 + 0.5) with the syn (WT-like) Gua38 base depicted in red and the unusual anti conformation in yellow.
DAP38 as a gain-of-function substitution
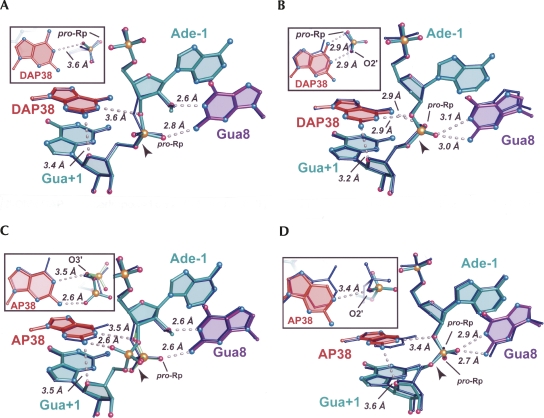
The DAP38 variant exhibited a 2.7-fold relative increase in cleavage rate and still exhibited a bell-shaped pH rate profile comparable to WT (Kuzmin et al. 2005). A superposition of the respective WT and DAP38 structures revealed minor differences between the respective precatalytic and reaction-intermediate structures (Fig. 4A,B). The distances between the N1 atom of DAP38 and the O5′ leaving group of Gua+1 were 4.3 Å and 3.3 Å for the respective precatalytic and reaction-intermediate structures; the comparable values were 4.0 Å and 3.0 Å in the Ade38 WT. Despite being within reasonable interaction distances for hydrogen bonding (3.3 Å and 3.0 Å) in both reaction-intermediate structures, the donor/acceptor geometry between atom N1 of position 38 and the O5′-leaving group was inappropriate for productive proton transfer, which is a general caveat associated with the use of the 2′,5′ reaction intermediate mimics. Comparison of the DAP38 base position relative to Ade38 of WT showed no significant deviations between these 2′-OMe structures (Fig. 4A, inset). Notably, the 2′,5′ structures showed a distance shift of 0.7 Å between the N6 atoms (Fig. 4B, inset), which exceeds the coordinate error of this model (Table 1). The origin of this difference is unclear, but may be associated with the presence of an ordered water near the N2 position of the DAP base (below). Despite the shift, the N6 exocyclic amine of DAP38 remained within 2.9 Å of the pro-Rp oxygen of Gua+1, as observed for the WT structure, thus suggesting the maintenance of a hydrogen bond at this position. Overall, the DAP38 structure appears very similar to WT in both precatalytic and reaction-intermediate conformations. This observation suggests that small k cat effects are intangible in RNA, as well as protein–enzyme structures, due to the paucity of observable binding effects that differentiate WT from mutant in representative reaction-intermediate and transition-state complexes.
FIGURE 4.
Structural superpositions of DAP38 and AP38 variants upon WT hairpin ribozymes in the context of 2′-OMe Ade-1 (precatalytic) and a 2′,5′-linkage (reaction intermediate). The WT structure is depicted in thin, dark blue bonds. Perspective for the full panel and inset are as shown in Figure 3; the scissile Gua+1 phosphate is included in the inset as well. Putative hydrogen bonds are indicated by dashed lines. (A) The DAP38 variant with a 2′-OMe Ade-1 superimposed on the WT structure. (B) The DAP38 variant with a 2′,5′ modification between Ade-1 and Gua+1 superimposed on the WT structure. (C) The AP38 variant with a 2′-OMe Ade-1 superimposed on the WT structure. The 2′-OMe modified AP38 variant exhibited two conformations of the scissile bond depicted in teal and light green. (D) The AP38 variant with a 2′,5′ modification between Ade-1 and Gua+1 superimposed on the WT structure.
AP38 does not productively align the reactive phosphate
Substitution of Ade38 with AP resulted in a 50-fold diminution of cleavage activity (Fig. 1B). The 2′-OMe-modifed structure of the AP38 variant solved here to 2.35 Å resolution showed that the relocation of the exocyclic amine to C2 on the purine Watson–Crick face was manifested primarily as the inability to localize the scissile phosphate in electron density maps. As such, this structure was consistent with at least two precatalytic conformations demonstrated by the bimodal electron density distribution (Fig. 3C). The local model was constructed in two forms with relative occupancies of 0.6 and 0.4 (Fig. 4C). In the dominant population, the pro-Rp oxygen engaged in a 2.6 Å hydrogen bond with the N2 exocyclic amine of Gua8; in the minor conformation, the pro-Sp oxygen engaged in a 2.6 Å hydrogen bond with the N2 exocyclic amine of AP38.
The reaction-intermediate variant of AP38 was solved to 2.75 Å resolution and does not indicate conformational heterogeneity at the cleavage site (Fig. 3D). Strong electron density exists for the Gua+1 phosphorus atom at the 12σ level. This result can be explained in terms of the restraints of the 2′,5′ phosphodiester linkage, which requires its nonbridging oxygens (i.e., pro-Rp and pro-Sp equivalents) to point toward the exocyclic amines of Gua8 and Ade38 in the WT. The geometric constriction and hydrogen bonding of this scissile-bond mimic provide insufficient rotational freedom to allow a hydrogen bond between the N2 of AP38 and the pro-Sp oxygen equivalent (Fig. 4D). Instead, this aspect of the 2′,5′ active site is similar to that of the oxo-vanadium complexes (Rupert et al. 2002; Torelli et al. 2007), which are widely accepted as transition-state mimics of the phosphoryl-transfer reaction (Davies and Hol 2004; Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006). Specifically, the pro-Sp oxygen equivalent of Gua+1 engaged in a bifurcated hydrogen bond with Gua8 at its N1 imino and N2 exocyclic amine (Fig. 4D), whereas the pro-Rp oxygen was in a hydrogen bond to N6 of Ade9 (not shown). The O2′ of Ade-1 was positioned 3.4 Å from the N1 imino group of AP38, which is a result of the 2′,5′ bond restraint as well. Despite apparent similarities between the AP38 2′,5′ structure compared to that of WT, the loss of a hydrogen bond between the pro-Rp oxygen equivalent of the scissile bond and the missing N6 amine of AP38 had tangible structural effects.
Absence of the AP38 N6 exocyclic amine resulted in a distinct shift of the scissile bond relative to WT. Gua+1 atoms P, O5′, and the pro-Rp oxygen of the AP38 variant exhibited the greatest deviation from WT among all 2′,5′ variants of this investigation (Fig. 4D, inset). The relative shift of the pro-Rp oxygen of AP38 was 1.3 Å; similarly, the distance between the O5′-leaving group of Gua+1 and the imino atom of AP38 was elongated from 3.0 Å in the WT to 4.0 Å in the AP38 structure. An all-atom, global superposition between the WT and AP38 structures revealed a 0.8-Å shift of the N1 atoms at position 38 (Fig. 4D, inset); a similar shift of 0.6 Å was observed for the DAP38 structure (Fig. 4B, inset). Removal of the exocyclic amine at position 38 from the major groove and its placement in the minor groove has ramifications for the stabilization of both precatalytic and reaction-intermediate states, due to the potential for new hydrogen bond formation between the N2 amine and O4′ of Gua+1 (Fig. 4). This interaction appears favorable for both DAP38 structures (3.4 and 3.2 Å), but comparable distances are elongated in each AP38 variant (3.5 and 3.6 Å). Overall, the results imply that the N6 amine of position 38 plays an important electrostatic anchoring role whose absence results in failure to localize the scissile bond, as well as the attached nucleobase, which can slide against its stacked neighbors. This phenomenon was examined further by substitution of the position 38 purine with a smaller pyrimidine that possesses a major groove exocyclic amine (Fig. 1B).
Cyt38 sequesters its imino group from the active site, but keeps N4 in an electrostatic pocket
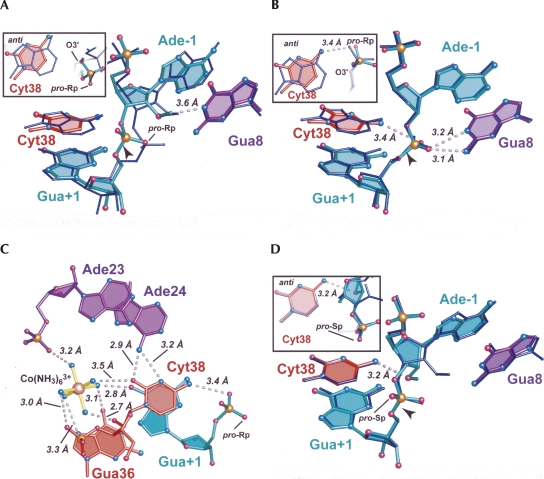
The electron density maps for Cyt38 indicated an unexpected anti conformation for both the 2′-OMe and 2′,5′-modified crystal structures (Fig. 3E,F), which were refined to 2.65 Å and 2.75 Å resolution, respectively. By comparison, Ade38 exhibited a syn conformation in all heretofore reported hairpin ribozymes (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Alam et al. 2005; Salter et al. 2006; MacElrevey et al. 2007; Torelli et al. 2007). Therefore, the anti conformers here were scrutinized closely, especially in light of the broadened electron density in simulated-annealing-omit maps calculated for the 2′-OMe structure, which was suggestive of a syn/anti conformational mixture. Broadened electron density was not observed for the 2′,5′ structure (Fig. 3, cf. E and F). Reduced bias mFo-DFc and 2mFo-DFc electron density maps were generated using phases calculated from the syn model, but these strongly supported the anti conformation. Importantly, modeling the syn orientation resulted in localized steric clashes; the O2 keto atom of the Cyt38 base was less than 3.0 Å from both the O2′ and O4′ atoms of its own ribose. Adjustment of the glycosidic linkage to correct this clash created a new steric collision between the C5 atom of Cyt38 and the N6 atom of Ade24. Lastly, the syn conformation placed the O2 keto atom of Cyt38 within 2.6 Å of a water molecule. Careful examination of this solvent atom indicated it is within hydrogen bonding distance of three acceptor groups, but only one donor. Therefore, Cyt38 was modeled in the heretofore undocumented anti conformation in both the 2′-OMe and 2′,5′ structures. Choice of the anti rotamer is also supported by reports that the syn conformation is less favorable energetically for pyrimidines (Saenger 1984).
The position of the N4 exocyclic amine of Cyt38 remained relatively unchanged regardless of whether the χ angle was modeled with a syn or anti rotamer. Strikingly, a pairwise atomic comparison of Cyt38 and WT models revealed that the N4 atom of Cyt38 superimposed well on the N6 atom of WT Ade38 in the anti conformation (Fig. 5A,B). Thus, the Cyt38 variants further corroborated the hairpin ribozyme propensity to position a hydrogen-bond donating group in the pocket occupied by N6 of Ade38, as observed in both the WT reaction-intermediate (2′,5′) and transition-state (oxo-vanadium) mimic structures (Torelli et al. 2007, 2008). In this manner, the N4 amine of Cyt38 was poised to engage in a 3.4 Å hydrogen-bond interaction with the pro-Rp oxygen (equivalent) of Gua+1 in the 2′,5′ structure (Fig. 5B, inset). This observation suggested the ability of this pyrimidine variant to contribute to transition-state stabilization despite its relatively diminutive size. Bifurcated hydrogen bonds comparable to WT were observed between the pro-Sp (equivalent) oxygen of Gua+1 and the N1 and N2 amines of Gua8 (Fig. 5B). This result demonstrated that Cyt38 had no deleterious effects on the fold of the active site.
FIGURE 5.
Structural superpositions of Cyt38 variants upon WT hairpin ribozymes and the coordination of cobalt hexamine. (A) Cyt38 2′-OMe structure superimposed on the WT structure. (B) Cyt38 with a 2′,5′ modification between Ade-1 and Gua+1 superimposed on WT structure. (C) Stick representation depicting stabilizing interactions for the unusual anti Cyt38 configuration and the novel Co(NH3)6 3+ ion observed when crystals grew from cobalt hexaamine. The 2′,5′ Cyt38 structure from B is shown, but is representative of that in A as well. (D) The stabilizing interactions observed for the anti configuration of Cyt38 in a crystal structure solved in the presence of Mg2+. The 2′-OMe group of Ade-1 adopts a novel C3′-endo conformation.
Other observations support the assignment of Cyt38 as an anti conformation. Three new stabilizing interactions appeared when contrasting this configuration to the alternative syn model. First, the N6 amine of Ade24 was positioned 3.2 Å from the imino group of Cyt38 (Fig. 5C). The N3 atom is expected to be deprotonated at the pH of crystallization (Table 1; Fig. 1B), and therefore capable of accepting a hydrogen bond from the Ade24 amine. The latter exocyclic amine was also poised to engage in a 2.9 Å hydrogen bond with the O2 atom of Cyt38. This keto group also interacted with an ammonia ligand contributed by a newly observed Co(NH3)6 3+ ion (Fig. 5C). This ion was present exclusively in electron density maps of the Cyt38 2′-OMe and 2′,5′ structures. Crystallization solutions for this variant included 1 mM Co(NH3)6Cl3, which was an identical quantity used for all other variants of this investigation. The same amount of cobalt hexaamine was shown previously to yield two other, separate Co(NH3)6 3+ binding sites for minimal hairpin ribozyme constructs harboring Ade38 (Alam et al. 2005). Thus, the Co(NH3)6 3+ ion observed here at Cyt38 constitutes a novel site that appears to have arisen only from Cyt substitution, regardless of whether the active site was cast into a precatalytic or reaction-intermediate configuration. B-factors for this ion were higher than those observed for the Co(NH3)6 3+ in the S-turn, but lower than those of the ion observed in the major groove of helix H2 (Alam et al. 2005). Importantly, all six amine groups engage in hydrogen bonds with the RNA molecule (Fig. 5C).
Cleavage rates measured for Cyt38 in the presence of Co(NH3)6 3+ were ∼10-fold lower at high pH than those measured in the presence of Mg2+ (Kuzmin et al. 2005). To investigate a possible structural basis for the deleterious influence of Co(NH3)6 3+, the Cyt38 variant was crystallized from a solution in which 10 mM MgCl2 was substituted for 1 mM Co(NH3)6Cl3. Again Cyt38 was observed to be in the anti configuration (Fig. 3G). Reduced bias mFo-DFc electron density maps showed no evidence of a Mg2+ ion coordinated to the Cyt38 residue, despite the fact that a Mg2+ ion was observed in an inner sphere coordination complex with the N7 atom of residue Gua21 in the S-turn (data not shown). This Mg2+ ion was located at a site near, but not identical to one previously coordinated to Co(NH3)6 3+ (Alam et al. 2005). Beyond this variation, no tangible structural differences were observed between these structures to suggest why Co(NH3)6 3+ diminished activity. The fact that a new ion binding site was observed at Cyt38 for Co(NH3)6 3+, but not Mg2+, suggested the potential of the trivalent ion to overstabilize a catalytically nonproductive conformation of Cyt38 in which the amidine moiety points away from the active site. The absence of a metal binding site at Cyt38 in the presence of Mg2+ supports this possibility and would be consistent with the notion that in an environment of Mg2+ the Watson–Crick face of Cyt38 is free to transiently face the scissile bond, thereby providing an essential functional group for the reaction.
A direct superposition of the 2′-OMe Cyt38 variants solved in the presence of Co(NH3)6 3+ versus Mg2+ revealed an interesting change in the sugar pucker of Ade-1 (Fig. 5D). All other structures to date, including those of the 4WJ variant, have exhibited a C3′-endo ribose pucker at Ade-1 in the presence of the 2′-OMe modification (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Alam et al. 2005; Salter et al. 2006). However, the Cyt38 variant solved in the presence of 10 mM Mg2+ exhibits a distinctly C3′-endo conformation (Fig. 5D). At 2.25 Å resolution, the electron density is clear (Fig. 3G), but the origin of this conformational variation is uncertain, since no Mg2+ was found that might contribute to this geometric switch. Prior 2′-OMe structures prepared from either Ca2+ or Co(NH3)6 3+ each exhibited C3′-endo conformations. One notable aspect of the Cyt38-Mg2+ structure was the formation of a new 3.2 Å hydrogen bond interaction between the Ade-1 2′-OMe group and the exocyclic amine of Cyt38 (Fig. 5D). This interaction is reminiscent of the nonproductive (i.e., non-in-line) conformations reported previously for Gua8 to Ade8 or Uri8 variants (Salter et al. 2006), although each of the latter constructs possessed a 2′-OH group at Ade-1. It was noted at that time that the hydroxyl groups of these mutants occupied the same position as the pro-Rp oxygen of Gua+1 in the transition state, which accepts electrostatic stabilization from the N6 amine of Ade38 (Salter et al. 2006). In contrast, the location of the pro-Rp oxygen atom in the transition-state mimic was occupied by a water, denoted wat52, in precatalytic crystal structures. Wat52 was hypothesized to enter a bulk-solvent-sequestered cavity in the transition state, thereby activating it to accept a proton from the Ade-1 nucleophile (Salter et al. 2006). As such, the results of the 2′-OMe Cyt38-Mg2+ structure suggest yet another means by which the loss of Ade38 could be deleterious. These possibilities include (1) overstabilization of a nonproductive sugar pucker in the ground state that blocks placement of wat52 (Fig. 5D) and (2) interruption of a potentially critical proton transfer network by exclusion of key solvent molecules from the active site. It is unknown whether the C3′-endo sugar pucker of Ade-1 is restricted to the Cyt38-Mg2+ variant or whether this is a more general phenomenon.
Gua38 is incompatible with formation of a transition-state conformation
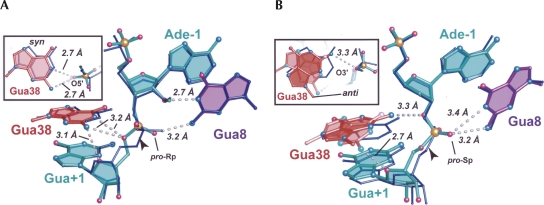
The keto group of Cyt38 was tolerated best by changing the base rotamer from syn to anti. Comparison of the Gua38 variant to Ade38 (WT) indicated that both bases adopted syn conformations in the precatalytic configuration (Figs. 3H, 6A). One minor but notable difference between Gua38 relative to the WT, DAP38, and AP38 structures was the relative location of the Gua+1 O5′ atoms. The 3.2-Å distance between the O5′ leaving group and the N2 amine of Gua38 was consistent with a hydrogen bond that is not present in either the DAP or AP model. In addition, the Gua38 precatalytic variant was the only structure determined so far in which the imino group of base 38 is within hydrogen bonding distance of the O5′ leaving group (Fig. 6A). Although the geometry for hydrogen bond formation was not ideal, a minor rearrangement could accommodate this interaction. One explanation for this disparity is that the Gua38 variant was crystallized at pH 8.6 where the N1 atom is likely protonated (Fig. 1B; Table 1). In contrast, the imino protonation state of DAP38 and AP38 (crystallized at pH 7.0 and 7.6; Table 1) suggested deprotonation. It may be of significance that N1 protonation of Gua yields a neutral nucleobase, whereas N1 protonation of DAP or AP induces a positive charge. Such results may have consequences for formation of precatalytic versus pre-reaction-intermediate structural conformations.
FIGURE 6.
Structural superpositions of Gua38 variants upon WT hairpin ribozymes in the context of 2′-OMe (precatalytic) and 2′,5′ (reaction-intermediate) modifications. (A) The Gua38 variant with a 2′-OMe Ade-1 superimposed on the WT structure. (B) The Gua38 variant with a 2′,5′ modification between Ade-1 and Gua+1 superimposed on the WT structure. The Gua38 base exhibited syn and anti conformations depicted as pink and red, respectively.
Introduction of a keto oxygen in lieu of an amino group produced a dramatic effect in terms of the local architecture for the reaction-intermediate Gua38 variant. The structure of the S-turn exhibited conformational heterogeneity not observed for any other hairpin ribozyme variant to date. Fortunately the Gua38 base was reasonably well defined in electron density maps (Fig. 3I). In the final analysis, two conformations of the S-turn were modeled that featured both syn and anti rotamers at Gua38. Each was assigned half atomic occupancy to optimize R work and R free. When we constructed the syn conformation the Gua38 base was positioned in the electron density in response to the perceived electrostatic clash between its O6 keto atom and the pro-Rp oxygen (equivalent) of Gua+1. A superposition between Gua38 and Ade38 (WT) structures revealed a 1.6-Å gap between the respective O6 and N6 atoms (Fig. 6B). Thus, the immediate consequence of substituting the N6 amine for oxygen was the repulsion of the pro-Rp oxygen at Gua+1, which was manifested as a lateral movement of the Gua38 base toward the S-turn. This repulsion also had the effect of shifting the N2 of Gua38 to engage in hydrogen bonds with the pro-Rp oxygen atom of its own phosphate (2.8 Å; not shown), the 2′-OH from U37 (2.8 Å; not shown) and the O4′ of Gua+1 (2.7 Å; Fig. 6B). These distances were shorter than those observed in the AP38 2′,5′ structure (3.7, 3.8, and 3.6 Å, respectively), and appear to constitute specific stabilizing interactions.
In the syn conformation of the Gua38 variant, the O6 atom recoils from the pro-Rp oxygen of the scissile bond. A second mode to ameliorate this electrostatic repulsion is adoption of an anti base conformation for Gua38 (Fig. 3I). In this manner, the Gua38 N2 amine was positioned closely (0.9 Å) to N6 of Ade38 WT (Fig. 6B). However, this base shift entailed a significant refolding of neighboring S-turn residues, which could not be modeled with high confidence in electron density maps. In the anti conformation of Gua38 its N2 atom projected toward the pro-Rp atom of the scissile bond, as well as O2′ of Ade-1 (Fig. 6B); this unusual orientation suggested a 3.3 Å hydrogen bond. This configuration was stabilized further by a 3.0 Å hydrogen bond between the O6 atom of Gua38 and the N6 atom of Ade24 (not shown). Overall, the Gua38 reaction-intermediate mimic represented the most dramatic change in the local structure and demonstrated the striking plasticity of the RNA backbone. These results have implications for the evolution of new RNA activities while preserving global architecture.
Waters in the precatalytic active site of position 38 variants
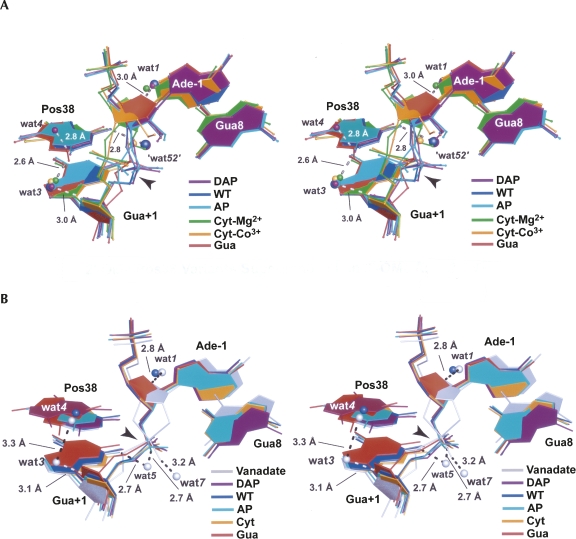
Experimental and computational evidence continues to suggest a crucial role for water in the mechanism of action of the hairpin ribozyme (Pinard et al. 2001; Kuzmin et al. 2005; Rhodes et al. 2006; Salter et al. 2006; Torelli et al. 2007, 2008). A summary of active site waters observed in the respective precatalytic and reaction-intermediate constructs of this investigation has been assembled for comparison to the previously reported WT coordinates (Fig. 7). Confidence in assignment of these waters is based on the diffraction quality of crystals. The precatalytic structure of the hairpin ribozyme was determined to 2.05 Å resolution and revealed ordered solvent molecules in and around the active site (Salter et al. 2006). The 2′-OMe variants of this study displayed lower resolutions (2.25 to 2.65 Å) compared to WT, possibly due to greater variation in the scissile-bond position resulting from nonoptimal hydrogen bonding (Fig. 7A). Nonetheless, their electron density was sufficient to model three solvent molecules identified in the WT 2′-OMe Ade-1 construct (wat1, “wat52,” and wat3), as well as wat4 observed in the 2′,5′ WT complex (Fig. 7A).
FIGURE 7.
Stereographic depiction of all Ade38-variant structures of this investigation superimposed onto representative WT hairpin ribozymes (dark blue). (A) Superposition of 2′-OMe variants onto the precatalytic ribozyme. Waters are numbered as in Torelli et al. (2007) with the exception of wat52 (Salter et al. 2006); waters are colored to match the corresponding variant. (B) Superposition of 2′,5′ variants onto the reaction-intermediate (dark blue) and transition-state (gray) mimics of the WT ribozyme. Waters are depicted only from the WT and vanadate structures.
Previously, wat1 in the WT 2′-OMe structure engaged in a hydrogen-bond network linking it to the exocyclic amine of Ade10 (not shown) and a 3.0 Å hydrogen bond to O4′ of Ade-1 (Fig. 7A; Salter et al. 2006). Similar waters were observed for the DAP38 and Cyt38-Mg2+ structures. Overall, this pocket was not significantly affected among the variants examined.
Waters located at “site 52” were present for DAP38 and Cyt38-Co3+, which were the highest resolution variants of this investigation (Table 1). These waters were within hydrogen bonding distance (∼2.8 Å) of N6 or N4 exocylic amines of the respective position 38 bases (Fig. 7A), as well as the N6 amine of Ade9 (not shown). Despite the superior 2.25 Å resolution of the Cyt38-Mg2+ structure, wat52 was not observed. Instead its 2′-OMe group occupied this site (Figs. 3G, 7A). This observation demonstrates the extreme favorability of accommodating a negatively charged atom in this pocket, which is filled by the pro-Rp oxygen equivalent of the scissile bond in reaction-intermediate and transition-state–analog complexes (Torelli et al. 2007, 2008). Interestingly, nucleobase variants AP38 and Gua38, which are devoid of an N6 exocylic amine equivalent, did not exhibit water molecules at site 52. This result may be attributable to changes in the electrostatic character of this pocket that would be predicted to destabilize the transition state.
Interest in the location of waters near the O5-leaving group of Gua+1, with the potential to serve as specific acid catalysts in cleavage or in activation of Ade38 as a general acid, have mandated a search for waters in this region (Kuzmin et al. 2005; Torelli et al. 2007; Walter 2007). Wat3 is most conserved among position 38 variants (Fig. 7A). Its conserved contacts included a 3.0 Å hydrogen bond distance to O4′ of Gua+1 (Fig. 7A) as well as the 2′-hydroxyl of Uri37 (not shown). These interactions appeared to stabilize an alternate conformation of Uri37 that promoted a more solvent-sequestered state for the scissile bond (Alam et al. 2005). Wat3 is also well represented in a number of minimal hairpin ribozyme structures and was observed before in a Gua8Ade variant that had a 2′-OH at the Ade-1 nucleophilic position (Salter et al. 2006). Unlike wat1 and wat52, the location of wat3 suggested it exchanges readily with bulk solvent (Torelli et al. 2007). Its accessibility and location were preserved in the DAP38, AP38, Cyt38-Co3+, and Cyt38-Mg2+ structures of this investigation (Fig. 7A).
Wat3 interactions with the N2 exocyclic amines of DAP38 and AP38 constitute a new hydrogen bond. In contrast, this solvent molecule was not observed in the Gua38 structure despite the presence of an N2 moiety. As described, this exocyclic amine formed a unique 3.2 Å hydrogen bond with O5′ of Gua+1 (Fig. 6A) that was not present in the DAP38 and AP38 variants. Differences in the presence of wat3 among variants may be attributable to changes in scissile-bond geometry or the presence of minor-groove amino groups. More work is required to document whether wat3 is relevant to catalysis.
Wat4 was also reported previously, but only in the context of the WT 2′,5′ and vanadium-oxide complexes of the minimal hairpin ribozyme (Torelli et al. 2007; Fig. 7B). Therefore, the DAP38 variant of this investigation represents the first report of wat4 in the context of a precatalytic conformation (Fig. 7A). An analysis of wat4 interactions revealed a direct 2.8 Å hydrogen bond to the N2 group of DAP38. Prior results from the WT enzyme showed a direct 3.3 Å wat3 to wat4 interaction (Fig. 7, cf. A and B) permitted by the absence of an N2 group for Ade38. Significantly, the precatalytic conformations herein provided no direct evidence for water coordinated at the N1 atom of position 38, which has implications for mechanistic proposals invoking water at this location (Kuzmin et al. 2005; Walter 2007). Nonetheless, the possibility remains that some rearrangement of the resident waters could occur in the transition state, as implied by the WT 2′,5′ and vanadium-oxide complexes (Torelli et al. 2007).
Effects of position 38 variants on waters in the context of a reaction-intermediate mimic
To examine the effect of position 38 variants on the hairpin ribozyme structure, as well as active site waters, a 2′,5′ reaction-intermediate analog was employed. Electron density maps for a WT construct provided prior evidence for several waters in the active site with wat1 and wat4 being resolved best (Torelli et al. 2007). These two sites were visible in a minimal, WT vanadium-oxide complex that revealed several other nearby solvent molecules (Fig. 7B). Significantly, no comparable waters for the 2′,5′, position 38 variants in the present investigation could be modeled with confidence. In the final analysis, the 2′,5′ reaction-intermediate analog structures provided important information regarding the contributions of position-38 variants to transition-state stabilization. However, nothing salient can be said regarding the identification and location of active-site waters in the reaction-intermediate state at this time.
DISCUSSION
Relating ribozyme structure to function represents a formidable challenge due to the paucity of high-resolution structures revealing the identity of functional groups relevant to proton transfer and transition-state stabilization (Bevilacqua and Yajima 2006). The goal of this investigation was to elucidate how nucleobase substitutions at a key active site residue of the hairpin ribozyme, Ade38, influenced the active site architecture and affected its hydration shell at the molecular level. Previous work on the hairpin ribozyme demonstrated a correlation between in-line geometry of the scissile bond and the catalytic fitness of position 8 base variants (Pinard et al. 2001; Salter et al. 2006); similar trends have been sought for the HDV ribozyme (Ke et al. 2004; Das and Piccirilli 2005). However, the prior hairpin ribozyme analysis was conducted in the context of “precatalytic” or “ground-state” complexes in which the pro-chiral, nonbridging oxygens of the labile phosphodiester were not poised to receive electrostatic stabilization from the exocylic amines of Gua8, Ade9, and Ade38 (for review, see Ferré-D'Amaré 2004). As such, the influence of various Gua8 base modifications on structures representative of the transition-state was not addressed. A similar problem was perceived as a shortcoming of a recent HDV ribozyme analysis (Strobel 2005).
Vanadium oxide has proven useful to induce a hairpin ribozyme geometry representative of catalysis (Rupert et al. 2002). One limitation of this strategy was the presence of conformational heterogeneity at base Gua8 of the active site, which was attributed to the partial atomic occupancy of the oxo-vanadium moiety in the context of the minimal ribozyme construct (Torelli et al. 2007). To overcome this problem, a 2′,5′-phosphodiester linkage was substituted at the scissile bond. This restraint produced a reasonable mimic of the reaction-intermediate geometry by emulating the positions of the pro-chiral oxygens in the trigonal-bipyramidal oxo-vanadium complex (Rupert et al. 2002; Torelli et al. 2007). A comparable 2′,5′ analog was shown recently to be consistent with reaction-intermediate geometry for the glmS ribozyme (Klein et al. 2007) and to exhibit features on the reaction coordinate of a protein ribonuclease (Torelli et al. 2008). As such, the current study set out to examine the structural effects resulting from various nucleobase substitutions at Ade38, a key catalytic residue, in the context of both the precatalytic (2′-OMe Ade-1) and reaction-intermediate (2′,5′) constructs. The results suggest that nucleobase substitutions at position 38 influence the hydrogen-bonding pattern of the precatalytic state and thus alter the propensity to adopt in-line conformations. Such overstabilization of ground-state conformations was observed for variations at Gua8 (Salter et al. 2006). Importantly, substitution of functional groups at position 38 has the potential to (1) deform the local structure of the S-turn, (2) alter the base conformation relative to WT from syn to anti, and (3) disrupt key hydrogen bonds essential for transition-state stabilization. To emphasize the various effects the discussion is framed on a “per-functional-group” basis that contrasts the perceived precatalytic and reaction-intermediate conformations.
Removal of the exocyclic amine at N6 and its addition at N2
The AP38 hairpin ribozyme structure provided a means to assess the influence of the N6 exocyclic amine on the local fold. Ade and AP display similar solution pKa values for the imino position (Fig. 1B), and each exhibits a single exocyclic amine attached to the purine ring. At the optimal pH for WT cleavage (i.e., 7.5, the condition under which the 2′-OMe construct was crystallized; Table 1), a 50-fold reduction in cleavage activity was reported for the AP38 variant (Kuzmin et al. 2005). From a structural vantage point, removal of the N6 amine was not especially disruptive to the local fold of the precatalytic state. However, AP38 lacked the capacity for hydrogen-bond donation at N6, which may have contributed to the absence of wat52 in contrast to the WT and DAP38 structures (Fig. 7A). In the 2.05 Å resolution WT structure, wat52 engaged in a hydrogen-bonding network sequestered from bulk solvent (Salter et al. 2006). It is conceivable that any single-atom or whole-base deletion at this position would disrupt the precatalytic state due to water reorganization, which has not been considered widely in the field.
The AP38 structure showed evidence for dual scissile bond conformations that were not observed for DAP38 (Fig. 4, cf. A and C), thus suggesting that the 2-amino group alone was insufficient to account for this heterogeneity. A comparison of the DAP38 and AP38 precatalytic structures revealed that the purine ring of the latter was not restrained by interactions with an N6 amine. Consequently, the AP base slid readily across its flanking, stacked neighbors to establish N2 charge neutralization via close positioning at the pro-S oxygen of Gua+1. This contact was observed for one of the AP38 conformations (Fig. 4C). In contrast, an equally favorable conformation placed the pro-Rp oxygen of Gua+1 within hydrogen bonding contact to the N2 amine of Gua8. The latter interaction was observed previously in the WT precatalytic state (Salter et al. 2006).
Perhaps the most significant structural effect resulting from the removal of the N6 amine was a failure to localize the pro-Rp oxygen of the reaction-intermediate mimic structure. The previously ascribed function of the exocyclic amine at position 38 was one of transition-state stabilization (Rupert et al. 2002; Ferré-D'Amaré 2004; Torelli et al. 2007). An additional proposed role for the Ade38 N6 protons is that they are transferred to and from the nonbridging oxygens to promote formation of the phosphorane intermediate (Bevilacqua et al. 2004). Functional studies in which Ade38 was replaced by purine resulted in only 4% residual catalytic efficiency compared to WT (Grasby et al. 1995). Nucleotide analog interference mapping demonstrated strong interference effects in ligation chemistry resulting from substitution with purine or AP (Ryder and Strobel 1999). Consistent with the precatalytic crystal structures (Rupert and Ferré-D'Amaré 2001; Alam et al. 2005), the N6 group of Ade38 was shown not to be involved in docking based on time resolved FRET, but instead appeared critical for catalysis (Ryder et al. 2001). Congruent with this fact was the observation that loss of the exocyclic amine in the AP38 structure resulted in the single largest deviation of the Gua+1 nonbridging oxygens among all reaction-intermediate analog structures reported herein (Fig. 7B, cyan). In fact if an N6 amino moiety were present at position 38 of AP38 2′,5′, the hydrogen bonding capability between these atoms would be negligible (∼4 Å).
Another ramification of the AP38 variant is that it does not contribute significantly to the positive electrostatic environment of the O5′-leaving group compared to Ade38 or DAP38. Prior consideration of the general acid properties of Ade38 suggested a hydrogen bond formed between its exocyclic amine and the pro-Rp oxygen of Gua+1. This interaction was observed only in the reaction-intermediate or transition-state structures and would abrogate the rotational freedom of the amine, resulting in electronic donation into the aromatic system (Bevilacqua 2003). Interestingly, a comparable interaction was present in the HDV ribozyme between the pro-Rp oxygen of γCyt22 and the N4 amino moiety of γC75 (Ferré-D'Amaré et al. 1998); the latter residue has been accepted widely as the general acid catalyst for cleavage. This electronic phenomenon would account for the elevated pKa of the base imino proton (Gong et al. 2007) and in the case of the hairpin and HDV ribozymes favor local positive charge placement in the vicinity of the 5′-oxyanion leaving group, as required for associative phosphoryl transfer (βleaving group ∼ −1.2) (Herschlag et al. 1993; Oivanen et al. 1998). Such positive charge localization is consistent with the work of Fedor and colleagues, whose data suggest a significant electrostatic role for Ade38 in catalysis (Fedor and Williamson 2005; Kuzmin et al. 2005). The importance of the N6 electrostatic contribution to structure and catalysis is realized further when the charge of the amine is reversed by substitution with a keto oxygen.
Replacement of the N6 amine with a keto oxygen
The Ade38Gua variant resulted in a 104-fold impairment of the cleavage rate (Fig. 1B). Whereas the precatalytic structure appeared to stabilize a potentially nonproductive ground state, the 2′,5′ modification destabilized the S-turn, resulting in a mixture of syn and anti conformations for the Gua38 base (Fig. 6B). In contrast, the AP38 and Cyt38 variants simply altered their scissile bond conformations to accommodate both precatalytic and reaction-intermediate analogs.
The presence of both syn and anti conformations for the Gua38 base in the context of the reaction-intermediate analog suggests this nucleobase interconverts its conformation in solution. This prospect seems somewhat remarkable given its close packing between Gua+1 and Ade40. In the WT-like syn conformation, the O6 of Gua38 recoils to avoid an electrostatic clash with the pro-Rp oxygen of the scissile bond. This observation implies that Gua38 contributes little or no electrostatic stabilization in the transition state. In contrast, the unusual anti-conformation of Gua38 refolds aptly to position its N2 amine near the pro-Rp group of Gua+1. The 3.8 Å distance between these atoms suggests only a modest electrostatic benefit, which does not appear to offset the resulting conformational heterogeneity observed in the associated S-turn. Moreover, the exocyclic amine to nonbridging oxygen inductive effect would move the pKa of the Gua38 imino group further from neutrality, producing little benefit. This situation is in contrast to Ade or Cyt, whose pKa values would shift closer to physiological pH (Fig. 1B).
The Ade38Gua variant in the context of the reaction-intermediate analog appeared somewhat reminiscent of the γCyt75Uri and Cyt76Uri variants of the HDV ribozyme. In the EDTA-treated HDV ribozyme crystal structure, the distance between γCyt75Uri O4 and the nearby γCyt22 pro-Rp oxygen is 4.4 Å, as compared to 2.9 Å in the WT, cleaved precursor (Ferré-D'Amaré et al. 1998; Ke et al. 2004). The comparable distances in the hairpin ribozyme are 4.5 Å for the syn Gua38 O6 keto group to the pro-Rp oxygen of Gua+1, and a distance of 2.9 Å for the 2′,5′ Ade38 WT construct. These observations suggested strikingly similar structural perturbations of each ribozyme in response to changes in the electrostatic character of a key catalytic base. One caveat in this interpretation, however, is that the HDV γCyt75Uri structure was not solved in the presence of a reaction-intermediate analog, making further structural rearrangements a distinct possibility (Das and Piccirilli 2005; Strobel 2005). Another important difference between the HDV and hairpin ribozymes is that the former enzyme “borrows” negative charge from the backbone of nearby γCyt22 to activate its general acid. In contrast, the hairpin ribozyme utilizes the negative charge of the scissile bond itself to perturb the pKa of Ade38. These differences in the source of base activation, as well as the potential for Gua38 to disrupt the S-turn and overstabilize a nonproductive ground state, provide insight into the 104-fold loss of cleavage activity reported for the Gua38 variant (Kuzmin et al. 2005), as compared to the 102- to 103-fold losses recorded for the γCyt75Uri and Cyt76Uri variants of the HDV ribozyme (Perrotta et al. 1999; Nakano et al. 2000).
Replacement of a purine base at position 38 with a pyrimidine
Substitution of Ade with Cyt resulted in an 8000-fold loss of cleavage activity (Fig. 1B). Without the benefit of structure, this dramatic disparity was attributed to replacement of a purine with a pyrimidine, which was posited to yield a rather significant gap between the Watson–Crick face of the smaller base and the O5′ leaving group of Gua+1 (Kuzmin et al. 2005). As such, the amidine moiety of Cyt, comprising atoms N3, C4, and N4, would be expected to be positioned similarly to that of Ade (comprising atoms N1, C6, and N6), albeit at a greater distance from the scissile phosphate (Kuzmin et al. 2005). Crystal structures of the Cyt38 variants of this investigation, in both 2′-OMe and 2′,5′ contexts, deviated significantly from these expectations, since both structures favored a distinctly non-WT, anti base conformation at Cyt38. Significantly, the global folds of each structure remained intact with no significant changes relative to WT.
The Cyt38 variant indicated a strong preference for an anti base conformation unlike Gua38, which displayed mixed conformers. The most significant effect of the Cyt38 anti conformation was sequestration of the N3 group from the active site. Although the N4 exocyclic amine was still within hydrogen bonding distance of the pro-Rp oxygen equivalent in the reaction-intermediate analog structure, the buried imino group suggested it would be incompetent to serve as a general acid in cleavage or to localize positive charge near the 5′-leaving group as suggested for WT (Bevilacqua 2003; Kuzmin et al. 2005). Nonetheless, the ability to adopt the in-line geometry would not be expected to be impaired.
Given the structural results it is tempting to speculate that the rate reported for Cyt38 at pH 6.0 (5 × 10−6 min−1) (Kuzmin et al. 2005) reflects catalysis in the absence of an imino proton. This rate is comparable to the Ade38dX (i.e., 2′-deoxy abasic) variant, which was disabled to a similar extent (i.e., a 5100-fold loss). Moreover, no evidence in electron density maps suggested the presence of an additional metal or solvent ion that might act in catalysis as proposed previously (Fedor and Williamson 2005; Walter 2007). As stated, the possibility that Cyt38 moves transiently from an anti to syn conformation cannot be dismissed, although the structure would require significant adjustments to circumvent the most serious steric clashes. The superior reactivity of the AP38 ribozyme (160-fold more reactive than Ade38Cyt) suggests that the N1 moiety of position 38 is more crucial to catalysis than the N6 amine.
Water in the active sites of variants with implications for proton shuttling in catalysis
Various roles for Ade38 in cleavage have been suggested that posit the existence of water between the Watson–Crick face of Ade38 and the scissile bond (Fedor and Williamson 2005; Kuzmin et al. 2005; Rhodes et al. 2006; Walter 2007). Importantly, no such novel solvent molecules were assigned in any of the precatalytic or reaction-intermediate structures of this investigation. This observation is especially significant since three of the variants, AP38, Gua38, and Cyt38, adopted conformations in which the nucleobase recoils slightly from the scissile bond, thus raising the possibility that water occupies the vacated space. Although the mixed scissile bond or base conformations of AP38 and Gua38 would appear to preclude the observation of a high-occupancy water molecule, Cyt38 variants were well defined. Indeed, Cyt38, despite being a pyrimidine, exposes only ∼10 Å2 of its scissile bond in the reaction-intermediate mimic relative to ∼11 Å2 for WT. Thus, it appears even the smallest base variant achieves a complementary fit between the position 38 base and the scissile bond. This close interaction suggests that the imino group contribution to scissile-bond cleavage is direct and not water mediated. This finding is significant in light of recent computational models that purport evidence to the contrary (Rhodes et al. 2006; Walter 2007).
Previously the role of water in hairpin ribozyme catalysis was evaluated by molecular dynamics (Rhodes et al. 2006). Starting from the precatalytic structure (Rupert and Ferré-D'Amaré 2001) and armed with the knowledge of active site waters derived from a high-resolution crystallographic analysis (Salter et al. 2006), a model was generated that suggested how a water-mediated proton-transfer network activates the 2′-nucleophile and protonates the 5′-leaving group in cleavage. Although this model has several desirable features, one weakness was that the in-line scissile-bond geometry differed significantly from oxo-vanadium structures reported from two different crystal forms of the hairpin ribozyme, which were virtually identical on a local level (Rupert et al. 2002; Torelli et al. 2007). In addition, the scissile bond of the molecular dynamics model made five fewer hydrogen bonds in its proposed transition state (Rhodes et al. 2006) than analogs solved crystallographically. One of the missing interactions was the hydrogen bond between N6 of Ade38 and the pro-Rp oxygen of Gua+1, whose importance has been reinforced by several observations herein. Specifically, this interaction (1) serves as an electrostatic anchor for the purine base and favors phosphorane formation; (2) it activates the position 38 imino group, thereby alleviating charge buildup in the transition state; and (3) it assures a close contact with the scissile bond, thus excluding water from the Watson–Crick face of position 38. These factors suggest that current models for hairpin ribozyme catalysis would be sensible to consider functional, computational, and experimentally derived structural observations.
Summary and Conclusions
Subtle modifications to RNA enzymes can have a profound effect on local folding that may convolute functional interpretation in the absence of molecular structure. The goal of this investigation was to provide a structural rationale for the loss of activity shown for four Ade38 variants of the hairpin ribozyme. The results demonstrated the importance of comparing precatalytic and reaction-intermediate analogs of each variant. In general, the precatalytic structures showed greatest deviation at the scissile bond due to differences in hydrogen bonding with variant functional groups. The locations of waters were well conserved relative to WT. Structures of reaction-intermediate analogs were associated with nonnative conformations of the local fold, such as Gua38, as well as syn to anti base alterations for Cyt38 and Gua38. In instances where activity was 103- to 104-fold lower than WT, the imino moiety faced away from the O5′-leaving group. Future efforts will focus on single-atom substitutions at the position 38 imino moiety, whose ablation is predicted to abolish enzymatic activity while leaving the global and local architectures intact.
MATERIALS AND METHODS
Hairpin ribozyme constructs
The 61-nt hinged hairpin ribozyme used for this investigation comprised three strands as described (MacElrevey et al. 2007). The sequence of the ribozyme used is depicted in Figure 2A. The nucleobase at position 38 was substituted with DAP, AP, Cyt, or Gua. The Uri39Cyt gain-of-function mutation (Joseph and Burke 1993) was included to reduce conformational heterogeneity (Alam et al. 2005). Position Ade14 was substituted with a 10-atom linker that preserved the phosphodiester bond (Fig. 2A). Two of the four helices that compose the 4WJ of the natural hairpin ribozyme (Hampel and Tritz 1989) were removed, although the regions of the hinged construct at H2 and H3 closely mimicked the 4WJ packing environment as a result of engineered intermolecular contacts afforded by crystallographic symmetry (MacElrevey et al. 2007).
Modifications to the substrate strand included a 2′-OMe Ade-1 or a 2′,5′-phosphodiester linkage between Ade-1 and Gua+1, which prevented cleavage. Oligonucleotide strands were purchased from Dharmacon with the exception of the 2′,5′ substrate 13-mer. The phosphoramidite for the latter modification was from Glen Research and was incorporated into the sequence at the W.M. Keck Facility (Yale University). With one exception, all RNA strands were subjected to deprotection, reverse phase HPLC purification, desalting, and domain docking as described (Wedekind and McKay 2000; Grum-Tokars et al. 2003); the Cyt38-Mg2+ structure was docked in the presence of 10 mM MgCl2 in lieu of Co(NH3)6Cl3.
Crystallization and X-ray diffraction experiments
Crystals grew as hexagonal rods over 5–6 wk to a size of 150 μm × 150 μm × 300 μm using vapor diffusion at 20°C. Crystallization conditions were based on those established (Alam et al. 2005; Salter et al. 2006); reagents common to all variants except the Cyt38-Mg2+ structure included 0.25 M Li2SO4, 1.0 mM Co(NH3)6Cl3, and 2.0 mM spermidine; for the Cyt38-Mg2+ structure, 10 mM MgCl2 was used in place of Co(NH3)6Cl3. Poly(ethylene)glycol 2000 monomethyl ether was the precipitant used in a range between 20% and 24% (w/v). Solutions were buffered using 0.10 M sodium cacodylate for pH ranges from 6.0 to 6.8, 0.10 M HEPES for pH ranges from 7.0 to 7.8, or 0.10 M TRIS for pH ranges from 8.0 to 8.8. Table 1 lists the pH value of each crystal.
All crystals were cryoprotected by transfers through four synthetic mother liquors containing 5% to 17.5% (v/v) glycerol. Crystals were captured in thin nylon loops (Hampton Research) and flash cooled in a −178°C stream of N2(g) (X-stream, Rigaku/MSC). Diffraction data for the AP38 2′-OMe structure were recorded on an R-Axis IV image plate system equipped with confocal optics (Rigaku/MSC). CuKα X-rays were generated using a Rigaku RUH3R rotating anode operated at 5.0 kW. A total of 130 images were collected at a crystal-to-detector distance of 15.0 cm at an exposure time of 25 min per 0.5° oscillation. All other crystals were stored in dewars of N2(l) until their retrieval for X-ray data collection using synchrotron radiation. Diffraction data for other 2′-OMe modified ribozymes were recorded at −178°C on a Quantum210 CCD (ADSC) at the A1 station of CHESS (λ = 0.977 Å). For each sample, high-resolution data were recorded from 150 to 250 images (depending on the c* orientation) at a crystal-to-detector distance of 21.0 cm and an exposure time of 15 to 20 sec per 0.5° rotation. Low resolution diffraction data were collected similarly, except the exposure time was ∼2 sec. Diffraction data for 2′,5′ hairpin ribozymes were recorded at −178°C using a Quantum270 CCD (ADSC) at the CHESS F1 station (λ = 0.918 Å). High-resolution data were collected from 200 to 300 images at a crystal-to-detector distance of 27.5 cm with exposure times ranging between 10 and 20 sec per 0.5° rotation. Low-resolution data were recorded similarly with a 1 sec exposure; these data were collected prior to high-resolution data except for DAP38 2′,5′.
X-ray data reduction, structure determination, and refinement
Diffraction data were reduced to intensities and scaled using Crystal Clear (Pflugrath 1999). High- and low-resolution data sets were merged after integration for all structures except Gua38 2′,5′, for which only the high-resolution scan was used. Data reduction statistics are provided in Table 1. Each construct crystallized in space group P6122 with a single molecule per asymmetric unit (Grum-Tokars et al. 2003; Alam et al. 2005; MacElrevey et al. 2007). Preliminary structures were determined by difference Fourier using refined models, such as the 2.05 Å resolution 2′-OMe WT structure (PDB ID 2OUE) or the 2.35 Å resolution 2′,5′ modified WT structure (PDB ID 2P7F) described by Salter et al. (2006) and Torelli et al. (2007). Refinement was performed using cross-validated maximum likelihood intensity targets as implemented in CNS (Adams et al. 1997; Brünger et al. 1998). In all cases, the R free test set was chosen to be coincident with that of the starting models, which was chosen initially at random to avoid the propagation of statistical bias associated with multiple difference Fourier analyses (Kleywegt and Brünger 1996). Rigid body refinement was performed first on the entire structure (devoid of ions and waters) and subsequently on the individual loop A and B domains. Cartesian simulated annealing, positional minimization, and individual B-factor refinement were conducted as necessary. Simulated-annealing omit maps were used to guide manual refinement and to reduce model bias (Brünger et al. 1997). Manual model building was conducted using the interactive graphics program O (Jones et al. 1991). Electron density maps were calculated using σA-coefficients throughout the refinement process (Pannu and Read 1996). Upon convergence, all data were recombined for a final round of positional minimization, during which a residual least squares target was employed, thereby giving a final overall R factor (Table 1).
The default CNS (v1.1) parameter and topology files for RNA were adapted to accommodate nonstandard nucleotides (MacElrevey et al. 2007). Bond angles, distances, and geometry were derived from high-resolution small molecule structures (MIUDAP, MTYDAP, WIPCAZ, BUREAP, FUREAP, and YACHAL) from the Cambridge Crystallographic Database (Allen 2002). A dual conformation was built at Uri-5 for all structures (MacElrevey et al. 2007). Alternate conformations were also modeled for the scissile phosphate of the AP38 2′-OMe structure, as well as S-turn residues Uri37, Gua38, and Cyt39 of the Gua38 2′,5′ structure. Occupancies of dual conformations were adjusted to minimize the R factor and R free statistics, peaks in difference electron density maps, and B-factor discrepancies between models of partial occupancy relative to those of uniform occupancy in the local structure. Sugar pucker restraints were derived from higher-resolution hairpin ribozyme structures (Salter et al. 2006; Torelli et al. 2007) and were evaluated further with simulated-annealing omit maps.
Waters were assigned manually to each structure based on the following criteria: peak height ≥ 3.0 σ in mFo-DFc maps and ≥ 0.8 σ electron density in 2mFo-DFc maps, reasonable geometry for hydrogen bonding with distances between donor and acceptor groups between 2.6 and 3.8 Å, and refined temperature factors comparable to surrounding RNA atoms. Two Co(NH3)6 3+ ions have been observed consistently in minimal hairpin ribozyme crystals located (1) near G21 and A40 of the S-turn and (2) within the major groove of H2 (Alam et al. 2005). New cobalt sites were observed in this investigation and were treated as described (Alam et al. 2005).
All-atom superpositions were generated using LSQKAB implemented in CCP4 (Collaborative Computational Project, Number 4 1994). Figures 2B–7 were generated in PyMOL (DeLano Scientific).
ACKNOWLEDGMENTS
The authors acknowledge Krystle Williams and Alena Janda for assistance with the refinement. The intellectual contributions of Andrew Torelli and Robert Spitale were appreciated greatly. Support for this project was derived from NIH grant GM-63162 to J.E.W. C.M. was supported in part by an Elon Huntington Hooker Graduate Fellowship. Gratitude is expressed to Dr. Richard Gillilan, Bill Miller, and the staff of the Cornell High Energy Synchrotron Source (CHESS, Ithaca, NY) for assistance. CHESS is supported by the NSF under award DMR-0225180 and the Public Health Services through NIH/NCRR award RR-01646.
Footnotes
Abbreviations: Ade, adenosine; AP, 2-aminopurine; Cyt, cytidine; Gua, guanosine; Uri, uridine; 2′-OMe, 2′-O-methyl modification of ribose; 2′,5′, 3′-deoxy, 2′,5′-phosphodiester linkage; WT, wild type with Ade38 intact; 4WJ, four-way helical junction hairpin ribozyme; γCyt75, nomenclature for the genomic polarity strand of the hepatitis δ virus ribozyme; Cyt76, the antigenomic polarity strand of the hepatitis δ virus ribozyme; CHESS, the Cornell High Energy Synchrotron Source, Ithaca, NY.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1055308.
REFERENCES
- Adams P.D., Pannu N.S., Read R.J., Brünger A.T. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S., Grum-Tokars V., Krucinska J., Kundracik M.L., Wedekind J.E. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Allen F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C. Mechanistic considerations for general acid–base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., Yajima R. Nucleobase catalysis in ribozyme mechanism. Curr. Opin. Chem. Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., Brown T.S., Nakano S., Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Rice L.M. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buzayan J.M., Hampel A., Bruening G. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 1986;14:9729–9743. doi: 10.1093/nar/14.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T.R., Zaug A.J., Grabowski P.J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: Involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Das S.R., Piccirilli J.A. General acid catalysis by the hepatitis δ virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Hol W.G. The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett. 2004;577:315–321. doi: 10.1016/j.febslet.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Doherty E.A., Doudna J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Lorsch J.R. Ribozyme catalysis: Not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- Fedor M.J., Williamson J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Ferat J.L., Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Gong B., Chen J.H., Chase E., Chadalavada D.M., Yajima R., Golden B.L., Bevilacqua P.C., Carey P.R. Direct measurement of a pKa near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J. Am. Chem. Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- Grabowski P.J., Zaug A.J., Cech T.R. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena . Cell. 1981;23:467–476. doi: 10.1016/0092-8674(81)90142-2. [DOI] [PubMed] [Google Scholar]
- Grasby J.A., Mersmann K., Singh M., Gait M.J. Purine functional groups in essential residues of the hairpin ribozyme required for catalytic cleavage of RNA. Biochemistry. 1995;34:4068–4076. doi: 10.1021/bi00012a025. [DOI] [PubMed] [Google Scholar]
- Grum-Tokars V., Milovanovic M., Wedekind J.E. Crystallization and X-ray diffraction analysis of an all-RNA U39C mutant of the minimal hairpin ribozyme. Acta Crystallogr. D Biol. Crystallogr. 2003;59:142–145. doi: 10.1107/s0907444902019066. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R. RNA catalytic properties of the minimum (−)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Eckstein F., Cech T.R. The importance of being ribose at the cleavage site in the Tetrahymena ribozyme reaction. Biochemistry. 1993;32:8312–8321. doi: 10.1021/bi00083a035. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Joseph S., Burke J.M. Optimization of an anti-HIV hairpin ribozyme by in vitro selection. J. Biol. Chem. 1993;268:24515–24518. [PubMed] [Google Scholar]
- Ke A., Zhou K., Ding F., Cate J.H., Doudna J.A. A conformational switch controls hepatitis δ virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Been M.D., Ferré-D'Amaré A.R. Essential role of an active-site guanine in glmS ribozyme catalysis. J. Am. Chem. Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J., Brünger A.T. Checking your imagination: Applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Fedor M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Cottrell J.W., Fedor M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lebruska L.L., Kuzmine I.I., Fedor M.J. Rescue of an abasic hairpin ribozyme by cationic nucleobases: Evidence for a novel mechanism of RNA catalysis. Chem. Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- Lilley D.M. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D.M., Uhlenbeck O.C. Self-cleaving catalytic RNA. FASEB J. 1993;7:25–30. doi: 10.1096/fasebj.7.1.8422971. [DOI] [PubMed] [Google Scholar]
- MacElrevey C., Spitale R.C., Krucinska J., Wedekind J.E. A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme. Acta Crystallogr. D Biol. Crystallogr. 2007;63:812–825. doi: 10.1107/S090744490702464X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D.B., Wedekind J.E. Small ribozymes. In: Cech T., editor. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 265–286. [Google Scholar]
- Nakano S., Chadalavada D.M., Bevilacqua P.C. General acid-base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller H.F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Oivanen M., Kuusela S., Lonnberg H. Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Brønsted acids and bases. Chem. Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- Pannu N.S., Read R.J. Improved structure refinement through maximum liklihood. Acta Crystallogr. A. 1996;52:659–668. [Google Scholar]
- Perrotta A.T., Been M.D. Cleavage of oligoribonucleotides by a ribozyme derived from the hepatitis delta virus RNA sequence. Biochemistry. 1992;31:16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- Perrotta A.T., Shih I., Been M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Pflugrath J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Pinard R., Hampel K.J., Heckman J.E., Lambert D., Chan P.A., Major F., Burke J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes M.M., Reblova K., Sponer J., Walter N.G. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert P.B., Ferré-D'Amaré A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Massey A.P., Sigurdsson S.T., Ferré-D'Amaré A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Ryder S.P., Strobel S.A. Nucleotide analog interference mapping of the hairpin ribozyme: Implications for secondary and tertiary structure formation. J. Mol. Biol. 1999;291:295–311. doi: 10.1006/jmbi.1999.2959. [DOI] [PubMed] [Google Scholar]
- Ryder S.P., Oyelere A.K., Padilla J.L., Klostermeier D., Millar D.P., Strobel S.A. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- Salter J., Krucinska J., Alam S., Grum-Tokars V., Wedekind J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S.A. Ribonucleic general acid. Nat. Chem. Biol. 2005;1:5–6. doi: 10.1038/nchembio0605-5. [DOI] [PubMed] [Google Scholar]
- Symons R.H. Small catalytic RNAs. Annu. Rev. Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Thomas J.M., Perrin D.M. Active site labeling of G8 in the hairpin ribozyme: Implications for structure and mechanism. J. Am. Chem. Soc. 2006;128:16540–16545. doi: 10.1021/ja063942y. [DOI] [PubMed] [Google Scholar]
- Torelli A.T., Krucinska J., Wedekind J.E. A comparison of vanadate to a 2′-5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli A.T., Spitale R.C., Krucinska J., Wedekind J.E. Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme. Biochem. Biophys. Res. Commun. 2008;371:154–158. doi: 10.1016/j.bbrc.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N.G. Ribozyme catalysis revisited: Is water involved? Mol. Cell. 2007;28:923–929. doi: 10.1016/j.molcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.C., Reich E., Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- Wedekind J.E., McKay D.B. Purification, crystallization, and X-ray diffraction analysis of small ribozymes. Methods Enzymol. 2000;317:149–168. doi: 10.1016/s0076-6879(00)17013-2. [DOI] [PubMed] [Google Scholar]
- Wilson T.J., Ouellet J., Zhao Z.Y., Harusawa S., Araki L., Kurihara T., Lilley D.M. Nucleobase catalysis in the hairpin ribozyme. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]