Abstract
A hallmark of mammalian embryonic development is the widespread induction of microRNA (miRNA) expression. Surprisingly, the transcription of many of these small, noncoding RNAs is unchanged through development; rather, a post-transcriptional regulatory event prevents accumulation of the mature miRNA species. Here, we present a biochemical framework for the regulated production of the Let-7 family of miRNAs. Embryonic cells contain a Drosha Inhibitor that prevents processing of the Let-7 primary transcript. This inhibitor specifically binds to conserved nucleotides in the loop region of the Let-7 precursor, and competitor RNAs that mimic the binding site restore Let-7 processing. We have identified the Drosha Inhibitor as the embryonic stem cell specific protein Lin-28. Lin-28 has been previously implicated in developmental regulatory pathways in Caenorhabditis elegans, and it promotes reprogramming of human somatic cells into pluripotent stem cells. Our findings outline a microRNA post-transcriptional regulatory network and establish a novel role for the miRNA precursor loop in the regulated production of mature Let-7.
Keywords: Let-7, Lin-28, miRNA, microRNA
INTRODUCTION
It is now generally appreciated that diverse forms of double-stranded RNA (dsRNA) can act as triggers of RNA interference (RNAi) or related homology-dependent gene silencing pathways (Chapman and Carrington 2007). Among these triggers are microRNAs (miRNAs), which are single stranded, but fold into stable stem–loop structures, providing the essential double-stranded feature. miRNAs are encoded in the genomes of most metazoans and function in a post-transcriptional layer of gene regulation (for review, see Bartel 2004).
The founding miRNA, lin-4, was discovered in Caenorhabditis elegans as a mutant that displayed heterochronic, or developmental timing, defects (Lee et al. 1993; Wightman et al. 1993). The activity of this small RNA is mediated largely through repression of two well established target mRNAs, lin-14 and lin-28 (Ambros 1989). A second miRNA, let-7, was later identified as a heterochronic mutant (Reinhart et al. 2000). Surprisingly, this miRNA has complete nucleotide conservation from C. elegans to humans, suggesting an ancient biological role (Pasquinelli et al. 2000). More recently, thousands of miRNAs have been identified across many phyla. While few validated mRNA targets have been assigned to these miRNAs, computational predictions suggest that each miRNA has tens to hundreds of targets, underscoring their immense potential for controlling gene expression (Bartel 2004).
The initiation of miRNA-dependent gene regulation is the transcription of a primary transcript, or pri-miRNA (for review, see Kim 2005). This RNA is typically thousands of nucleotides long and is often capped, spliced, and polyadenylated. The stem–loop structure is excised by the ribonuclease enzyme Drosha, liberating the precursor, or pre-miRNA. After export out of the nucleus, the precursor is further processed by the enzyme Dicer. The resultant siRNA-like molecule is loaded into the RNAi effector complex RISC, where it directs nucleolytic degradation and translational repression of target mRNAs.
While hundreds of miRNAs have been identified in the human and mouse genomes, the biological role of most miRNAs is unknown. However, numerous studies have linked miRNA function to the regulation of cell growth and differentiation (for review, see Esquela-Kerscher and Slack 2006). For example, the miRNA cluster miR-17-92 is highly expressed in a wide range of cancers, and ectopic expression of this candidate oncogene promotes cancer in several mouse models. This miRNA cluster is also highly expressed in early mouse development, and at least one study has confirmed its role in preventing the differentiation of progenitor cells (Lu et al. 2007). In contrast to these “oncomiRs,” a large number of miRNAs, including the Let-7 family, are depleted in cancer. Let-7 itself has been shown to target the oncogenes Ras, Myc, and HMGA2, and expression of this miRNA inversely correlates with disease severity (Esquela-Kerscher and Slack 2006; Lee and Dutta 2007; Mayr et al. 2007). Interestingly, global down-regulation of all miRNAs promoted disease in a mouse model for lung carcinoma, suggesting that the overall tumor-suppressive functions of miRNAs are more important than the oncogenic functions of, for example, miR-17-92 (Kumar et al. 2007).
RESULTS
Regulated microRNA biogenesis
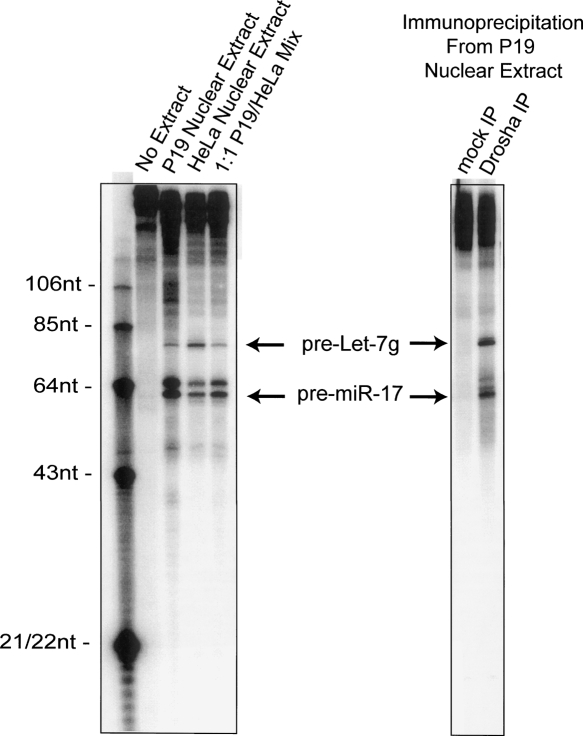
The widespread alterations in miRNA expression in cancer prompted us to investigate the regulatory mechanisms responsible for their production. Our initial studies focused on the Let-7 family, where we previously demonstrated that induction of this miRNA family during mouse embryogenesis occurs at a post-transcriptional stage (Thomson et al. 2006). Since we observed a large amount of primary transcript in the absence of precursor and mature species, we predicted that a block was in place at the Drosha endonuclease processing step. We tested this directly using an established cell-free assay that reports Drosha activity (Lee et al. 2002). We compared the processing efficiency of a regulated miRNA, Let-7g, with a miRNA that is readily processed in all known cell types, miR-17. We incubated radiolabeled primary transcripts for these miRNAs in nuclear extracts prepared from the mouse embryonal carcinoma cell line P19. We had previously shown that this cell line exhibits regulated processing of Let-7, as it has abundant pri-Let-7 but no detectable mature Let-7 (Thomson et al. 2006). As a control, we performed assays in extracts derived from HeLa cells, which have abundant mature Let-7, and thus are competent for Let-7 processing. Nuclear extracts derived from HeLa cells were efficient at processing both Let-7g and miR-17, with approximately equal amounts of each precursor product being generated (Fig. 1). While several RNA species are generated after the Drosha reaction, we were able to confirm the identity of the correct precursor reaction products by Northern blot hybridization of unlabeled products (Supplemental Fig. 1).
Figure 1.
Embryonic cells contain a Drosha Inhibitor that specifically regulates Let-7. (Left) Radiolabeled pri-miRNA substrates corresponding to Let-7g and miR-17 were incubated in P19 or HeLa nuclear extracts, as indicated. (Right) Drosha protein, or mock, was immunoprecipitated from P19 nuclear extracts with a polyclonal antibody. Immobilized protein was incubated with Let-7g and miR-17 pri-miRNA substrates, as indicated. Drosha endonuclease products were resolved on a denaturing polyacrylamide gel. (Arrows) Production of Let-7g and miR-17 precursors. A labeled RNA oligonucleotide ladder is shown for size reference.
While the control nuclear extract was competent for both Let-7g and miR-17 processing, extracts derived from undifferentiated P19 cells were inefficient at processing Let-7g (Fig. 1). We calculated the ratio of Let-7g product to miR-17 product to generate a Let-7 processing efficiency. Undifferentiated P19 cells were ∼10-fold less efficient at processing Let-7g compared with miR-17.
These data demonstrate that Drosha processing of Let-7 is less efficient in embryonic cells, but they do not discriminate between an activator present in the HeLa extract or an inhibitor present in the P19 extract. Therefore, we performed the same assay in a 1:1 mixed extract of P19 and HeLa cells. The mixed extract assay yielded product ratios similar to the P19 extract alone, indicating that the regulatory factor is dominant in P19 cells and is therefore an inhibitor of Let-7 processing (Fig. 1). To rule out the possibility that the regulatory event was due to the modification of Drosha itself, we immunoprecipitated the Drosha protein from undifferentiated P19 nuclear extracts. This purified protein was no longer subject to regulated processing, as it was fully competent for Let-7 processing (Fig. 1).
While our data demonstrate a regulatory point at the Drosha processing step, this does not preclude regulation at other steps in miRNA biogenesis. Specifically, regulation of Let-7 biogenesis at the Dicer processing step has been reported (Wulczyn et al. 2007).
microRNA regulatory sequences
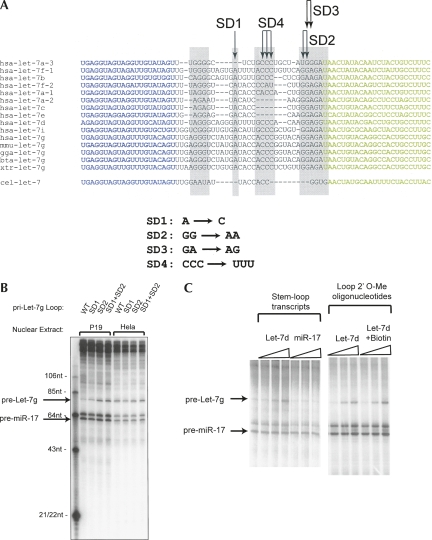
Since Drosha protein that had been immunopurified had lost the regulatory factor, the most logical conclusion of our data was that a Drosha Inhibitor, present in the P19 extract, was interacting directly with the Let-7 primary transcript. Alignment of the stem–loop precursors for the Let-7 family revealed highly conserved nucleotides within the loop region (Fig. 2A). These sequence elements do not contribute to mRNA targeting; therefore, we reasoned they maintained conservation due to a regulatory role. We undertook several approaches to test whether the loop sequences of Let-7 were essential for regulated Drosha processing. Our first strategy was to build chimeric Let-7g primary transcripts that contained loop regions from other, unregulated miRNAs. We did detect weak processing in P19 cells, though it was unclear if these altered stem–loops were interacting properly with the processing machinery (data not shown). As an alternative, we employed site-directed mutagenesis to alter conserved residues in the loop (for sites, see Fig. 2A). Importantly, these mutations did not affect folding of the stem–loop structure based on computational folding algorithms (mFOLD). Pri-Let-7g transcripts containing the SD1 or SD2 mutations were partially released from the Drosha processing block as evidenced by increased pri-Let-7g cleavage in P19 nuclear extracts (Fig. 2B). The SD3 and SD4 mutations had no effect (data not shown). Mutation at both the SD1 and SD2 sites further released the processing block. Importantly, these mutations did not impair general Drosha cleavage, as processing was unaffected in HeLa extracts.
Figure 2.
The loop region of Let-7 interacts with the Drosha Inhibitor. (A) The sequence alignment of Let-7 family members is shown. (Blue text) Mature miRNA sequence; (green text) the complementary stem strand (not exactly the star strand); (gray boxes) regions of homology; (arrows) nucleotides that were mutagenized. Sequence changes are indicated below the alignment. (B) Wild-type (WT) or mutant pri-Let-7g substrates were combined with the pri-miR-17 substrate and incubated in a P19 or HeLa nuclear extract. Drosha products were resolved and are indicated. (C) Pri-miRNA substrates for Let-7g and miR-17 were combined and incubated in P19 nuclear extracts. Competitor RNA transcripts corresponding to the loop plus 12 nt of each stem, or competitor 2′-O-methyl oligonucleotides, were included at 10, 50, and 250 nM final concentration. In one case, the oligonucleotide had a 3′ Biotin moiety. The left lane had no competitor. Drosha products were resolved on a denaturing polyacrylamide gel. (Arrows) Precursor products.
While these data suggest the requirement for these loop nucleotides, this does not formally prove that the Drosha Inhibitor interacts with the loop. To establish this, we designed competitor RNAs based on Let-7 stem–loop sequences. If the Drosha Inhibitor was binding to the loop region, we reasoned that these competitor RNAs should divert the Inhibitor away from the Let-7 primary transcript, restoring processing in P19 cells. We tested competitors corresponding to all 11 Let-7 family members (not including miR-98). Competitors that matched Let-7 stem–loop sequences restored Drosha processing of Let-7g without affecting production of miR-17 (data not shown). The most effective competitor was based on Let-7d. Figure 2C illustrates restoration of Let-7g processing in P19 nuclear extracts in the presence of Let-7d stem–loop competitor. Competitors based on an unregulated miRNA, miR-17, had no effect on Let-7 processing (Fig. 2C). Interestingly, an oligonucleotide corresponding to the loop alone was highly effective, suggesting that a stem–loop structure is not essential for interaction with regulatory proteins (Fig. 2C).
The Drosha Inhibitor
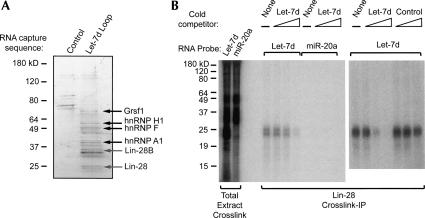
We next investigated the nature of the Drosha Inhibitor. We designed an affinity purification strategy using the Let-7d loop as a capture reagent. We confirmed that a linear sequence, comprised of loop sequence only, fully 2′-O-methyl modified (for stability), and linked to a biotin moiety, was able to restore Drosha processing of Let-7g when used as a competitor (Fig. 2C). We bound this capture probe to streptavidin-agarose and isolated binding proteins from P19 nuclear extracts. Proteins were identified by peptide mass fingerprinting. A large number of proteins specifically bound to the Let-7 loop probe, as shown in Figure 3A. Many of the proteins belong to the hnRNP family. This family of RNA binding proteins has diverse roles in RNA splicing and has been implicated in the regulation of miR-18a processing (Guil and Caceres 2007). Interestingly, we also captured the RNA binding proteins Lin-28 and Lin-28B. This protein family was originally identified in C. elegans as a genetic mutant that exhibited abnormal cell lineage (Ambros and Horvitz 1984). Family members are characterized as having a cold shock domain and two zinc finger domains, a unique domain organization among all known RNA binding proteins (Moss et al. 1997). The biochemical activity of LIN-28 is not well characterized, though it has been suggested to regulate translation of specific mRNAs (Polesskaya et al. 2007).
Figure 3.
The RNA binding protein Lin-28 specifically binds to the Let-7 loop region. (A) Oligonucleotide capture probes corresponding to the Let-7d loop or a random (control) sequence, fully 2′-O-methyl modified, 3′ Biotin linked, were bound to streptavidin agarose. Proteins were captured from P19 nuclear extracts, were resolved on a 4%–20% polyacrylamide gel, and were stained with coomassie blue. Proteins were isolated and identified by MALDI-TOF fingerprinting. (B) Radiolabeled RNA probes corresponding to the Let-7d stem–loop or miR-20a stem–loop were incubated with P19 nuclear extracts. Unlabeled Let-7d loop or control oligonucleotide competitors were included as indicated at 12.5, 125, or 1250 nM. Proteins were crosslinked to probes with UV light. Lin-28 was immunoprecipitated from crosslink reactions with a polyclonal antibody. Total extract (IP input) and immunoprecipitates were resolved, as indicated, on a polyacrylamide gel.
We confirmed the interaction between Lin-28 and the Let-7 loop by UV-crosslink analysis. Radiolabeled Let-7 stem–loop RNA specifically crosslinked a protein in P19 nuclear extracts that matched the size of Lin-28 (Fig. 3B). We confirmed the identity of the protein by immunoprecipitation of crosslink reactions with an antibody to Lin-28. As evident in Figure 3B, one labeled protein of the correct size was recovered from Let-7 crosslink reactions, but no detectable protein was immunoprecipitated from the control reaction. Importantly, the Let-7/Lin-28 interaction was sensitive to the presence of excess competitor Let-7 stem–loop RNA. Control competitors did not disrupt the Lin-28/Let-7 interaction.
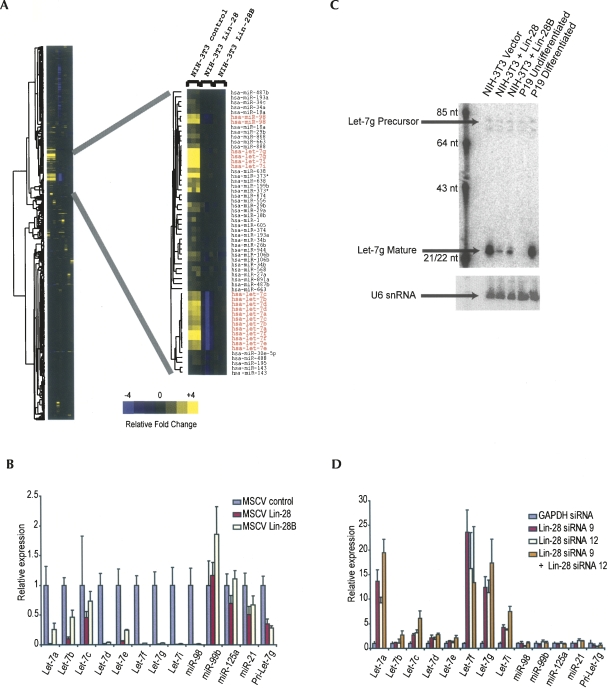
While these data demonstrated interaction of Lin-28 with the Let-7 stem–loop, they did not demonstrate a regulatory role. To test this, we introduced Lin-28 and Lin-28B into NIH-3T3 cells by retroviral transduction and measured steady-state levels of mature miRNAs. Microarray analysis indicated that most miRNAs were unchanged, except for a cluster of miRNAs reduced in cells expressing either Lin-28 or Lin-28B (Fig. 4A). As predicted, this cluster included most Let-7 family members. We confirmed miRNA expression changes by RT-PCR (Fig. 4B). This method is more sensitive and provides greater discrimination among the highly related Let-7 family members. Let-7f, Let-7g, and Let-7i were repressed 240-, 90-, and 195-fold, respectively, with other family members repressed to a lesser degree. We measured steady-state levels of unrelated miRNAs and found no significant change. Importantly, the reduction in Let-7g was at a post-transcriptional step, as the level of the primary transcript was not decreased to the same magnitude (2.8-fold). Northern blot analysis indicated no accumulation of Let-7g precursor, consistent with a block at the Drosha processing step (Fig. 4C). While it is formally possible that Lin-28 promotes a block at the Dicer processing step, this would require an increase in precursor turnover at the same rate as precursor production, since no alteration in precursor steady state levels is detected.
Figure 4.
Lin-28 expression blocks production of Let-7. (A) NIH-3T3 cells were transduced with MSCV retroviral constructs that drive expression of Lin-28, Lin-28B, or control, as indicated. Steady-state miRNA expression levels were quantitated using a custom microarray platform 10 d post-infection. Normalized measurements were hierarchically clustered and are plotted as a heat map. (Yellow) High expression, (blue) low expression, relative to the mean. (Red font) Let-7 family members. (B) Steady-state miRNA expression levels from NIH-3T3 cells expressing Lin-28, Lin-28B, or control were quantitated by real-time RT-PCR. U6 snRNA was used as the reference. Expression of pri-Let-7g was also quantitated by real-time RT-PCR. β2-microglobulin was used as the reference. (C) Let-7g precursor and mature species in NIH-3T3 cells expressing Lin-28, Lin-28B, or control were analyzed by Northern blotting. (D) P19 cells were transfected with siRNAs targeting Lin-28. Two effective siRNAs were used alone or in combination. Five days post-transfection, mature miRNA levels were measured by real-time RT-PCR. U6 was used as a reference. Expression of pri-Let-7g was also quantitated by real-time RT-PCR. β2-microglobulin was used as the reference.
To further confirm a role for Lin-28 in the repression of Let-7 expression, we performed RNAi knockdown of Lin-28 in P19 embryonic cells. Two siRNAs that reduced protein levels >90% released the processing block and led to increased mature Let-7 (Fig. 4D; Supplemental Fig. 3). As above, Let-7 family members had differential response to siRNA knockdown, with Let-7g, Let-7i, Let-7a, and Let-7f the most affected.
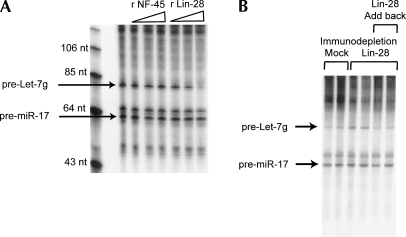
Our final goal was to demonstrate the direct regulation of Let-7 processing by Lin-28. We generated recombinant Lin-28 and an unrelated RNA binding protein NF-45 in Escherichia coli and introduced these purified proteins into our cell free Drosha assay (for recombinant proteins, see Supplemental Fig. 2). As described above, HeLa extracts will process Let-7g with similar efficiency as miR-17. As shown in Figure 5A, Lin-28 inhibited processing of Let-7g without affecting processing of miR-17. This was not a nonspecific effect of RNA binding as the control protein (NF-45) had no effect of processing of either substrate. To further support our conclusions, we removed endogenous Lin-28 from the P19 embryonic cell extract by immunodepletion. This enabled processing of Let-7 (Fig. 5B).
Figure 5.
Lin-28 is necessary and sufficient for regulated Let-7 processing. (A) Pri-miRNA substrates for Let-7g and miR-17 were combined and incubated in HeLa nuclear extracts. Purified, recombinant NF-45 (control) or Lin-28 were included at 2, 20, and 200 ng per reaction. Drosha products were resolved on a denaturing polyacrylamide gel. (Arrows) Precursor products. Recombinant protein was produced in E. coli. (B) (Left lanes) Polyclonal Lin-28 antibody, or mock, was bound to protein A sepharose. P19 nuclear extracts were incubated with immobilized antibody. Resultant immunodepleted extracts were incubated with pri-miRNA substrates for Let-7g and miR-17. (Right lanes) Recombinant Lin-28 was added back to immunodepleted reactions at 100 ng/μL final concentration. Drosha products were resolved on a denaturing polyacrylamide gel. (Arrows) Precursor products. Recombinant protein was produced in HEK-293 cells.
DISCUSSION
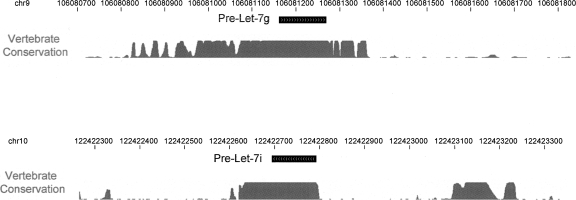
Our results outline a regulatory mechanism for the production of the Let-7 family of miRNAs. The primary transcripts for these miRNAs are uniformly expressed during embryonic development. The interaction of a Drosha Inhibitor with the loop structure of the pri-miRNAs leads to a block at the Drosha processing step. We have identified conserved nucleotides within the loop that are essential for the Drosha processing block. This is the first report of cis-regulatory elements within the Let-7 primary transcript. While our work focused on the loop region of Let-7, it is interesting that sequence conservation extends outside the stem–loop precursor. Figure 6 illustrates that extensive vertebrate conservation is present in sequences flanking the precursor. This sequence may be important for splicing of the host gene Wdr82. However, the proximity to the stem–loop suggests these elements play a regulatory role in some aspect of miRNA function. In further support of this, Let-7 family members that are not intronic (for example, Let-7i, as shown in Fig. 6) have conserved sequence elements flanking the stem–loop.
Figure 6.
Extensive sequence conservation of pri-Let-7. The genomic locus of the mouse pre-Let-7g and pre-Let-7i stem–loop sequence are shown, with 600 nt flanking on each side included. The conservation plot of 30 vertebrate species is shown. Data were obtained from the UCSC genome browser, July 2007 release.
We identify Lin-28 as a protein that interacts with the loop region of Let-7. This interaction is specific for Let-7 family members and has a dissociation constant in the high nanomolar range, based on competitor assays (Fig. 3B) and fluorescent anisotropy binding assays (not shown). The interaction of Lin-28 with Let-7 inhibits Drosha processing. This RNA binding protein is highly expressed in early development and has been proposed as a marker for ES cells (Yang and Moss 2003; Richards et al. 2004). Differentiation of P19 cells with retinoic acid leads to robust induction of Let-7; Lin-28 protein decays at the time point that Let-7 processing is enabled (Lee et al. 2005; Wu and Belasco 2005). We propose that a similar decay of this protein during embryonic development is what allows Drosha processing of Let-7 and eventual production of the mature species.
While it is clear that Let-7 processing is negatively regulated by Lin-28, is this the primary mode of regulation for the production of this miRNA family? We have previously reported that Let-7 processing efficiency increases >1000-fold during mammalian development (Thomson et al. 2006). Our overexpression studies, however, perturb Let-7 levels ∼100-fold. It is probable that we are not achieving high enough expression levels to fully recapitulate embryonic cells, and Western blot analysis confirms this (Supplemental Fig. 4). Similarly, our siRNA knockdown studies do not fully eliminate Lin-28 expression, and it is possible that remaining Lin-28 protein has some repressive function (Supplemental Fig. 3). These points suggest we have identified the major regulatory point in Let-7 production, though we cannot rule out contributions at Dicer processing or at pri-Let-7 transcription, both of which have been reported (Wulczyn et al. 2007; Chang et al. 2008).
Recently, an independent study has demonstrated that Lin-28 selectively inhibits processing of Let-7 (Viswanathan et al. 2008). Similarly, this work suggested inhibition at the Drosha processing step, lending further support to our model proposed herein.
The exact mechanism whereby Lin-28 inhibits Drosha processing is not known. Its high affinity binding to Let-7 may simply block access of Drosha or DGCR8 to the pri-miRNA. This binding event may promote turnover of the pri-miRNA, as there is no accumulation of this RNA species under conditions where processing is inhibited. Alternatively, pri-miRNAs may have a short half-life under all conditions and therefore are degraded if not rapidly processed by the Drosha machinery. A final possibility is that the Lin-28/Let-7 complex is exported out of the nucleus and shuttled to P-bodies for degradation. Lin-28 is known to shuttle between the nucleus and cytoplasm and has been localized to P-bodies, lending support to this hypothesis (Balzer and Moss 2007).
The regulation and developmental expression of the mammalian Lin-28 closely parallels that of the C. elegans ortholog, suggesting it performs a similar function in that organism. Expression of let-7 during C. elegans larval development is known to be regulated at transcription (Johnson et al. 2003). In lin-28 mutants, however, the production of mature let-7 is accelerated by one larval stage. Similarly, in lin-4 mutants, which have elevated expression of lin-28, the production of mature let-7 is delayed (Johnson et al. 2003). While it is not known if these effects are a result of direct interaction between the lin-28 protein and the let-7 primary transcript, it is possible that the mechanism we report here is conserved from C. elegans to mammals.
The role of Lin-28 in mammalian development is not known. However, a combination of Lin-28, Nanog, Oct-3/4, and Sox2 are sufficient to reprogram human somatic cells to pluripotent stem cells (Yu et al. 2007). The role of Nanog, Oct-3/4, and Sox2 in maintaining pluripotency is well established (Pan and Thomson 2007). The additional requirement for Lin-28 in these studies suggests that restoration of the Let-7 processing block is an essential step for reprogramming stem cells. These findings strongly implicate miRNA expression patterns as important determinants of stem cell fate.
MATERIALS AND METHODS
Pri-microRNA substrates
Substrate constructs (stem–loop with ∼10 nucleotides [nt] of ssRNA flanking region) were created by the polymerase chain reaction (PCR) using oligonucleotides as the DNA templates:
pri-Let-7g: 5′-TGCCTGATTCCAGGCTGAGGTAGTCGTTTGTACAGTTTGAGGGTCTATGATACCACCCGGTACAGGAGATAACTGTACAGGCAACTGCCTTGCCAGGAACAGCGCG-3′; and
pri-miR-17: 5′-GTCAGAATAATGTCAAAGTGCTTACAGTGCAGGTAGTGATATGTGCATCTACTGCAGTGAAGGCACTTGTAGCATTATGGTGAC-3′.
Forward and reverse primers added ssRNA flanks as well as 5′ BglII and 3′ XhoI restriction sites.
Primers for pri-Let-7g:
Forward: 5′-GTCAAGATCTCGTTTCCTTTTGCCTGATTCCAGGCTGA-3′; and
Reverse: 5′-GTCACTCGAGGGCAGCTGGCGCGCTGTTCCTGGC-3′.
Primers for pri-miR-17:
Forward: 5′-GTCAAGATCTATTGTGACCAGTCAGAATAATGTCAAAGTGCTTACAG-3′; and
Reverse: 5′-GTCACTCGAGCGAGGCAGCTGTCACCATAATGCTACAAGTGCCT-3′.
The resulting PCR products were digested and cloned into a MSCV-splice-donor/splice-acceptor vector (SDSA3.0) (J.M. Thomson and S.M. Hammond, unpubl.) based on the MSCV-puro vector (Clontech).
Pri-microRNA transcription templates were created from the pri-microRNA constructs described above; PCR was employed to add on a 5′ T7 promoter (5′-TCGTAATACGACTCACTATAGGGTCCGCTAGCCTAGCTACTACCA-3′ and 5′- ATAAGTATGATATTGTCAAGGAAACCC-3′). Radiolabeled pri-microRNAs were created using T7 RNA polymerase (NEB) in accordance with the manufacturer's instructions. The resulting pri-microRNA substrates were (pri-microRNA sequences are in bold type; flanking vector sequences are italicized):
Pri-Let-7g: TCCGCTAGCCTAGCTACTACCAGGTGAGTGGAGATCT CGTTTCCTTTTGCCTGATTCCAGGCTGAGGTAGTCGTTTGTACAGTTTGAGGGTCTATGATACCACCCGGTACAGGAGATAACTGTACAGGCAACTGCCTTGCCAGGAACAGCGCGCCAGCTGCC CTCGAGGTTTAAACGAATTCAGGGTTTCCTTGACAATATCATACTTAT; and
Pri-miR-17: TCCGCTAGCCTAGCTACTACCAGGTGAGTGGAGATCT ATTGTGACCAGTCAGAATAATGTCAAAGTGCTTACAGTGCAGGTAGTGATATGTGCATCTACTGCAGTGAAGGCACTTGTAGCATTATGGTGACAGCTGCCTCG CTCGAGGTTTAAACGAATTCAGGGTTTCCTTGACAATATCATACTTAT.
Cell culture and nuclear extract preparation
P19 cells were differentiated for 10 d in retinoic acid, as described (Thomson et al. 2006). Undifferentiated and differentiated P19 cells and HeLa S3 nuclear extracts were prepared as previously described (Dignam 1990). In brief, cell pellets, washed once in phosphate buffered saline (PBS), were resuspended in 2.5 volumes of Buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiotreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 20% glycerol, and Complete protease inhibitors [CPI, Roche]) and incubated for 10 min on ice. Cells were then lysed using 5–10 passes in a dounce homogenizer. Nuclei were centrifuged at 1200 rpm for 5 min, followed by centrifugation at 15,000g for 15 min, and the resulting supernatant was discarded. Pelleted nuclei were extracted in buffer C (20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, 20% glycerol, and CPI) for 30 min at 4°C (3 mL of buffer C was used per 1 × 109 cells). Nuclear debris was then pelleted by centrifugation at 15,000 rpm for 30 min, and the resulting nuclear extract was dialyzed for 5 h against >50 volumes of buffer D (20 mM HEPES, pH 7.9, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, and 20% glycerol).
Cell-free Drosha processing assay
Drosha assay reactions were carried out as previously described with some modifications (Lee et al. 2002). 1 × 105 cpm each of pri-Let-7g and pri-miR-17 transcripts were combined and incubated with 100 μg of nuclear extract (where indicated, either 100 μg of P19 extract, 100 μg of differentiated P19 extract, 100 μg of HeLa extract, or 50 μg P19 + 50 μg HeLa extract) + 0.1μL of RNasin (Promega) for 30 min at 37°C. RNA was isolated using Trizol and precipitated in cold isopropanol. RNA pellets were washed once in 70% ethanol and resuspended in 15 μL of formamide loading buffer. Processed pri-microRNA products were resolved on a 12.5% acrylamide–8 M urea sequencing gel and visualized by autoradiography using a Storm PhosphorImager (GE Life Sciences); pre-microRNA band intensities were quantitated using Imagequant software (GE Life Sciences). A molecular weight ladder of radiolabeled, concatamerized RNA oligos was used to determine the size of the pre-microRNA products.
Drosha immunoprecipitation
For Drosha assays using immunoprecipitated Drosha, we first generated rabbit polyclonal anti-Drosha antibody using the peptide sequence CPEEAEDIKK. 250 μL of P19 nuclear extract was pre-cleared with 25 μL of protein-A sepharose and incubated either with 40 μL of affinity-purified Drosha antibody and 45 μL of packed protein-A sepharose or with protein-A sepharose alone. Igepal CA-630 (Sigma) was added to 0.5% v/v, and immunoprecipitation reactions were incubated for 1 h at 4°C. Beads were then washed three times in buffer D + 0.5% Igepal.
Individual Drosha reactions were carried out as described above using 10 μL of immobilized protein from each immunoprecipitation reaction, pri-miRNA substrates individually or combined, and buffer D and 1 μL of RNasin.
Drosha assay Northern blot
To identify pre-microRNA products, Drosha assays were carried out as described above except that 0.15 μg of unlabeled pri-microRNA was individually incubated in HeLa extract. Separated RNA products were then transferred onto Nylon (Amersham) which was then UV crosslinked and hybridized to locked nucleic acid (LNA) probes (Integrated DNA Technologies) that were labeled on the 5′ end with T4 polynucleotide kinase (NEB). Hybridizations occurred overnight at 65°C, were washed three times at 65°C in 0.4× SSC + 0.2% SDS, and cross-reacted bands were visualized by autoradiography.
Northern probe sequences are listed below (lowercase denotes LNA; uppercase denotes DNA):
Let-7g: 5′-ACTgTaCaAaCgAcTaCcTcA-3′; and
miR-17-3p: 5′-ACAaGtGcCtTcAcTgCaGt-3′.
Stem–loop RNA competitor assay
Drosha reactions were pre-incubated on ice for 30 min with unlabeled microRNA stem–loop RNAs added to final concentrations of 12.5 nM, 125 nM, and 1250 nM, respectively. The competitor RNAs were either 2′-O-methlyated RNA oligos (Dharmacon) or were in vitro transcribed from long double-stranded DNAs (dsDNAs) encoding a T7 promoter, the upper 10 base pairs of stem, and the loop of the indicated pri-microRNA. The dsDNAs were created by annealing a T7 adapter primer to a longer microRNA primer, followed by a brief PCR extension step with Pfu DNA polymerase (Promega) (5 min, 95°C; 1 min, 59°C; 10 min, 72°C 1 cycle).
T7 adapter: 5′-gatgTAATACGACTCACTATAGGG-3′;
hsa-let-7d: 5′-caggtcgtatagttacctccttgtgggcaaaatccctgccctaaaactatgcaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7a-2: 5′-gaggctgtacagttatctcccttgatgtaattctaaactatacaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7f-1: 5′-agattgtatagttatctcctgaacagggtaaaatcactaccccacaactatacaatcccctatagtgagtcgtattaCATC-3′;
hsa-let-7c: 5′-aaggttgtacagttaactcccagggtgtaactctaaaccatacaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7a-3: 5′-tagattgtatagttatcccatagcagggcagagccccaaactatacaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7i: 5′-agcttgcgcagttatctccacagcgggcaatgtcacaacccgaccaacagcacaaacccctatagtgagtcgtattaCATC-3′;
hsa-let-7f-2: 5′-tagactgtatagttatctccaagatggggtatgaccctaaaactatacaatcccctatagtgagtcgtattaCATC-3′;
hsa-let-7g: 5′-ggcctgtacagttatctcctgtaccgggtggtatcatagaccctcaaactgtacaaacccctatagtgagtcgtattaCATC-3′;
hsa-let-7a-1: 5′-tagattgtatagttatctcccagtggtgggtgtgaccctaaaactatacaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7e: 5′-gaggccgtatagtgatctccttgggtgtcctcctcaactatacaaccccctatagtgagtcgtattaCATC-3′;
hsa-let-7b: 5′-aggttgtatagttatcttccgaggggcaacatcactgccctgaaaccacacaaccccctatagtgagtcgtattaCATC-3′;
hsa-mir-17: 5′-tcactgcagtagatgcacatatcactacctgcactgtccctatagtgagtcgtattaCATC-3′; and
hsa-mir-20a: 5′-ctcataatgcagtagataactaaacactacctgcactatccctatagtgagtcgtattaCATC-3′.
Lin28 immunodepletion Drosha assay
Protein A-sepharose beads were washed three times in “Hi DTT” immunoprecipitation (IP) buffer (20 mM HEPES, pH 7.6, 2 mM MgCl2, 150 mM NaCl, 10 mM DTT, 1 mM PMSF, 0.5% NP-40) followed by five washes in IP buffer (20 mM HEPES, pH 7.6, 2 mM MgCl2, 150 mM NaCl, 1 mM DTT, 1 mM PMSF, 0.5% NP-40). 15 μL of anti-Lin-28 antibody (Abcam) was prebound to 30 μL of protein A sepharose beads (Sigma) for 1 h at 25°C in IP buffer, followed by extensive washing with buffer D + 0.5% NP-40. 100 μL of P19 nucelar lysate (5 mg/mL) was diluted twofold in buffer D + 2 μL of RNasin and rotated overnight at 4°C with either the antibody–beads mixture or beads alone as a mock.
Recombinant protein competitor assay
Drosha assays were carried out as described above except that, where indicated, 2 ng, 20 ng, and 200 ng of recombinant protein was added to HeLa extract/pri-microRNA mixtures before the 37°C incubation step.
RNA affinity pulldown
30 μL of 100 μM Let-7d Loop biotinylated 2′-O-methylated RNA oligonucleotide, (5′-UUmAmGmGmGmCmAmGmGmGmAmUmUmUmUmGmCmCmCmAmCmAmAmGmGmAmGmGmU-18s-Bi-3′, Dharmacon) or a nonspecific control oligo (5′-Bi-18S-18S-mAmUmAmAmGmUmAmUmGmAmUmAmUmUmGmUmC-3′, Dharmacon) was bound to 150 μL of streptdavidin-agarose beads (Fluka) for 1 h at 37°C in “high salt” buffer D (+1 M KCl). Bead–oligo mixtures were washed three times in buffer D+T. 15 mg of P19 cell nuclear extract (equivalent to 1 × 109 cells) was diluted up to 20 mL in buffer D+T, pre-cleared with streptavidin agarose beads, and incubated with bead–oligo mixtures for 3 h at 25°C. Beads were then washed six times in buffer D+T, and bound proteins were eluted using SDS protein loading buffer + 50 mM DTT. Proteins were separated on a 4%–20% pre-cast polyacrylamide gel (Jule Biotechnologies) and stained with Coomassie brilliant blue G-250 (Bio-Rad); appropriate bands were excised and submitted to the UNC–Duke Michael Hooker Proteomics Facility for tryptic digestion and MALDI-TOF footprinting.
UV crosslinking
Crosslinking experiments were carried out essentially as previously described (Myer et al. 1997). In brief, 50 μg of P19 nuclear lysate, 3 μg of yeast tRNA, and 5 × 105 cpm of hsa-Let-7d or hsa-miR-20a loop probes (same competitor RNA sequences as in the “stem–loop RNA competitor assay” above) were combined and adjusted to a volume of 14 μL with buffer D (20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF). Reaction mixtures were incubated for 30 min at 25°C, followed by irradiation with 1 Joule of UV light. Each sample was then incubated with 10 μg of RNase A for 30 min at 37°C. The samples were then either prepared for immunoprecipitation (see below) or boiled in SDS protein loading buffer for 5 min (“input” samples). Where indicated, 2′-O-methyl oligonucleotides were added to the final concentration of 12.5 nM, 125 nM, or 1250 nM to the reaction mixture and pre-incubated for 30 min at 25°C prior to the addition of the labeled probe. 2′-O-methyl competitor sequences are given below:
Let-7d loop 2′-O-methyl: 5′-mUmUmAmGmGmGmCmAmGmGmGmAmUmUmUmUmGmCmCmCmAmCmAmAmGmGmAmGmGmU-3′; and
Nonspecific 2′-O-methyl: 5′-mAmCmCmAmAmCmAmGmGmCmCmGmGmGmAmCmAmAmGmUmGmCmAmmAUmAmAmC-3′.
Crosslinking immunoprecipitations
RNase A-treated samples were boiled in TSD buffer (50 mM Tris-Cl, pH 7.5, 1% SDS, 5 mM DTT) for 10 min and diluted 10-fold in TNN buffer (50 mM Tris-Cl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, and complete protease inhibitor [Roche]). Diluted lysates were pre-cleared with protein-A sepharose beads (Sigma) followed by immunoprecipitation overnight at 4°C with anti-Lin28 antibody (Abcam) and protein A-beads or with protein-A beads alone (“mock” immunoprecipitation). Protein-bound beads were washed extensively with cold TNN buffer; proteins were eluted by boiling SDS protein loading buffer and were separated on a 12% SDS-PAGE gel, which was subsequently fixed and dried down. Radiolabeled proteins were visualized by autoradiography.
Recombinant proteins
cDNAs were amplified from full-length ESTs coding for NFAT-45 and Lin-28 as well as a partial EST for Lin28B (Open Biosystems) using the following primers:
Lin-28:
5′-tacgaattcACCATGGACTACAAAGACGATGACGACAAGGGCTCGGTGTCCAACCA-3′; and
5′-catgcggccgcTCAATTCTGGGCTTCTGGG-3′.
Lin28B:
5′-AGCCAGAAAAACTGCCCGGGCTGGCAGAGGACGAACCCCAGGTTCTGCATGGC-3′; and
5′-TGACGACAAGGCCGAAGGCGGGGCAAGCAAAGGTGAAGAGCCAGAAAAACTGCCCG-3′.
To rebuild the 5′ end of the Lin28B gene by overlap extension PCR:
5′-tacgaattcACCATGGACTACAAAGACGATGACGACAAGGCCGAAG-3′; and
5′- catgcggccgcCTAAGTCTTTTTCCGTTTCTGAATCA-3′.
NFAT 45:
5′-tacgaattcACCATGGACTACAAAGACGATGACGACAAGAGGGGTGACAGAGGAC-3′; and
5′-catgcggccgcTCACTCCTGAGTCTCCATGC-3′.
Flag-tagged cDNAs were cloned into pcDNA 3.0 using 5′ EcoRI and 3′ NotI restriction sites. cDNAs were subsequently subcloned into pMSCV-puro-IRES-GFP (He et al. 2005). For overexpression studies, NIH-3T3 cells were transduced using virus generated with the LinXE ecotropic packaging line. Cells were selected with puromycin. Ten days post-infection, total RNA and protein samples were isolated using Trizol and SDS loading buffer, respectively. MicroRNA expression levels were examined by quantitative real-time PCR (qRT-PCR) as well as by microRNA microarray as previously reported (Thomson et al. 2004, 2006). Protein overexpression was verified by Western blot with a Lin-28 antibody (Abcam).
Purification of His-tagged Lin-28 and NF-45
The following primers were used to amplify mouse coding sequences for NF-45 and Lin-28:
6×His-NF45: 5′-ggccatcatcatcatcatcacaggggtgacagagga-3′;
BsmFI_6×His_NF45_push: 5′-cagtgggacgctgtctcaccatgggccatcatcatc-3′;
XhoI_NF45: 5′-catgctcgagtcactcctgagtctccatgc-3′;
6×_His_Lin28: 5′-ggccatcatcatcatcatcacggctcggtgtccaac-3′;
NcoI_6×His_Lin28_push: 5′-ataccatgggccatcatcatcatcac-3′; and
Lin28_XhoI: 5′-catctcgagtcaattctgggcttctggg-3′.
PCR products were inserted into the pET28b vector (Novagen) after NcoI/XhoI (vector; Lin28) or BsmFI/XhoI (NF45) digestion. BL21 (DE3) pRIL E. coli (Stratagene) served as the host for protein expression. Briefly, positive clones were grown in 1 L of LB to a density of 0.6 OD600 at which time recombinant protein was induced by the addition of 0.25 mM IPTG and allowed to grow for an additional 3 h. Cells were harvested by centrifugation, and cell pellets were suspended in 30 mL of Talon resin equilibration/wash buffer, 50 mM sodium phosphate, pH 7.0, 300 mM NaCl (Clontech). Cells were lysed by sonication and cleared by centrifugation. The cell lysate was incubated with 5 mL of equilibrated Talon resin beads and allowed to bind for 1 h at 4°C. The resin was batch washed 3 × 20 min in 30 mL of equilibration/wash buffer and applied to a disposable column. The protein was eluted along a 50-mL gradient of equilibration buffer containing 0–200 mM imidazole, and fractions were collected. Pure fractions were combined and dialyzed in buffer D (20 mM Hepes, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 1.5 mM MgCl2, 1 mM PMSF, 1 mM DTT, and 20% glycerol). Aliquots were stored at −80°C.
Purification of Flag-tagged recombinant Lin-28
Twenty 10-cm plates of HEK293 cells were transiently transfected with either Flag-tagged NFAT-45, Lin28, or Lin28B overexpression constructs using Fugene 6 (Roche) according to the manufacturer's instructions. After 48 h, cells were harvested by scraping in cold PBS, centrifuged at 1200 rpm for 5 min, and lysed in IP buffer. Lysates were pre-cleared with 50 μL of protein A beads for 1 h followed by immunoprecipitation with 50 μL of M2 Flag agarose beads (Sigma) overnight at 4°C. Beads were then pelleted and washed 10 times in cold buffer D followed by elution with Flag peptide (Sigma) at 400 μg/mL in buffer D. Purity of eluted proteins was verified by SDS-PAGE followed by coomassie blue staining of the gel.
Lin-28 knockdown/overexpression studies
P19 cells were seeded in six-well plates at ∼40% confluency and 2 h later were transfected with siRNAs to either GAPDH or Lin-28 (Dharmacon) using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer. Total RNA and protein was isolated ∼72 h later using Trizol and SDS loading buffer, respectively. microRNA expression levels were examined by quantitative real-time PCR (qRT-PCR) (Thomson et al. 2006). The siRNA sequences were:
si #2: 5′-ggagacaggugcuacaacuuu-3′;
si #9: 5′-ugacguaucuugugcguuuuu-3′; and
si #12: 5′-aaaugugucucacggguuuuu-3′.
Lin-28 and Lin-28B were ectopically expressed using MSCV retroviral expression constructs. MSCV retrovirus was transduced into NIH-3T3 cells and cells were selected with puromycin. Ten days post-infection, cells were harvested and miRNA expression levels were characterized by microarray analysis and real time RT-PCR essentially as described (Thomson et al. 2004, 2006). Northern blot analysis was performed as described, using a radiolabeled LNA probe for Let-7g (Thomson et al. 2006).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Keith Woods for technical support. We thank Zbigniew Dominski and William Marzluff for advice and reagents. We thank members of the Hammond Laboratory for discussions and comments on the manuscript. Treeview and Cluster software was obtained from Stanford University. This research was funded by the National Institutes of Health, the American Cancer Society, and by funds from the Department of Cell and Developmental Biology, UNC-CH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1155108.
REFERENCES
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans . Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H.R. Heterochronic mutants of the nematode Caenorhabditis elegans . Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Balzer E., Moss E.G. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Carrington J.C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Dignam J.D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Guil S., Caceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.M., Lin S.Y., Slack F.J. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Kim H.K., Chung S., Kim K.S., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Lu Y., Thomson J.M., Wong H.Y., Hammond S.M., Hogan B.L. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Myer V.E., Fan X.C., Steitz J.A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes & Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Richards M., Tan S.P., Tan J.H., Chan W.K., Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Post-transcriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn F.G., Smirnova L., Rybak A., Brandt C., Kwidzinski E., Ninnemann O., Strehle M., Seiler A., Schumacher S., Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang D.H., Moss E.G. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Brain Res. Gene Expr. Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]