Abstract
Pre-mRNA splicing proceeds through assembly of the spliceosome complex, catalysis, and recycling. During each cycle the U4/U6.U5 tri-snRNP is disrupted and U4/U6 snRNA base-pairing unwound, releasing separate post-spliceosomal U4, U5, and U6 snRNPs, which have to be recycled to the splicing-competent tri-snRNP. Previous work implicated p110—the human ortholog of the yeast Prp24 protein—and the LSm2-8 proteins of the U6 snRNP in U4/U6 recycling. Here we show in vitro that these proteins bind synergistically to U6 snRNA: Both purified and recombinant LSm2-8 proteins are able to recruit p110 protein to U6 snRNA via interaction with the highly conserved C-terminal region of p110. Furthermore, the presence of a 2′,3′-cyclic phosphate enhances the affinity of U6 snRNA for the LSm2-8 proteins and inversely reduces La protein binding, suggesting a direct role of the 3′-terminal phosphorylation in RNP remodeling during U6 biogenesis.
Keywords: splicing, snRNP recycling, U6 snRNA, Sm-like proteins, La antigen
INTRODUCTION
The removal of introns from eukaryotic pre-mRNAs is catalyzed by a large macromolecular complex, the spliceosome, which consists of more than 100 different proteins and five small nuclear RNAs (snRNAs). It assembles on the pre-mRNA in a highly ordered and stepwise process, involving the U1, U2, and the U4/U6.U5 snRNPs as well as many non-snRNP protein factors. Prior to the first catalytic step of the splicing reaction, dynamic structural rearrangements occur that lead to the formation of the active site of this macromolecular enzyme. This includes the destabilization and early release of the U1 and U4 snRNPs from the activated spliceosome. During this initial step of the splicing reaction the U4 and U6 snRNAs that are extensively base-paired to each other in the U4/U6.U5 tri-snRNP are unwound by the RNA helicase Brr2/U5-200K. This enables the U6 snRNA to participate in a mutually exclusive RNA base-pairing interaction with the U2 snRNA and the pre-mRNA in the catalytic core of the spliceosome. After completion of both steps of the splicing reaction, the machinery is dismantled, releasing mRNA and intron-lariat products as well as the U2, U5, and U6 snRNPs as single entities (for review, see Brow 2002).
For participating in further rounds of splicing the postspliceosomal single U4, U5, and U6 snRNPs have to be recycled to the U4/U6.U5 tri-snRNP. First, the U4/U6 di-snRNP with base-paired U4 and U6 snRNAs reforms, which subsequently associates with the U5 snRNP via protein–protein interactions yielding the splicing-competent U4/U6.U5 tri-snRNP. We have recently identified and characterized the human protein p110, which is closely related to the yeast Prp24 protein (Shannon and Guthrie 1991); p110 binds specifically to an internal U6 sequence, remains only transiently associated with U6 snRNA, and plays a major role in recycling the U4/U6 di-snRNP, a prerequisite for generation of the splicing-competent U4/U6.U5 tri-snRNP (Raghunathan and Guthrie 1998; Bell et al. 2002; Damianov et al. 2004). p110 escorts U6 to the Cajal bodies, where U4/U6 snRNP recycling takes place (Stanek et al. 2003).
The p110-mediated interaction of the U4 and U6 snRNAs is most likely also involved in the de novo assembly of the U4/U6 snRNP. The biogenesis of the spliceosomal U6 snRNP differs in many ways from the biogenesis of the other spliceosomal snRNPs. In contrast to U1, U2, U4, and U5 snRNAs that are RNA polymerase II transcripts and have a cytoplasmic maturation phase, U6 snRNA is transcribed by RNA polymerase III, receives a γ-monomethyl cap (Shimba and Reddy 1994), and probably never leaves the nucleus (Will and Lührmann 2001 and references therein). Furthermore, U6 snRNA contains a 3′-oligouridine stretch, the typical RNA polymerase III termination signal, that is initially bound by the La protein, an RNA chaperone and stability factor. Subsequently 2′-O-methyl ribose and pseudouridine modifications are introduced into U6 snRNA by guide modification RNPs, the snoRNPs (Tycowski et al. 1998; Tycowski et al. 2004), a process taking place in the nucleoli, where U6 snRNA transiently travels through (Lange and Gerbi 2000).
Surprisingly this 3′-terminal uridine stretch of U6 is not static, but noncoded residues are post-transcriptionally incorporated into U6 snRNA by a U6-specific terminal uridyl transferase (TUTase; Trippe et al. 1998, 2006), and therefore U6 variants with up to 12 uridines are found that can assemble into di- and tri-snRNPs (Lund and Dahlberg 1992; Tazi et al. 1993). The majority of mature U6 snRNA carries a 2′,3′-cyclic phosphate end (Lund and Dahlberg 1992), and is associated with a set of seven LSm proteins, LSm2-8, instead of the La protein (Séraphin 1995; Achsel et al. 1999; Mayes et al. 1999; Salgado-Garrido et al. 1999; Vidal et al. 1999; for review, see Khusial et al. 2005). The LSm2-8 proteins are organized in form of a heteroheptameric ring structure, analogously to the classical Sm ring, and LSm binding requires the 3′-terminal uridine stretch, the same region where La is initially bound. Therefore, it is assumed that after formation of the 2′,3′-cyclic phosphate at the 3′ end of U6 snRNA, the La protein dissociates from U6 snRNA. Once the LSm2-8 complex is bound, it keeps U6 in the nucleus (Spiller et al. 2007).
Interestingly, LSm2-8 proteins purified from HeLa nuclear extract interact with U6 snRNA in vitro and promote U4-U6 snRNA annealing without any additional factors (Achsel et al. 1999). Moreover, a direct interaction between the LSm2-8 proteins and the Prp24 recycling factor was demonstrated in S. cerevisiae; deletion of the highly conserved C-terminal sequence motif in Prp24 abolished the interaction with LSm2-8 in yeast-two-hybrid assays; it also resulted in a cold-sensitive growth phenotype, due to inefficient U4/U6 snRNP recycling based on reduced association of Prp24 with the U6 snRNP (Rader and Guthrie 2002). Furthermore, extracts prepared from yeast strains lacking functional LSm6 or -7 proteins—although viable—were unable to maintain splicing activity over several sequential incubations either due to a reduced stability of the U4/U6 di-snRNP and U4/U6.U5 tri-snRNP or—more likely—a defect in U4/U6 recycling (Verdone et al. 2004). Finally, the LSm site of U6 snRNA becomes accessible for crosslinking during splicing, and thus the LSm proteins appear to dissociate during activation of the spliceosome (Chan et al. 2003; Bessonov et al. 2008). Taken together, these data imply a direct role for LSm proteins in U4/U6 snRNP recycling with an association/dissociation cycle similar to what we have proposed for the function of p110 (Bell et al. 2002).
Here we report that LSm2-8 and p110 proteins bind synergistically to U6 snRNA. This effect depends both on the presence of the C-terminal, highly conserved region of p110 and an intact LSm2-8 binding site in U6 snRNA. Furthermore, compared to a 3′-hydroxyl, the presence of a U6 2′,3′-cyclic phosphate end enhances the affinity to recombinant LSm2-8 proteins. Inversely, the interaction with recombinant La protein is impaired by the 2′,3′-cyclic phosphate end, implying a direct role of 3′-terminal modification in RNP remodeling during U6 snRNP biogenesis.
RESULTS
3′-terminal modifications of U6 snRNA modulate binding of LSm2-8 and La proteins
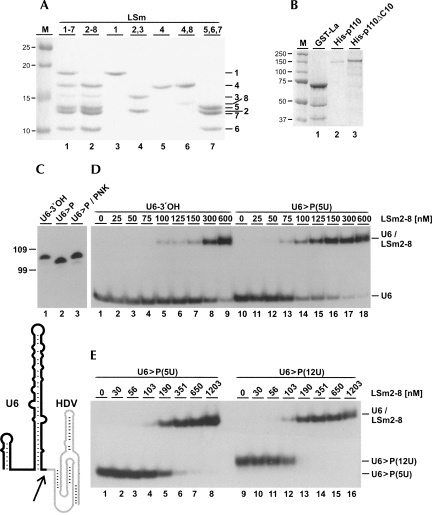
Mature U6 snRNA carries an unusual 3′ terminus with a 2′,3′-cyclic phosphate (>P), in contrast to newly synthesized U6 snRNA with a uridine stretch of variable length and a 3′-hydroxyl end (Lund and Dahlberg 1992). Therefore we compared binding of the LSm2-8 complex to U6 snRNAs with a 3′-hydroxyl or a 2′,3′-cyclic phosphate end (Fig. 1C,D; see Fig. 1A for a protein analysis). U6 snRNA was in vitro transcribed, either by DraI runoff transcription (U6-3′OH) or by Hepatitis delta virus (HDV) ribozyme-mediated endonucleolytic cleavage (U6>P), followed by gel purification. As expected, U6 snRNA with a 2′,3′-cyclic phosphate end had a slightly faster gel mobility; polynucleotide kinase treatment converted the 2′,3′-cyclic phosphate end to 2′- and 3′-hydroxyl termini (Fig. 1C, cf. lanes 1–3; Cameron and Uhlenbeck 1977).
FIGURE 1.
3′-terminal modifications of the U6 snRNA modulate LSm2-8 binding. Analysis of recombinant proteins and of purified human LSm2-8 complex. (A) Recombinant LSm1-7 (lane 1) and LSm2-8 (lane 2) complexes, as well as subcomponents LSm1, 2/3, 4, 4/8, and 5/6/7 (lanes 3–7) (Zaric et al. 2005), were analyzed by SDS-PAGE and detected by Coomassie blue staining (identities on the right). M, molecular weight markers (in kDa). (B) Bacterially expressed GST-tagged La protein (GST-La [lane 1]), and baculovirus-expressed His-tagged p110 (His-p110 [lane 2]) and p110 lacking the 10 C-terminal amino acids (His-p110 ΔC10 [lane 3]). The proteins were analyzed as in A. (C) In vitro transcribed, 32P labeled U6 snRNAs with different 3′ ends were analyzed by denaturing gel electrophoresis (lane 1, 3′-hydroxyl end (U6-3′OH); lane 2, 2′,3′-cyclic phosphate end (U6>P); lane 3, 2′,3′-cyclic phosphate end after polynucleotide kinase treatment (U6>P/PNK). Below, the secondary structure model of human U6 snRNA with the 3′-terminal HDV domain (in gray), which is autocatalytically cleaved (arrow), generating U6 snRNA with a 2′,3′-cyclic phosphate end. Marker positions are indicated on the left (in nucleotides). (D,E) In vitro transcribed, 32P labeled U6 snRNAs U6-3′OH (A, lanes 1–9) or U6>P(5U) (A, lanes 10–18; B, lanes 1–8), or U6>P(12U) (B, lanes 9–16) were incubated with increasing concentrations of recombinant LSm2-8 proteins (nanomolar concentrations as indicated above the lanes). Complex formation was analyzed by native gel electrophoresis. The positions of free U6 snRNAs and the U6/LSm2-8 complexes are marked on the right.
32P-labeled U6 snRNA with either the 3′-hydroxyl or the 2′,3′-cyclic phosphate end were incubated with recombinant LSm2-8 protein complex in different molar ratios. Complex formation was followed by native gel electrophoresis, and binding affinities were determined by quantitation. We found that U6 snRNA with the cyclic phosphate exhibited an approximately twofold higher affinity for LSm2-8 proteins compared to U6 snRNA with the 3′-hydroxyl [U6-3′OH: KD 518 ± 30 nM, n = 3; U6>P(5U): KD 274 ± 34 nM, n = 5] (see Fig. 1D). To rule out that 3′-non-templated nucleotides and 3′-end heterogeneity in the runoff transcripts (Milligan et al. 1987) are responsible for this effect, we converted the 2′,3′-cyclic phosphate to a 2′- or 3′-hydroxyl group using polynucleotide kinase, resulting in decreased binding affinity (data not shown). This shows clearly that the observed effect is indeed due to the presence or absence of the cyclic phosphate at the 3′ end of U6 snRNA.
The U6 snRNA is also heterogeneous in the length of its 3′-terminal uridine stretch (Lund and Dahlberg 1992). Longer forms, with up to 12 uridines, are thought to be precursors to the shorter forms and U6 with a 2′,3′-cyclic phosphate end. Therefore we compared the LSm2-8 affinity to U6 snRNA with five uridines (as found in the majority of U6 snRNA) or 12 uridines, either one with a 2′,3′-cyclic phosphate end. Bandshift assays revealed that the affinity to LSm2-8 was significantly higher for the 3′-extended uridine tails [U6>P(12U): KD 150 ± 10 nM, n = 3, versus U6>P(5U): KD 274 ± 34 nM, n = 5] (see Fig. 1E).
In sum, we have shown that 3′-terminal modifications (both the 3′ terminus itself and the length of the 3′-uridine stretch) can modulate the LSm2-8 binding affinity.
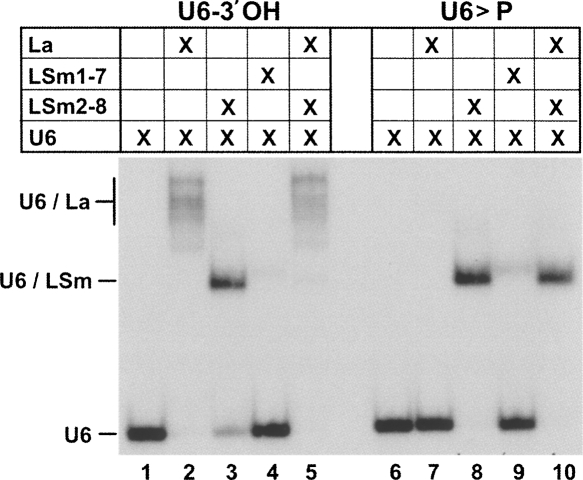
In addition to the LSm2-8 proteins, the La protein is involved in the early stages of U6 snRNP biogenesis. Therefore we tested next how 3′-terminal modifications (2′,3′-cyclic phosphate versus 3′-hydroxyl end) and LSm2-8 binding affect La association (Fig. 2). As followed by native gel electrophoresis, La bound specifically to U6 snRNA with a 3′-hydroxyl end, but not to U6 with a cyclic phosphate 3′ end (Fig. 2, lanes 2,7). The LSm2-8 complex bound both forms, but U6 snRNA with a cyclic phosphate 3′ end with a higher affinity (Fig. 2, lanes 3,8, compare with Fig. 1). In contrast, the LSm1-7 complex showed only background binding activity to both U6 snRNA forms (Fig. 2, lanes 4,9). The strong selectivity of LSm2-8 versus La binding was underlined, when either of the two U6 snRNA forms was incubated first with the LSm2-8 complex, then with La protein: La was able to completely displace the LSm2-8 complex from U6-3′OH; this depended on the 3′OH end, since with U6>P the same sequential binding resulted in the exclusive formation of the U6/LSm2-8 complex (Fig. 2, lanes 5,10).
FIGURE 2.
La protein binds exclusively to U6-3′OH, displacing prebound LSm2-8 proteins. Recombinant La protein (lanes 2,7), LSm2-8 proteins (lanes 3,8), or LSm1-7 proteins (lanes 4,9) were incubated with in vitro transcribed U6-3′OH or U6>P snRNAs (left and right panels; for inputs, see lanes 1,6). In addition, LSm2-8 proteins were pre-incubated with U6 snRNA, followed by the addition of La protein (lanes 5,10). Complex formation was analyzed by native gel electrophoresis.
LSm2-8 and p110 proteins bind synergistically to U6 snRNA, depending on the p110 C terminus and the LSm binding site of U6 snRNA
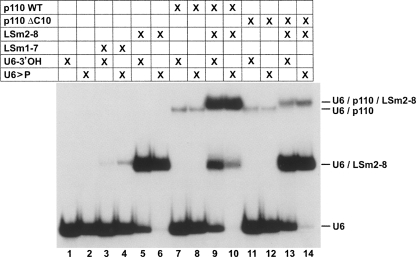
Since our previous work has shown that p110 binds specifically to U6 snRNA (Bell et al. 2002), we next determined whether binding of both p110 and LSm proteins can be detected by native gel electrophoresis and how they affect each other (Fig. 3). Recombinant LSm2-8 proteins (and as a control for specificity, recombinant LSm1-7) were used, as well as recombinant baculoviral-expressed p110 and a truncated form lacking the highly conserved C-terminal 10-amino acid sequence (Medenbach et al. 2004; for a protein analysis, see Fig. 1A,B). We tested combinations of the individual proteins for their ability to form stable complexes with in vitro transcribed 32P-labeled U6 snRNA carrying either a 3′-terminal hydroxyl group (U6-3′OH) or the mature 2′,3′-cyclic phosphate (U6>P).
FIGURE 3.
LSm2-8 proteins promote p110 binding to U6 snRNA, depending on conserved C-terminal p110 region. The interaction of recombinant proteins LSm1-7, LSm2-8, p110, p110ΔC10, and combinations thereof (lanes 3–14) with in vitro transcribed, 32P labeled U6 snRNAs (U6-3′OH or U6>P; for inputs, see lanes 1,2) was analyzed by native gel electrophoresis. The mobilities of free RNAs and the different complexes are indicated on the right.
The LSm1-7 complex showed no significant binding activity with U6 snRNA, independently of its 3′ end (Fig. 3, lanes 1–4), whereas LSm2-8 very efficiently formed under the same conditions a U6 snRNP, at higher efficiency with U6>P than with U6-OH (Fig. 3, cf. lanes 5 and 6), consistent with our binding experiments described above (see Fig. 1D). On the other hand, p110 bound both U6 snRNAs at similar, weak efficiency under these conditions (Fig. 3, lanes 7,8). However, the addition of p110 and the LSm2-8 complex resulted in efficient formation of a ternary complex (U6/p110/LSm2-8), which ran slightly above the U6/p110 complex (Fig. 3, cf. lanes 8 and 9) and which formed more quantitatively with U6>P than with U6-OH (Fig. 3, cf. lanes 9 and 10). Ternary complex assembly required the C-terminal short conserved region of p110, since the complete assembly reaction with p110ΔC10 yielded mostly U6/LSm2-8 complex and only low levels of the complete U6/LSm2-8/p110 complex (Fig. 3, lanes 11–14). In sum, we conclude that LSm2-8 and p110 proteins bind synergistically to U6 snRNA; in initial experiments we had used the natural LSm2-8 complex purified from human U4/U6.U5 tri-snRNPs; this revealed the same binding behavior and dependence on the intact C terminus of p110, validating our use of the recombinant, in vitro assembled LSm2-8 complex (data not shown).
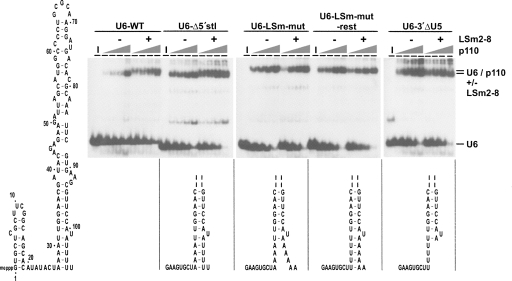
To address the question of whether the LSm-U6 interaction affects p110 integration, we next introduced several mutations in the LSm binding site of U6 snRNA that leave the normal p110 interaction site intact. For ease of cloning they all lacked the 5′ hairpin of U6 (U6-Δ5′stl), since it is not essential for p110 nor LSm2-8 binding (p110: Bell et al. 2002; LSm2-8: Achsel et al. 1999): U6-LSm-mut and U6-3′ΔU5, in which the 3′-terminal five uridines were replaced by adenosines or deleted, respectively. In addition, in the U6-LSm-mut rest mutant the normal secondary structure of the 3′-terminal region of U6 was restored, by introducing four compensatory A→U mutations in the 5′ part of the U6 intramolecular stem-loop (Fig. 4, see secondary structures).
FIGURE 4.
LSm2-8 enhancement of p110 binding to U6 snRNA depends on an intact LSm binding site. Synergistic LSm2-8/p110 binding to 32P-labeled mutant U6 snRNAs were assayed by bandshift assays, using increasing concentrations of recombinant p110 (2, 4, 8, and 20 nM), either in the absence or presence of LSm2-8 complex purified from human tri-snRNPs (–/+). Lanes I, input RNA. The identities of free RNAs and the complexes are given on the right. Below the corresponding autoradiograms, mutations are schematically represented and—except for the full-length wild-type U6 snRNA (U6-WT)—all have the 5′-terminal stem-loop deleted (U6-Δ5′stl), and in addition the LSm site mutated (U6-LSm-mut), the LSm site mutated and base-pairing restored (U6-LSm-mut rest), or the LSm site deleted (U6-3′ΔU5). Shown is only the bottom part of the 3′ stem-loop), except for the full-length U6 snRNA secondary structure model (left side).
For each 32P-labeled U6 snRNA derivative, bandshift assays were carried out, titrating in p110 protein in the absence or presence of the LSm2-8 complex (Fig. 4). Surprisingly, p110 had a higher affinity for the 5′-truncated version of U6 snRNA, which may reflect folding of part of the full-length U6 snRNA into a structure that is not active in p110 binding. However, the LSm effect on p110 binding was still observed (Fig. 4, cf. U6-WT and U6-Δ5′stl). Deletion or substitution of the 3′-terminal uridine stretch completely abolished LSm2-8 binding (data not shown). Titration of the p110 protein revealed that p110 association with the mutant RNAs that are impaired in LSm binding could not be enhanced any more by the addition of purified LSm2-8 complex (panels U6-LSm-mut and U6-3′ΔU5, respectively). This suggests that it is not a p110/LSm association in the absence of RNA that increases the affinity of p110 to U6 snRNA per se; instead, the synergistic effect of p110 and LSm2-8 proteins appears to require prior LSm binding to the 3′ end of U6 snRNA.
In addition, a mutant U6 snRNA was tested, U6-LSm-mut rest, in which the LSm binding site was inactivated, and compensatory mutations were introduced that were designed to mimic the 3′-terminal stem: Again, no significant synergistic effect was observed, indicating that the 3′-terminal uridines are required rather than a secondary structure unit.
DISCUSSION
U6 snRNA plays a central role in spliceosome catalysis and is engaged in several dynamic changes (see Introduction for references). Here we have used an in vitro reconstitution approach to investigate the contributions of the LSm2-8 protein complex and the p110 recycling factor to U6 snRNP dynamics. Initially we have demonstrated that the association with the LSm2-8 complex and La protein is modulated by modifications at the 3′-terminus of U6 snRNA: Not only the chemical nature of its 3′ end (3′-hydroxyl versus 2′,3′-cyclic phosphate), but also the length of its 3′-terminal uridines is important.
As the LSm2-8 complex and La bind to the 3′ end of U6 snRNA in a mutually exclusive fashion, the cyclic phosphorylation of the RNA provides a molecular switch for the exchange of these protein factors. Thus, 3′-end modification would convert the U6 snRNA from an initial, La-bound state (U6 3′OH) to the mature, LSm-bound state (U6-2′,3′-cyclic phosphate), which is found predominantly in snRNPs. It is known that the La protein binds specifically to RNAs carrying a 3′-hydroxyl group, and the recently published crystal structure of La showed that the 2′ and 3′ hydroxyl groups of the terminal uridine are contacted by D33 of the La domain (Teplova et al. 2006). Therefore one can envision the 2′,3′-cyclic phosphate of mature U6 snRNA to sterically interfere with La interaction, explaining the disruption of the La-U6 interaction after 3′-end modification of U6 snRNA. Enzymatic activities involved may be the 3′-terminal phosphate cyclase, which localizes to the nucleoplasm and is excluded from nucleoli (Genschik et al. 1997; Gu et al. 1997) and the nucleolar U6-specific terminal uridylyl transferase (TUTase) (Trippe et al. 1998, 2006). The exact order of all these differentially localized events is not clear.
Our data argue for a model whereby LSm2-8 binding to U6 snRNA rapidly recruits the p110 protein. This is based on in vitro binding assays, which consistently showed that p110 binds much more efficiently to the U6-LSm complex than to naked U6 snRNA (see Fig. 3, cf. lanes 1,8 and 6,10). Moreover, efficient recruitment of p110 onto a U6/LSm2-8 complex strongly depends on the C-terminal conserved motif of p110, an effect that we have clearly shown here in a mammalian in vitro system (Fig. 3), but was previously based only on two-hybrid assays in yeast (Rader and Guthrie 2002). Yet the C-terminal truncation of p110 (p110 ΔC10) still is incorporated in a U6 snRNP, although at a much lower efficiency; this may explain why we had previously found the p110 C-terminal sequence motif to be dispensable for recycling activity in vitro (Medenbach et al. 2004). This rather surprising substrate specificity of the p110 protein (binding of the U6-LSm RNP versus U6 snRNA) probably reflects different U6 snRNA structures in the free and protein-bound forms. This has been characterized in detail only for the yeast U6 snRNP, carrying both LSm2-8 and Prp24, but not distinguishing between the contributions of both protein components (Karaduman et al. 2006).
MATERIALS AND METHODS
RNA synthesis
U6 snRNA mutants were PCR-amplified from pUC19-T7-U6 (Wolff and Bindereif 1993) and cloned into pCR2.1-TOPO vector (Invitrogen). SP6 promotor sequence, mutations, and restriction sites for runoff transcription (using DraI or NsiI) were introduced by the PCR primers. For U4 snRNA an MscI runoff plasmid template was created in a similar fashion from pUC13-U4c (Wersig and Bindereif 1990) to generate the correct 3′ end of the RNA after transcription. To create the 2′,3′-cyclic phosphate of mature U6 snRNA, the Hepatitis Delta Virus (HDV) ribozyme sequence was cloned 3′ of the U6 snRNA sequence essentially as described (Mörl et al. 2005). Restriction sites and T7 promotor were introduced by the PCR primers, and the fragment was inserted into pUC19, using the EcoRI and BamHI restriction sites. The construct for the 3′-extended U6 snRNA [U6>P(12U)] was created accordingly. Ribozyme cleavage occurred under transcription conditions (see below). To remove the 2′,3′-cyclic phosphate, U6>P was incubated with 10 U of T4 polynucleotide kinase (NEB) for 6 h at 37°C in a buffer containing 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, and 5 mM dithiothreitol, essentially as described (Mörl et al. 2005).
SnRNAs and mutant derivatives were generated by in vitro runoff transcription from linearized plasmid templates, in the case of 32P-labeled RNAs in the presence of [α-32P]UTP or -ATP. Subsequently RNAs were purified by denaturing PAGE and gel elution.
Purification of HeLa LSm2-8 proteins
Endogenous human LSm proteins were purified from HeLa nuclear extract essentially as described before (Achsel et al. 1999). In brief, total snRNPs were immunopurified using an H20 monoclonal anti-m3G-antibody column and eluted with m7-guanosine followed by fractionation on a preparative linear 10%–30% (w/w) glycerol gradient in buffer G (20 mM HEPES/KOH at pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and 0.5 mM PMSF). After concentration of the tri-snRNP fractions by ultracentrifugation (265,000g, 10 h, 4°C in 10%–30% glycerol gradient), tri-snRNP particles were partially disrupted by 700 mM NaCl and fractionated by yet another round of linear 5%–20% (w/w) glycerol gradient ultracentrifugation (118,000g, 7 h, 4°C). Fractions containing LSm, but no Sm proteins were dialyzed against a buffer containing 20 mM HEPES/KOH pH 7.6, 1.5 mM MgCl2 and 125 mM NaCl, bound to a MonoS column (0.1 mL) and eluted by a 2-mL linear salt gradient (100–400 mM NaCl in 20 mM HEPES/KOH at pH 7.6, and 1.5 mM MgCl2).
Recombinant proteins
Recombinant LSm1-7 and 2-8 proteins were obtained from C. Kambach (Zaric et al. 2005), recombinant p110 protein and the truncated version ΔC10 were expressed in baculovirus-infected insect cells and purified as described (Bell et al. 2002; Medenbach et al. 2004). For the GST-La construct the cDNA of human La (kindly obtained from G. Pruijn [Pruijn et al. 1991]) was amplified by PCR, thereby introducing two restriction sites for KpnI and NcoI and removing an internal restriction site for KpnI. The PCR fragment was cloned into pETM-30, which contains an N-terminal GST domain, using the KpnI and NcoI sites. The protein was expressed in E. coli BL21 Star (Invitrogen) containing the Rosetta 2 plasmid (Novagen) and purified via glutathione Sepharose beads (GE Healthcare).
Complex formation and bandshift assays
p110 and/or LSm proteins (concentrations, see below) were incubated together with 50 fmol 32P-labeled snRNAs in a 10-μl reaction for 30 min at 30°C or at room temperature in a buffer containing 12 mM HEPES/KOH pH 7.6, 1.5 mM MgCl2, 100 mM NaCl, 10% glycerol, 0.1% Triton X-100, and 1 μg of RNase-free yeast tRNA (Roche). The following concentrations of recombinant/purified LSm proteins were used: Figure 1, 40 nM of LSm proteins; Figure 2, 1 μM of LSm proteins and 150 nM of La protein; Figure 3, 0.5 μM of LSm proteins and 50 nM of p110 or p110ΔC10. For titration experiments with recombinant LSm proteins the concentrations are indicated above the lanes (Fig. 1). In the absence of the LSm proteins, LSm binding buffer containing 20 mM HEPES/KOH pH 7.6, 1.5 mM MgCl2, and 200 mM NaCl was used to compensate the volume, in the absence of p110 buffer D (Dignam et al. 1983) was used. After the incubation complexes were separated on a native 6% polyacrylamide gel (acrylamide:N,N′-methylene bisacrylamide 80:1 in 0.5 × TBE) for 10 h at 2.5 Volts/cm and 4°C (Fig. 4) or for 3–5 h at 7.6 volts/cm and 4°C (Figs. 1–3). Complexes were visualized by autoradiography and quantified using an Instant Imager (Packard) or phosphorimager (Bio-Rad).
For the titration experiments, in which U6 mutant snRNAs were analyzed (Fig. 4), LSm-U6 complexes were preformed for 15 min under the conditions described above (mock reactions with buffer D only), then a constant volume of buffer D containing the indicated amount of recombinant p110 protein was added, and incubation was continued for 15 min. Complexes were separated and analyzed as described above.
ACKNOWLEDGMENTS
We thank Stephanie Nottrott and Annemarie Schulz (Göttingen) for essential help in the preparation of LSm2-8 protein from purified tri-snRNPs, G. Pruijn (Nijmegen) for the gift of a La cDNA construct, as well as G. Meiss (Giessen) for help in modeling of the La protein-RNA interaction. We also thank Silke Schreiner for p110 protein expression, Heiko Andrea for plasmid construction and expression of GST-La protein, Tanja Rösel for cloning of the original U6-HDV construct, Zsofia Palfi for anti-m3G antiserum, and other members of our group for discussions and comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the European-Commission-funded Network of Excellence EURASNET, and the Fonds der Chemischen Industrie.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1129608.
REFERENCES
- Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C.L., Urlaub H., Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Cameron V., Uhlenbeck O.C. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977;16:5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- Chan S.P., Kao D.I., Tsai W.Y., Cheng S.C. The prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Damianov A., Schreiner S., Bindereif A. Recycling of the U12-type spliceosome requires p110, a component of the U6atac snRNP. Mol. Cell. Biol. 2004;24:1700–1708. doi: 10.1128/MCB.24.4.1700-1708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P., Billy E., Swianiewicz M., Filipowicz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria, and Archaea. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Shumyatsky G., Makan N., Reddy R. Formation of 2′,3′-cyclic phosphates at the 3′ end of human U6 small nuclear RNA in vitro. Identification of 2′,3′-cyclic phosphates at the 3′ ends of human signal recognition particle and mitochondrial RNA processing RNAs. J. Biol. Chem. 1997;272:21989–21993. doi: 10.1074/jbc.272.35.21989. [DOI] [PubMed] [Google Scholar]
- Karaduman R., Fabrizi P., Hartmuth K., Urlaub H., Lührmann R. RNA structure and RNA-protein interactions in purified yeast U6 snRNPs. J. Mol. Biol. 2006;356:1248–1262. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Khusial P., Plaag R., Zieve G.W. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Lange T.S., Gerbi S.A. Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell. 2000;11:2419–2428. doi: 10.1091/mbc.11.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J.E. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- Mayes A.E., Verdone L., Legrain P., Beggs J.D. Chracterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J., Schreiner S., Liu S., Lührmann R., Bindereif A. Human U4/U6 snRNP recycling factor p110: Mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol. Cell. Biol. 2004;24:7392–7401. doi: 10.1128/MCB.24.17.7392-7401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M., Lizano E., Willkomm D.K., Hartmann R.K. Production of RNAs with homogeneous 5′ and 3′ ends. In: Hartmann R.K., et al., editors. Handbook of RNA biochemistry. Wiley-VCH, Weinheim; Germany: 2005. pp. 22–35. [Google Scholar]
- Pruijn G.J., Slobbe R.L., van Venrooij W.J. Analysis of protein-RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–5180. doi: 10.1093/nar/19.19.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader S.D., Guthrie C. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA. 2002;8:1378–1392. doi: 10.1017/s1355838202020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan P.L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K.W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes & Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Shimba S., Reddy R. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J. Biol. Chem. 1994;269:12419–12423. [PubMed] [Google Scholar]
- Spiller M.P., Boon K.L., Reijns M.A., Beggs J.D. The Lsm2-8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–929. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D., Rader S.D., Klingauf M., Neugebauer K.M. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Forne T., Jeanteur P., Cathala G., Brunel C. Mammalian U6 small nuclear RNA undergoes 3′-end modifications within the spliceosome. Mol. Cell. Biol. 1993;13:1641–1650. doi: 10.1128/mcb.13.3.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M., Yuan Y.R., Phan A.T., Malinina L., Ilin S., Teplov A., Patel D.J. Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R., Sandrock B., Benecke B.J. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R., Guschina E., Hossbach M., Urlaub H., Lührmann R., Benecke B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Aab A., Steitz J.A. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., You Z.H., Graham P.J., Steitz J.A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Verdone L., Galardi S., Page D., Beggs J.D. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 2004;14:1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Vidal V.P., Verdone L., Mayes A.E., Beggs J.D. Characterization of U6 snRNA–protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersig C., Bindereif A. Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res. 1990;18:6223–6229. doi: 10.1093/nar/18.21.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Wolff T., Bindereif A. Conformational changes of U6 RNA during the spliceosome cycle: An intramolecular helix is essential both for initiating the U4–U6 interaction and for the first step of splicing. Genes & Dev. 1993;7:1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- Zaric B., Chami M., Remigy H., Engel A., Ballmer-Hofer K., Winkler F.K., Kambach C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J. Biol. Chem. 2005;280:16066–16075. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]