Abstract
The initiation of protein synthesis on mRNAs within eukaryotic cells is achieved either by a 5′ cap-dependent mechanism or through internal initiation directed by an internal ribosome entry site (IRES). Picornavirus IRES elements, located in the 5′ untranslated region (5′UTR), contain extensive secondary structure and multiple upstream AUG codons. These features can be expected to inhibit cap-dependent initiation of translation. However, we have now shown that certain mutant hepatitis C virus-like picornavirus IRES elements (from porcine teschovirus-1 and avian encephalomyelitis virus), which are unable to direct internal initiation, are not significant barriers to efficient translation of capped monocistronic mRNAs that contain these defective elements within their 5′UTRs. Moreover, the translation of these mRNAs is highly sensitive to the expression of an enterovirus 2A protease (which induces cleavage of eIF4G) and is also inhibited by hippuristanol, a specific inhibitor of eIF4A function, in contrast to their parental wild-type IRES elements. These results provide a possible basis for the evolution of viral IRES elements within the context of functional mRNAs that are translated by a cap-dependent mechanism.
Keywords: translation, protein synthesis, picornavirus, IRES, hippuristanol
INTRODUCTION
The initiation of protein synthesis on mRNAs within eukaryotic cells is usually achieved as a consequence of recognition of the 5′ terminal cap-structure (m7GpppG…), which is present on all cytoplasmic cellular mRNAs. This cap-structure is bound by the translation initiation factor eIF4E, which is part of a complex (eIF4F) containing eIF4A (an RNA helicase) and eIF4G (a scaffold protein that interacts with several other proteins including eIF3 and the poly(A) binding protein [PABP]) (for review, see Gingras et al. 1999). These interactions allow the 40S ribosomal subunit to bind and then “scan” along the 5′ untranslated region (UTR) of the mRNA until an initiation codon in a favorable context is reached. At this point, the 60S ribosomal subunit joins, initiation factors are released, and polypeptide chain formation commences. Generally, cellular mRNAs have relatively short 5′UTRs (100–200 nucleotides [nt]); it is believed that the presence of significant secondary structure or upstream AUG (uAUG) codons is detrimental to the efficiency of translation initiation at the correct point (Kozak 1989a,b; Svitkin et al. 2001). However, a significant proportion of cellular mRNAs are now known to contain uAUG codons and upstream open reading frames (see Yamashita et al. 2003; Iacono et al. 2005).
A second, distinct mechanism of translation initiation within eukaryotic cells has been identified that is dependent on an element termed an internal ribosome entry site (IRES), and this mechanism is independent of the 5′ terminal cap structure. The IRES elements were first identified within picornavirus RNAs (for review, see Belsham and Jackson 2000). These viral RNAs are positive sense and function like mRNAs, but they have very long 5′UTRs (generally ca. 650–1300 nt), which contain extensive secondary structure and multiple AUG codons preceding the initiation codon (e.g., the initiation codon on encephalomyocarditis virus [EMCV] RNA is AUG-11) (Kaminski et al. 1994). Furthermore, the picornavirus RNAs are uncapped; indeed, the genomic viral RNA is linked at its 5′ terminus to a short, virus-encoded peptide, termed VPg, although this is rapidly lost within cells. The RNAs from most picornaviruses (except for hepatitis A virus [HAV]) (Borman and Kean 1997; Ali et al. 2001) can be translated without any requirement for eIF4E (the cap-binding protein) and when eIF4G is cleaved (which releases its N-terminal domain including the eIF4E binding site). Certain picornaviruses, for example, the enteroviruses (such as poliovirus [PV]) and foot-and-mouth disease virus [FMDV]), produce proteases (2A and L, respectively), which induce the cleavage of eIF4G and hence inhibit cellular cap-dependent protein synthesis while the IRES-dependent viral protein synthesis is maintained (or even enhanced) under these conditions (e.g., Jang et al. 1988; Borman et al. 1997; Roberts et al. 1998).
IRES elements have also been found in the RNA genomes of other viruses including hepatitis C virus (HCV) and the pestiviruses (which are all members of the flaviviridae) plus the dicistroviridae, for example, cricket paralysis virus (Wilson et al. 2000) and Rhopalosiphum padi virus (Woolaway et al. 2001; Terenin et al. 2005). Different types of viral IRES element have been shown to have distinct requirements for cellular proteins to allow translation initiation to occur efficiently (Doudna and Sarnow 2007).
There are now known to be four different classes of picornavirus IRES that differ in their secondary structures and their requirements for translation initiation factors plus other cellular proteins. The various picornavirus IRES elements range in size from about 450 nt (e.g., from PV, EMCV, and FMDV) down to about 280 nt (porcine teschovirus-1, PTV-1). The PV IRES and EMCV IRES elements are representatives of two separate classes, while the HAV IRES represents a third type that uniquely requires eIF4E plus intact eIF4G (Borman and Kean 1997; Ali et al. 2001). Unexpectedly, the fourth class of picornavirus IRES element has a strong resemblance to the IRES found in the HCV and pestivirus RNAs. Initially this class of picornavirus IRES elements was identified within the PTV-1 RNA (Kaku et al. 2002; Pisarev et al. 2004; Chard et al. 2006a). However, a number of other picornaviruses have now also been shown to contain this type of IRES, including porcine enterovirus-8 (Chard et al. 2006b), certain simian picornaviruses (e.g., SV2, Chard et al. 2006b, and SPV9, de Breyne et al. 2008), and avian encephalomyelitis virus (AEV, Bakhshesh et al. 2008). Sequence alignments also suggest that other picornaviruses, including the duck hepatitis virus-1 (Ding and Zhang, 2007), Seneca Valley virus (M. Willcocks, M. Bakhshesh, Z. Gomwalk, G.J. Belsham, and L.O. Roberts, unpubl.), and a seal picornavirus (Kapoor et al. 2008), also contain this class of IRES element (see Hellen and de Breyne 2007). A distinctive feature of this class of IRES elements is their resistance to the action of hippuristanol (Bordeleau et al. 2006; Chard et al. 2006b; Bakhshesh et al. 2008), a specific inhibitor of eIF4A, which is consistent with their lack of requirement for any eIF4 initiation factors for 48S initiation complex assembly in vitro (Pestova et al. 1998; Pisarev et al. 2004).
There have been numerous reports of IRES elements within the long 5′UTRs of certain cellular mRNAs, including those encoding c-myc, Apaf-1, and XIAP (for reviews, see Stoneley and Willis 2004; Baird et al. 2006). Such reports have been controversial (Kozak 2005). In part, this reflects the fact that the use of dicistronic reporter gene constructs can readily detect a quite small apparent stimulation of translation of the second open reading frame (ORF) resulting from the presence of an inserted 5′UTR between the ORFs while the detection of truncated RNA transcripts only including the second ORF (e.g., derived from cryptic promoter activity) is less sensitive.
Recently, Dmitriev et al. (2007) have examined the properties of the 900-nt long 5′UTR of the human LINE-1 mRNA. In these studies, no significant evidence for an IRES element within this highly structured GC-rich region was obtained, and the translation of this mRNA was strictly cap dependent. This work is at variance with previous studies on the mouse LINE-1 mRNA, which suggested the presence of two IRES elements within this natural dicistronic mRNA (Li et al. 2006). The results of Dmitriev et al. (2007) suggested that long, structured, and AUG-burdened sequences did not necessarily act as severe barriers to cap-dependent translation initiation. Previous studies with capped monocistronic mRNAs containing defective forms of the PV IRES showed that these RNAs could be translated in vitro (Trono et al. 1988); however, in contrast, capping of similar transcripts containing the HAV IRES was inhibitory to translation (Brown et al. 1994). With this background, we have now examined the properties of some defective, mutant, picornavirus IRES elements when assayed within the context of capped monocistronic mRNAs produced in the cytoplasm of cells. We have shown that some of these highly structured viral RNA elements, lacking IRES activity, do not block translation initiation on these mRNAs, although the translation is mediated by a cap-dependent mechanism. Thus, it is possible to envisage how IRES elements could evolve within the context of functional capped mRNAs.
RESULTS
Translation of capped monocistronic mRNAs containing wild-type and mutant EMCV IRES elements within cells
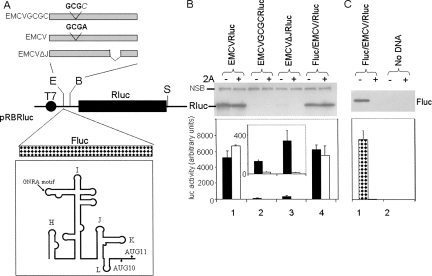
A monocistronic reporter gene vector, pRBRluc (see Fig. 1A), was constructed for the analysis of IRES activity within the context of capped monocistronic mRNAs. This plasmid contains a T7 promoter, which is recognized by the T7 RNA polymerase expressed from the recombinant vaccinia virus, vTF7-3 (Fuerst et al. 1986). The transcripts are produced and then capped in the cytoplasm by the vaccinia virus capping enzyme. The cap dependence of these transcripts is readily demonstrated by the coexpression of an enterovirus 2A protease, which induces the cleavage of eIF4G and inhibits cap-dependent protein synthesis. In the following experiments, we have used the plasmid pGEM3Z/J1, which expresses the 1D-2A region of the swine vesicular disease virus (SVDV) polyprotein, for this purpose as described previously (Sakoda et al. 2001; Chard et al. 2006a; Bakhshesh et al. 2008). Derivatives of the pRBRluc vector were produced containing the wild-type and defective forms of the EMCV IRES (Fig. 1A). The EMCV cDNA used was about 550 bp in length (nt 280–830 of the EMCV genome). The IRES mutant GCGC contains a point mutation within the highly conserved GNRA tetraloop within domain I, which strongly inhibits its activity (see Roberts and Belsham 1997; Robertson et al. 1999). The ΔJ mutant has the domain J sequence (nt 696–730) precisely deleted, which also destroys the activity of the IRES when assayed within the context of a dicistronic reporter construct. However, it is noteworthy that we did observe some translation from monocistronic transcripts containing these two mutant elements previously (Robertson et al. 1999). We have now assayed these elements alone and in the presence of the SVDV 2A protease. As expected, the wild-type EMCV IRES directed very efficient expression of Rluc, as detected either by immunoblotting or enzyme assay, both in the presence and absence of 2A expression (Fig. 1B). The expression of Rluc from the monocistronic transcripts containing the GCGC or ΔJ mutant forms of the EMCV IRES was much lower (less than 10% of wild type), although this was still easily quantified in the enzyme assays (the activities of 100–400 compare with a signal of about 20 in these Rluc assays, as obtained from an Fluc/Rluc dicistronic RNA lacking an IRES, see below). In the presence of the 2A protease, the Rluc expression from the mutant EMCV IRES elements was strongly reduced (to about 20 in each case, see inset in Fig. 1B, note the Rluc activity from the Fluc/Rluc RNA in the presence of 2A was reduced to about 1 under these conditions), suggesting that the residual translation (seen in the absence of 2A) was occurring principally by a cap-dependent mechanism. When a dicistronic vector (pFluc/EMCV/Rluc) containing the Fluc coding sequence upstream of the wild-type EMCV IRES (see Fig. 1A) was assayed, it was again shown that the EMCV IRES-directed Rluc expression was unaffected by the coexpression of the 2A protease (Fig. 1B), but the expression of the upstream ORF (Fluc) was strongly inhibited (>95%) under these conditions as expected (Fig. 1C). Thus, the wild-type EMCV IRES displayed the same properties within both the monocistronic and dicistronic mRNAs.
FIGURE 1.
Cap-dependent and cap-independent translation within cells. (A) The structure of the basic pRBRluc reporter plasmid used in this study. Derivatives containing the wild-type or mutant EMCV IRES cDNA were made by insertion into the EcoRI (E) and BamHI (B) sites while the blunt-ended Fluc fragment (BamHI–XhoI fragment from pGEM-luc, see Materials and Methods) was inserted into the blunt-ended EcoRI site within pEMCVRluc to produce pFluc/EMCV/Rluc. The 5′UTR upstream of the Fluc coding region is 16 nt long. The SalI (S) site downstream from the Rluc coding sequence is indicated. It should be noted that the BamHI site located at the 3′ end of the EMCV cDNA was introduced at the position of AUG-10, modifying the ATG to ATC within this site (GGATCC); thus, AUG-10 is not present in these constructs. The line diagram of the EMCV IRES secondary structure shows the location of the GCGC sequence in place of the GNRA motif and the J domain as deleted in the ΔJ mutant. (B) Transient expression assays were performed by transfecting the indicated plasmids alone (−) or with (+) pGEM3Z/J1, which expresses the SVDV 2A protease, into vTF7-3 infected BHK cells. Cell extracts prepared after 20 h were analyzed by SDS-PAGE plus immunoblotting using anti-Rluc antibodies; detection was achieved using peroxidise labeled secondary antibodies plus chemiluminescence reagents. A nonspecific background band (labeled NSB) demonstrates the loading of cell extracts in each case. Expression levels within the cell extracts were quantified by Renilla luciferase assays. Results are presented as the mean (+S.D.) from four independent transfections. The inset shows the same data but on a different vertical scale for clarity. (C) The extracts from transfected cells were analyzed for Fluc expression by immunoblotting with anti-Fluc antibodies and by firefly luciferase enzyme assays.
Defective PTV IRES elements do not inhibit translation of monocistronic reporter mRNAs
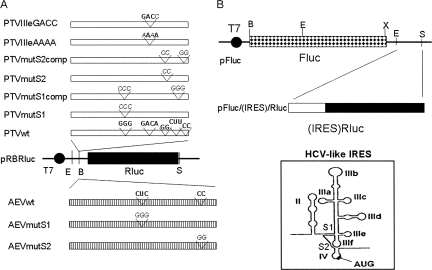
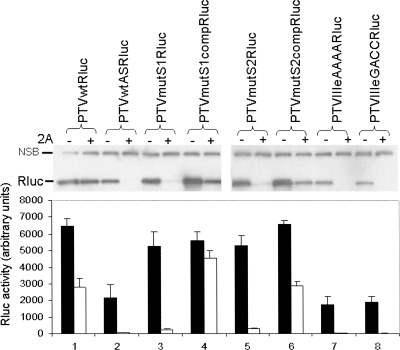
In previous studies, we have characterized a number of defective mutants of the IRES from PTV-1 Talfan (Chard et al. 2006a). This IRES shares many characteristics with the HCV IRES, including the presence of a pseudoknot (domain IIIf) and a highly conserved stem–loop (domain IIIe), which plays a critical role in the interaction with the 40S ribosomal subunit (Psaridi et al. 1999; Lukavsky et al. 2000; Kieft et al. 2001; Laletina et al. 2006). We have now constructed a series of plasmids containing some of these variants of the PTV-1 IRES; the cDNA (∼300 nt) was inserted as a BamHI fragment into the pRBRluc vector (Fig. 1A) upstream of the Rluc coding sequence (see Fig. 2A). This strategy enabled plasmids with the IRES cDNA in either the sense or antisense (AS) orientations to be obtained. The mutations analyzed were in the pseudoknot stem regions (mutS1 and mutS2) and within domain IIIe (changing the terminal loop from GACA to either AAAA or GACC). These mutations have been shown previously to severely restrict the activity of the IRES within dicistronic reporter mRNAs (Chard et al. 2006a). The plasmids were each tested within cells alone and in the presence of pGEM3Z/J1 as above. In the context of the capped monocistronic mRNAs, the wild-type PTV IRES and the (pseudo-wild-type) variants containing compensatory mutations for the S1 and S2 modifications (termed mutS1comp and mutS2comp) all worked very efficiently alone and also expressed Rluc at high levels in the presence of the 2A protease (Fig. 3), as expected (see Pisarev et al. 2004). There was a moderate drop in the activity of the wild-type PTV IRES (and the mutS2comp element) observed in the presence of 2A (by about 60%) and the reason for this is not known. It was also apparent that the drop in IRES activity under these conditions from the PTVmutS1comp (pseudo-wild-type) IRES was rather less (by only about 15%) than that observed with either the wild-type or mutS2comp elements (see Discussion). The expression of Rluc seen from the similar plasmids containing the defective or inverted IRES elements (PTVwtAS, PTVmutS1, PTVmutS2, PTVIIIeAAAA, and PTVIIIeGACC) was unexpectedly high (see Fig. 3). Indeed, in some cases (e.g., PTVmutS1 and PTVmutS2), it was about 80% of that observed with the wild-type PTV IRES, whereas for the AS version of the wild-type PTV IRES and the domain IIIe mutants the expression was about 30% of the level seen with the wild-type IRES in the sense orientation. However, when these plasmids were coexpressed with the SVDV 2A protease the Rluc expression was severely decreased (by well over 90%) in each case. Thus, the translation of these monocistronic mRNAs containing the defective PTV IRES elements appeared to be occurring using the cap-dependent initiation mechanism.
FIGURE 2.
Structure of monocistronic and dicistronic reporter plasmids containing wild-type and mutant forms of the PTV and AEV IRES elements. (A) Monocistronic derivatives of pRBRluc (Fig. 1) were prepared by inserting the indicated wild-type or mutant PTV-1 or AEV cDNA fragments into the BamHI (B) site. The wild-type and mutant sequences (in italics) for each derivative are indicated (the production of the mutants has been described previously, Chard et al. 2006a; Bakhshesh et al. 2008). Derivatives containing the IRES elements in the sense and antisense (AS) orientations were obtained by this procedure. (B) Dicistronic derivatives of pFluc were prepared by inserting EcoRI (E)–SalI (S) fragments from the monocistronic plasmids shown in A into the same sites of the vector downstream from the Fluc coding sequence. Note that the vector was prepared using a partial digestion with EcoRI and complete digestion with SalI. This construction therefore includes just nt 136–405 of the PTV cDNA, which clearly retains full IRES activity (as observed for nt 125–405). A line diagram of the secondary structure of the HCV-like IRES elements is shown to identify the locations of the individual IRES domains and the stems (S1 and S2), which are part of the pseudoknot (domain IIIf).
FIGURE 3.
Cap-dependent translation of monocistronic mRNAs containing defective PTV IRES elements. Plasmids encoding monocistronic mRNAs containing the wild-type or mutant forms of the PTV IRES were transfected alone (−) or with (+) pGEM3Z/J1 into vTF7-3 infected BHK cells (as for Fig. 1B). Cell extracts were analyzed for Rluc expression by immunoblotting and by enzyme assay (results are the mean +S.D. for three or four independent transfections). The Rluc and a nonspecific background band (NSB) are indicated on the blots.
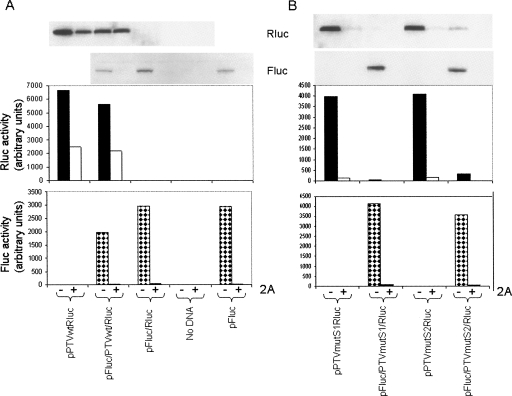
To confirm that the PTV IRES elements within these plasmids were functioning as reported previously within a dicistronic mRNA context, the IRES-Rluc cassettes were placed downstream from the Fluc coding sequence (Fig. 2B), and the resultant plasmids, which express dicistronic mRNAs, were assayed within cells as before (Fig. 4). As anticipated, the wild-type PTV IRES was active both in the monocistronic and dicistronic plasmids. In each case, the activity was inhibited by about 50% in the presence of the 2A protease, whereas the expression of the upstream Fluc from the dicistronic plasmid was inhibited by over 95% under these conditions (Fig. 4A). In contrast, the PTVmutS1 and PTVmutS2 elements displayed less than 5% of wild-type PTV IRES activity when assayed within the context of the dicistronic cassette, whereas from the monocistronic plasmids Rluc expression was about 60% of the wild-type level (Fig. 4A,B). As observed in Figure 3, the Rluc expression from the PTVmutS1 and PTVmutS2 monocistronic plasmids was strongly inhibited (>95%) by the coexpression of the 2A protease, and the low-level Rluc expression observed from the corresponding dicistronic plasmids was also strongly inhibited (>95%) under these conditions. Similarly, the Fluc expression from these dicistronic plasmids was also greatly suppressed (>95%) in the presence of the 2A protease (Fig. 4B).
FIGURE 4.
Comparison of translation from defective PTV IRES elements in mono- and dicistronic plasmids. The indicated monocistronic (Rluc alone) and dicistronic plasmids (Fluc/Rluc) containing the wild-type (A) or mutant (B) forms of the PTV IRES were assayed in parallel alone (−) or with (+) pGEM3Z/J1 (as for Fig. 1B). Cell extracts were prepared and analyzed for Fluc and Rluc expression as shown. Results are from one experiment but very similar results were observed in a second assay of the same set of plasmids.
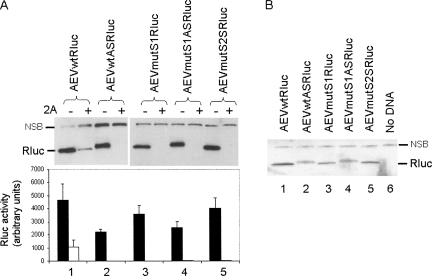
Influence of defective AEV IRES elements on the translation of monocistronic Rluc mRNAs
The AEV IRES is contained within nt 100–494 from the 5′UTR of the virus genome (Bakhshesh et al. 2008). Like the PTV-1 IRES, the AEV IRES has strong similarities to the HCV IRES and contains a pseudoknot (domain IIIf). We have shown previously that this IRES is also resistant to hippuristanol (an inhibitor of eIF4A function) and still functions when eIF4G cleavage is induced, albeit with reduced efficiency (Bakhshesh et al. 2008). Mutations within the pseudoknot strongly inhibited the activity of this IRES when assayed within the context of dicistronic reporter RNAs. Wild-type and mutant forms of the AEV IRES (within cDNA fragments including nt 1–494) were inserted as BamHI fragments into the pRBRluc vector (Fig. 2A), and the resultant monocistronic plasmids were assayed, as above, in the presence and absence of the SVDV 2A protease. As expected, the wild-type AEV IRES (in the sense orientation) efficiently directed the expression of Rluc (Fig. 5A) and, in accord with previous observations (Bakhshesh et al. 2008), this expression was significantly inhibited in the presence of 2A (by about 75%–80%) in these BHK cells. The monocistronic RNAs containing the AEV IRES elements with mutations in the stem 1 (AEVmutS1) or stem 2 (AEVmutS2) of the pseudoknot were also very efficiently translated when assayed alone, but this expression was essentially eliminated in the presence of 2A (Fig. 5A). When the AEVwt and AEVmutS1 elements were inserted in the AS-orientation upstream of Rluc, efficient expression of Rluc was again observed (Fig. 5A) both by immunoblotting and by enzyme assay (at about 50% of the sense orientation). In the presence of the 2A protease this expression was essentially eliminated (Fig. 5A). The Rluc protein produced from the plasmids containing the AS form of the AEV IRES appeared to migrate slightly more slowly (and was slightly heterogeneous) compared with the wild-type protein (Fig. 5A). To confirm this observation, the extracts from cells transfected with the reporter plasmids alone were examined side by side (Fig. 5B), and the slower migration of Rluc produced from the RNAs containing the AS form of the AEV IRES was clearly evident. This indicated that at least one AUG codon present in the AS form of the AEV 5′UTR was being recognized as an initiation codon and hence a fusion protein containing an N-terminal extension to the Rluc was produced. This N-terminal extension may have reduced the enzymatic activity of the protein slightly since the luciferase assays suggested some diminution of expression, which was not readily apparent from the immunoblots although these are not considered quantitative (Fig. 5A). The sense version of the AEV 5′UTR contains 9 upstream AUG codons, whereas the antisense version of this sequence contains 12 AUG codons. The two 3′ proximal AUGs within this AS sequence are in-frame with the Rluc coding sequence and, if used, would add either 20 or 36 amino acid residues to the N terminus of Rluc, which is consistent with the observed products (Fig. 5B). The previous 5 AUGs are not in-frame with the Rluc sequence, and, although other upstream AUG codons are in-frame with the Rluc sequence, any initiation from these codons would be terminated by the presence of a stop-codon and hence would not modify the Rluc protein.
FIGURE 5.
Cap-dependent translation of monocistronic mRNAs containing defective AEV IRES elements. (A) Plasmids encoding monocistronic mRNAs containing the wild-type or mutant forms of the AEV IRES were transfected alone (−) or with (+) pGEM3Z/J1 into vTF7-3 infected BHK cells (as for Fig. 1B). Cell extracts were analyzed for Rluc expression by immunoblotting and by enzyme assay (results are the mean +S.D. for three or four independent transfections). (B) The indicated cell extracts were reanalyzed to facilitate comparison of the Rluc products expressed from plasmids containing the wild-type or mutant AEV IRES in the sense or antisense (AS) orientation. The Rluc and a nonspecific background band (NSB) are indicated on the blots.
Influence of hippuristanol on the translation of monocistronic mRNAs
Hippuristanol is a potent inhibitor of eIF4A; it blocks cap-dependent translation initiation and the activity of the poliovirus and EMCV IRES elements; however, the PTV, HCV, and AEV IRES elements are relatively resistant to this agent (Bordeleau et al. 2006; Chard et al. 2006b; Bakhshesh et al. 2008). Hence, it is a second useful tool to distinguish between the possible mechanisms of translation initiation that may be occurring on the defective PTV and AEV IRES elements. Since the translation of RNAs containing the defective IRES elements was highly sensitive to cleavage of eIF4G, it was expected that their translation should also be sensitive to hippuristanol. To test this prediction, selected plasmids were assayed in a coupled transcription/translation (TNT) system containing rabbit reticulocyte lysate and [35S]methionine in the presence and absence of 10 μM hippuristanol, and the products were analyzed by SDS-PAGE and autoradiography. The results are shown in Figure 6. In accord with previous results (Bordeleau et al. 2006), translation directed by the wild-type PTV-1 IRES was relatively resistant to the presence of hippuristanol. In contrast, the efficient translation of the PTVmutS1Rluc and PTVmutS2Rluc RNAs was much more sensitive to the presence of this inhibitor of eIF4A activity. These results are consistent with the sensitivity of translation of these RNAs to the cleavage of eIF4G (as above) and confirm that translation initiation on these defective IRES elements is occurring by a different mechanism.
FIGURE 6.
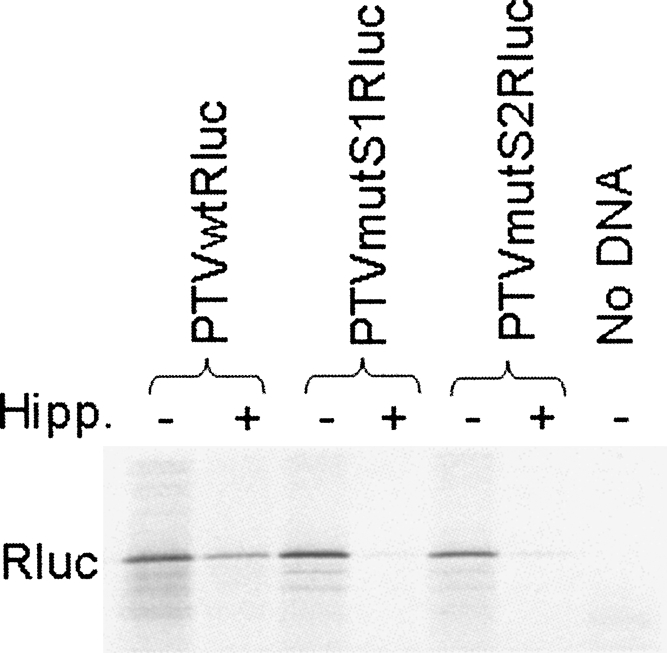
Influence of the eIF4A inhibitor, hippuristanol, on the translation of monocistronic mRNAs containing wild-type and defective PTV IRES elements. The indicated plasmids were assayed in the presence or absence of hippuristanol (10 μM), as indicated, using an in vitro TNT system containing [35S]methionine, and the products were analyzed by SDS-PAGE and autoradiography.
DISCUSSION
The studies presented here have demonstrated that monocistronic mRNAs can be translated in mammalian cells very efficiently despite the presence within their 5′UTRs of defective HCV-like picornavirus IRES elements, which include extensive secondary structure and multiple uAUGs. These defective viral IRES elements displayed minimal IRES activity when assayed within the context of dicistronic mRNAs (see Fig. 4). Furthermore, the translation of the monocistronic mRNAs containing these defective IRES elements was almost totally dependent on an intact cap-binding complex, eIF4F. The cleavage of eIF4G induced by the enterovirus 2A protease produced a dramatic loss of translation from these mRNAs. In contrast, translation from the wild-type EMCV IRES was unaffected under these conditions, as expected (e.g., Jang et al. 1988; Borman et al. 1997; Roberts et al. 1998). These results are consistent with the recent report by Dmitriev et al. (2007) indicating that a long, highly structured 5′UTR containing multiple uAUGs is not inhibitory to efficient translation of the human LINE-1 mRNA.
In this study, we have found that translation from the PTV IRES was inhibited to some degree (about 60%) by the expression of the 2A protease while the AEV IRES activity was inhibited even more (by about 75%) whereas cap-dependent translation was inhibited by over 95% under these conditions. It is not known why there is some sensitivity of the PTV and AEV IRES elements to the loss of intact eIF4G, but this effect on the AEV IRES has been noted previously (Bakhshesh et al. 2008). The elements are closely related to the HCV IRES element, and within in vitro 48S assembly assays neither the HCV IRES nor the PTV IRES displays a requirement for any eIF4 initiation factors (Pestova et al. 1998; Pisarev et al. 2004); however, this clearly does not mean that the presence of these factors may not be beneficial for optimal activity within cells when many other mRNAs are also being translated. Furthermore, recent studies on a simian picornavirus IRES, which also shares characteristics with the HCV IRES, have demonstrated some sensitivity of translation within the RRL translation system to a dominant negative mutant of eIF4A. Indeed, initiation complex formation on this RNA was also enhanced in the presence of eIF4F, and this effect was influenced by RNA secondary structure around the initiation codon (de Breyne et al. 2008). These data are consistent with the results presented here; however, the role of eIF4F in the function of these HCV-like IRES elements is not clear. It is interesting to note that the PTVmutS1comp (pseudo-wild-type) IRES (containing 6-nt substitutions compared with the wild-type sequence, see Fig. 2) appeared significantly less sensitive to coexpression of the 2A protease (Fig. 3). Indeed, in five separate experiments, the wild-type PTV IRES was inhibited by about 60% in the presence of 2A while the PTVmutS1comp IRES was only inhibited by 15%. This may suggest that minor changes in the structure/sequence of the pseudoknot (domain IIIf) may also influence the sensitivity of the PTV IRES (and its relatives) to the presence of eIF4F. The use of a different reporter gene in previous studies with the PTV IRES (Chard et al. 2006a) may explain the small apparent disparity in sensitivity of the PTV IRES to eIF4G cleavage since this will result in a different secondary structure around the initiation codon. Consistent with these results, we also observed some sensitivity of the wild-type PTV IRES to hippuristanol (a potent inhibitor of eIF4A), but this was much less than the effect on the translation of the RNAs containing defective versions of the PTV IRES (see Fig. 6).
In each of the mRNAs containing the mutant PTV or AEV 5′UTR sequences, there was either ca. 300 nt or ca. 500 nt of sequence upstream of the initiation codon, and clearly the presence of these sequences, even in AS orientation, did not significantly block translation initiation. So how was translation initiation on these mRNAs achieved? It is apparent that the mechanism of translation initiation on these mRNAs is quite different from that which occurs with the functional wild-type IRES elements since the process was very sensitive to expression of the 2A protease and, where tested, also to hippuristanol. It seems most probable that, following cap-dependent recognition of these mRNAs, the 40S ribosomal subunits will have migrated along the 5′UTR. Furthermore, it would appear that at least some uAUG codons were accessed since from two versions of the AS form of the AEV 5′UTR we obtained evidence for translation initiation occurring upstream of the standard Rluc initiation codon, which resulted in the production of enlarged polypeptides (see Fig. 5B). However, it is not known whether all of the upstream AUGs were potentially available for translation initiation. It is possible that some form of discontinuous migration of the ribosomal subunit along the 5′UTR occurs, possibly circumventing highly structured regions. This, of course, could also result in the avoidance of some uAUGs too. Some specific examples of ribosome shunting have been reported (Futterer et al. 1993; Yueh and Schneider 2000; Sherrill and Lloyd 2008), but it is very unlikely that the process of ribosome migration across the ∼500-nt AS form of the AEV 5′UTR sequence is achieved by such a specific mechanism since this sequence is never normally present within translated mRNAs. However, a more general mechanism of ribosomal shunting mediated by base-pairing to ribosomal RNA has been reported (Chappell et al. 2006), which could be relevant.
It is apparent that just 2- or 3-nt substitutions (as in the mutS1 and mutS2 constructs) within the wild-type PTV and AEV IRES sequences was sufficient to change the mode of translation initiation from IRES-directed to cap dependent, but this had very little effect in terms of the efficiency of translation of these monocistronic mRNAs (see Figs. 3–6). In contrast, the mutations within the EMCV IRES did significantly block the efficiency of translation (Fig. 1B); however, it was interesting to note that the residual translation (but not the wild-type EMCV IRES activity) was highly sensitive to the expression of the 2A protease and thus appeared to be achieved through a different initiation mechanism than the IRES-directed cap-independent process occurring on the wild-type EMCV IRES.
Earlier studies on picornavirus IRES elements had shown that capped monocistronic RNA transcripts containing the PV 5′UTR could be translated in a cell-free translation system, and this applied to mutants that were unable to direct cap-independent translation. However, the efficiency of translation appeared quite low compared with the cap-independent translation from the wild-type PV 5′UTR (Trono et al. 1988). In contrast, capping of transcripts containing the HAV IRES inhibited translation in vitro (Brown et al. 1994). The results presented here showed that the EMCV IRES, when mutated to block IRES activity, did act as a barrier to translation. This raises the question of what underlies the difference in behavior between the EMCV IRES and the HCV-like IRES elements. In each case, highly structured RNA sequences are present containing multiple AUG codons, and the sizes are comparable (e.g., the EMCV sequence used was 550 nt while the AEV 5′UTR is 494 nt). There is not a clear answer at present, and it may reflect a number of factors including the contexts of the various uAUGs, the stability of particular RNA structures, and the presence of bound cellular proteins (e.g., it is known that the EMCV IRES has high-affinity binding sites for the polypyrimidine tract binding protein, PTB) (see Belsham and Jackson 2000).
One consequence of the results presented here is that it is possible to envisage how a region of RNA that lacks significant IRES activity could be tolerated within the 5′UTR of a capped mRNA and then evolve, within that context (either of cellular or viral origin), to a functional IRES element without necessarily inhibiting translation initiation on the mRNA by the standard cap-dependent mechanism.
MATERIALS AND METHODS
Reporter plasmids
DNA preparations and manipulations were performed using standard methods as described (Sambrook and Russell, 2006) or as stated in manufacturer's instructions. The Rluc sequence was amplified in a PCR using pRLnull (Promega) as a template and the primers RLucEBfor (dCCGAATTCAAAGGATCCACCATGACTTCGAAAGTTTATGATCC) and Rlucrev (dCCGTCGACTTATTGTTCATTTTTGAGAACTCG), which introduced EcoRI and BamHI sites at the 5′ end of the gene and a SalI site near the 3′ terminus (these sites are indicated within the primer sequences in italics and underlined). The product was cloned into pCR-XL-TOPO (Invitrogen), which was then digested with EcoRI and SalI, and the released fragment was inserted into similarly digested pGEM3Z (Promega) to place the Rluc coding sequence under control of the T7 promoter; the plasmid was termed pRBRluc (Fig. 1A). Derivatives of this vector containing the wild-type or mutant forms of the EMCV IRES cDNA were generated by insertion of EcoRI–BamHI fragments into the same sites of pRBLuc (Fig. 1). The dicistronic plasmid pFluc/EMCV/Rluc was made by insertion of the Fluc sequence, as a blunt-ended BamHI-XhoI fragment from pGEM-luc (Promega), into the blunt ended EcoRI-linearized and phosphatased pEMCVRluc vector (Fig. 1A). The wild-type and mutant forms of the PTV-1 and AEV cDNAs corresponding to the IRES elements (as described previously, Chard et al. 2006a; Bakhshesh et al. 2008) were inserted as BamHI fragments into the same site of the pRBLuc vector (see Fig. 2).
A derivative of pGEM3Z (Promega) was generated by digesting it with EcoRI and SalI and introducing two annealed oligonucleotides (dAATTGGATCCCGGGCTCGAGAATTCGGGCCCG and dTCGACGGGCCCGAATTCTCGAGCCCGGGATCC) to produce a new polylinker (T7-BamHI-SmaI-XhoI-EcoRI-ApaI-SalI). Into this vector the fLuc coding sequence (BamHI-XhoI fragment from pGEM-luc [Promega]) was inserted to produce the plasmid pFluc (Fig. 2B), and dicistronic derivatives of this plasmid were obtained by the introduction of EcoRI–SalI fragments containing the Rluc or IRES–Rluc sequences downstream from the fLuc coding region (see Fig. 2A,B).
Transient expression assays
The various reporter plasmids (2 μg) were transfected into recombinant vaccinia virus (vTF7-3)-infected BHK cells alone or with the plasmid pGEM3Z/J1 (0.5μg), which expresses the swine vesicular disease virus (SVDV) 2A protease essentially as previously described (Roberts et al. 1998; Sakoda et al. 2001; Bakhshesh et al. 2008). Briefly, the plasmids were transfected using FuGene6 (3 μL; Roche) into cells (35-mm dishes) previously infected with the vaccinia virus vTF7-3 (Fuerst et al. 1986), which expresses the T7 RNA polymerase. Cell lysates were prepared 20 h after transfection and were analyzed for reporter gene expression by SDS-PAGE and immunoblotting. Detection was achieved with mouse anti-Rluc (clone 1D5.2, Chemicon) or goat anti-Fluc (Promega) antibodies and peroxidase-labeled anti-mouse or anti-goat (Dako Cytomation) antibodies, respectively, using chemiluminescence reagents (GE Amersham). Images were either obtained on X-ray film and then scanned or captured directly with a Bio-Rad Chemi-Doc XRS system. The activities of Rluc and Fluc present within the cell lysates were also quantified using the separate Renilla and firefly luciferase assay kits (Promega) with a luminometer. The results are presented as the mean (+S.D.) from three or four independent transfection experiments.
In vitro transcription/translation assays
In vitro TNT assays on selected plasmids were performed in the presence or absence of 10 μM hippuristanol (Bordeleau et al. 2006) (kindly provided by J. Pelletier, McGill University, Montreal, Canada) in rabbit reticulocyte lysate (RRL) using a TNT T7 Quick coupled RRL system (Promega) with [35S]methionine essentially as described by the manufacturer. Samples were analyzed by SDS-PAGE and autoradiography.
ACKNOWLEDGMENTS
We thank Jerry Pelletier (McGill University, Montreal) for the kind provision of hippuristanol. This work was supported in part by grants from the Danish Research Council (no. 274-07-0104) (to G.J.B.) and by the BBSRC (to L.O.R.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1039708.
REFERENCES
- Ali I.K., McKendrick L., Morley S.J., Jackson R.J. Activity of the Hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 2001;75:7854–7863. doi: 10.1128/JVI.75.17.7854-7863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S.D., Turcotte M., Korneluk R.G., Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshesh M., Groppelli E., Willcocks M.M., Royall E., Belsham G.J., Roberts L.O. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site (IRES) element. J. Virol. 2008;82:1993–2003. doi: 10.1128/JVI.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G.J., Jackson R.J. Translation initiation on picornavirus RNA. In: Sonenberg N., editor. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 869–900. Cold Spring Harbor Monograph 39, [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nature Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Borman A.M., Kean K.M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- Borman A.M., LeMercier P., Girard M., Kean K.M. Comparison of picornaviral IRES-driven internal initiation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.A., Zajac A.J., Lemon S.M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell S.A., Dresios J., Edelman G.E., Mauro V.P. Ribosomal shunting mediated by a translational enhancer element that base pairs to 18S rRNA. Proc. Natl. Acad. Sci. 2006;103:9488–9493. doi: 10.1073/pnas.0603597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard L.S., Kaku Y., Jones B., Nayak A., Belsham G.J. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006a;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard L.S., Bordeleau M.E., Pelletier J., Tanaka J., Belsham G.J. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 2006b;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- de Breyne S., Yu Y., Pestova T.V., Hellen C.U.T. Factor requirements for translation initiation on the simian picornavirus internal ribosome entry site. RNA. 2008;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Zhang D. Molecular analysis of duck hepatitis virus type 1. Virology. 2007;361:9–17. doi: 10.1016/j.virol.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Dmitriev S.E., Andreev D.E., Terenin I.M., Olovnikov I.A., Prassolov V.S., Merrick W.C., Shatsky I.N. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap-dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J., Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews M.B., editor. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. Cold Spring Harbor Monograph 48, [Google Scholar]
- Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer J., Kiss-László Z., Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Hellen C.U.T., de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono M., Mignone F., Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Jang S.K., Kräusslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku Y., Chard L.S., Inoue T., Belsham G.J. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 Talfan. J. Virol. 2002;76:11721–11728. doi: 10.1128/JVI.76.22.11721-11728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A., Belsham G.J., Jackson R.J. Translation of encephalomyocarditis virus RNA: Parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Victoria J., Simmonds P., Wang C., Shafer R.W., Nims R., Nielsen O., Delwart E. A highly divergent picornavirus in a marine mammal. J. Virol. 2008;82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S., Zhou K., Jubin R., Doudna J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: An update. J. Cell Biol. 1989a;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989b;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laletina E., Graifer D., Malygin A., Ivanov A., Shatsky I., Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.W., Li J., Timmerman S.L., Krushel L.A., Martin S.L. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: Implications for retrotransposition. Nucleic Acids Res. 2006;34:853–864. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky P.J., Otto G.A., Lancaster A.M., Sarnow P., Puglisi J.D. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNA. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Chard L.S., Kaku Y., Johns H.L., Shatsky I.N., Belsham G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaridi L., Georgopoulou U., Varaklioti A., Mavromara O. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 1999;453:49–53. doi: 10.1016/s0014-5793(99)00662-6. [DOI] [PubMed] [Google Scholar]
- Roberts L.O., Belsham G.J. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the co-expression of fragments of the IRES. Virology. 1997;227:53–62. doi: 10.1006/viro.1996.8312. [DOI] [PubMed] [Google Scholar]
- Roberts L.O., Seamons R.A., Belsham G.J. Recognition of picornavirus internal ribosome entry sites within infected cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M.E.M., Seamons R.A., Belsham G.J. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoda Y., Ross-Smith N., Inoue T., Belsham G.J. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 2001;75:10643–10650. doi: 10.1128/JVI.75.22.10643-10650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. The condensed protocols. [Google Scholar]
- Sherrill K.W., Lloyd R.E. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol. Cell. Biol. 2008;28:2011–2022. doi: 10.1128/MCB.01446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Willis A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is directly proportional to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin I.M., Dmitriev S.E., Andreev D.E., Royall E., Belsham G.J., Roberts L.O., Shatsky I.N. A “cross-kingdom” IRES reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005;25:7879–7888. doi: 10.1128/MCB.25.17.7879-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D., Pelletier J., Sonenberg N., Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science. 1988;241:445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Powell M.J., Hoover S.E., Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolaway K.E., Lazaridis K., Belsham G.J., Carter M.J., Roberts L.O. The 5′UTR of Rhopalosiphum padi virus (RhPV) contains an internal ribosome entry site (IRES) which functions efficiently in mammalian, insect and plant translation systems. J. Virol. 2001;75:10244–10249. doi: 10.1128/JVI.75.21.10244-10249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R., Suzuki Y., Nakai K., Sugano S. Small open reading frames in 5′ untranslated regions of mRNAs. C. R. Biol. 2003;326:987–991. doi: 10.1016/j.crvi.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Yueh A., Schneider R.J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes & Dev. 2000;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]